Fumagillin, a Mycotoxin of Aspergillus fumigatus: Biosynthesis, Biological Activities, Detection, and Applications
Abstract
1. Introduction
2. Fumagillin from a Chemical/Analytical Point of View
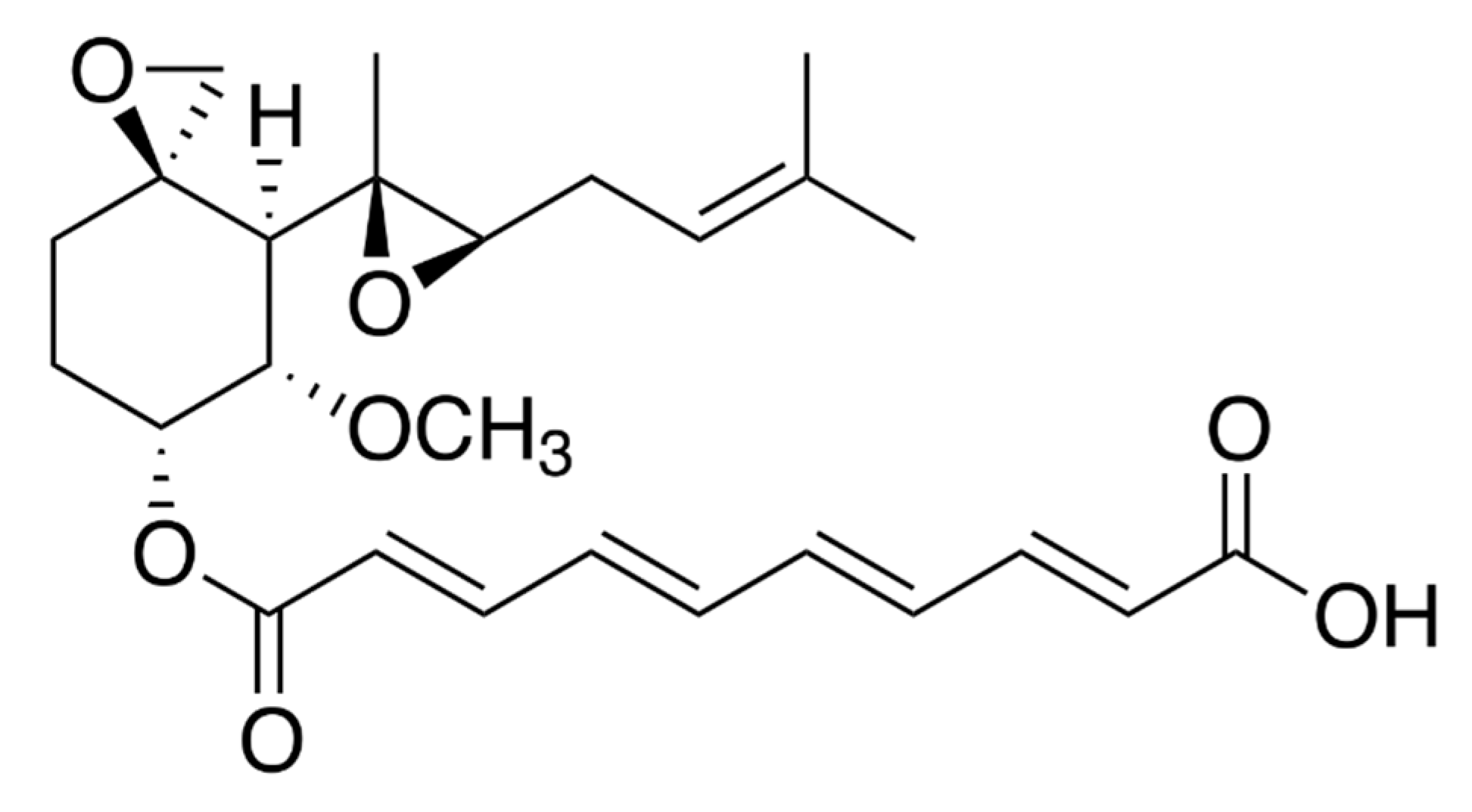
2.1. Fumagillin Physichochemical Properties
2.2. Degradation of Fumagillin
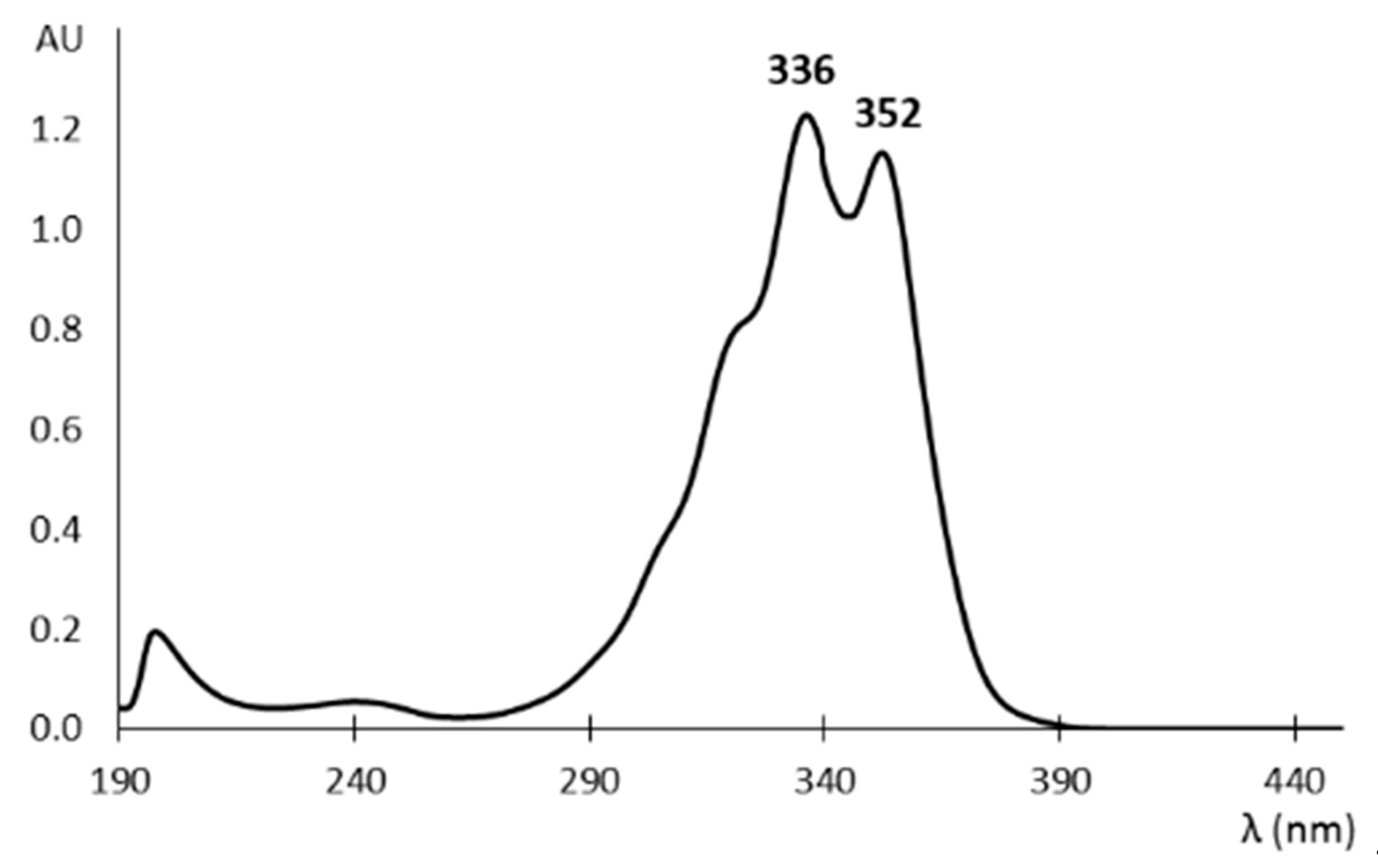
2.3. Analytical Methods for the Determination of Fumagillin
3. Fumagillin from a Biological Point of View
3.1. Fumagillin Biosynthetic Gene Cluster
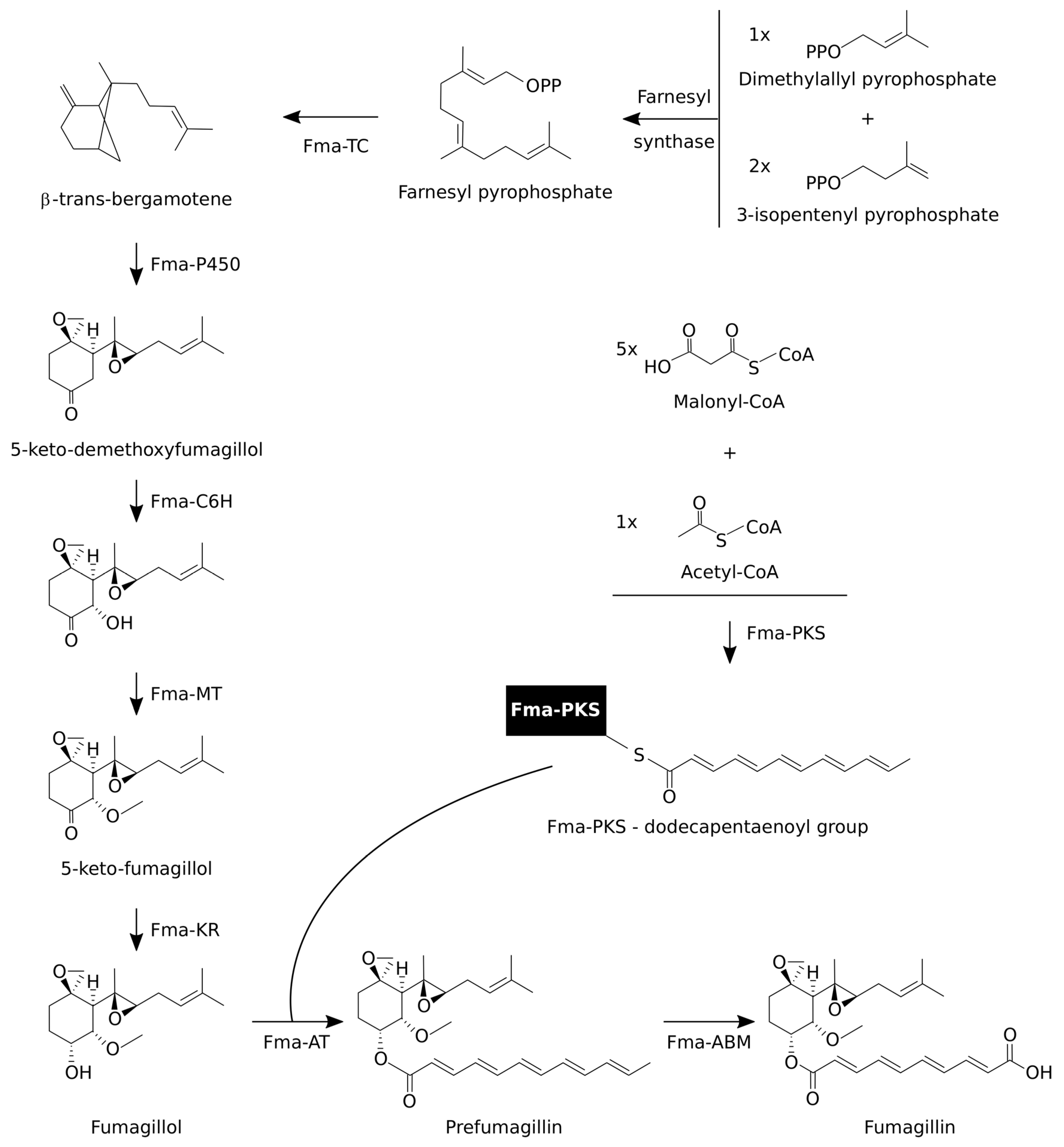
3.2. Fumagillin Biosynthetic Pathways
3.3. Regulation of Fumagillin Cluster
4. Biological Activities of Fumagillin
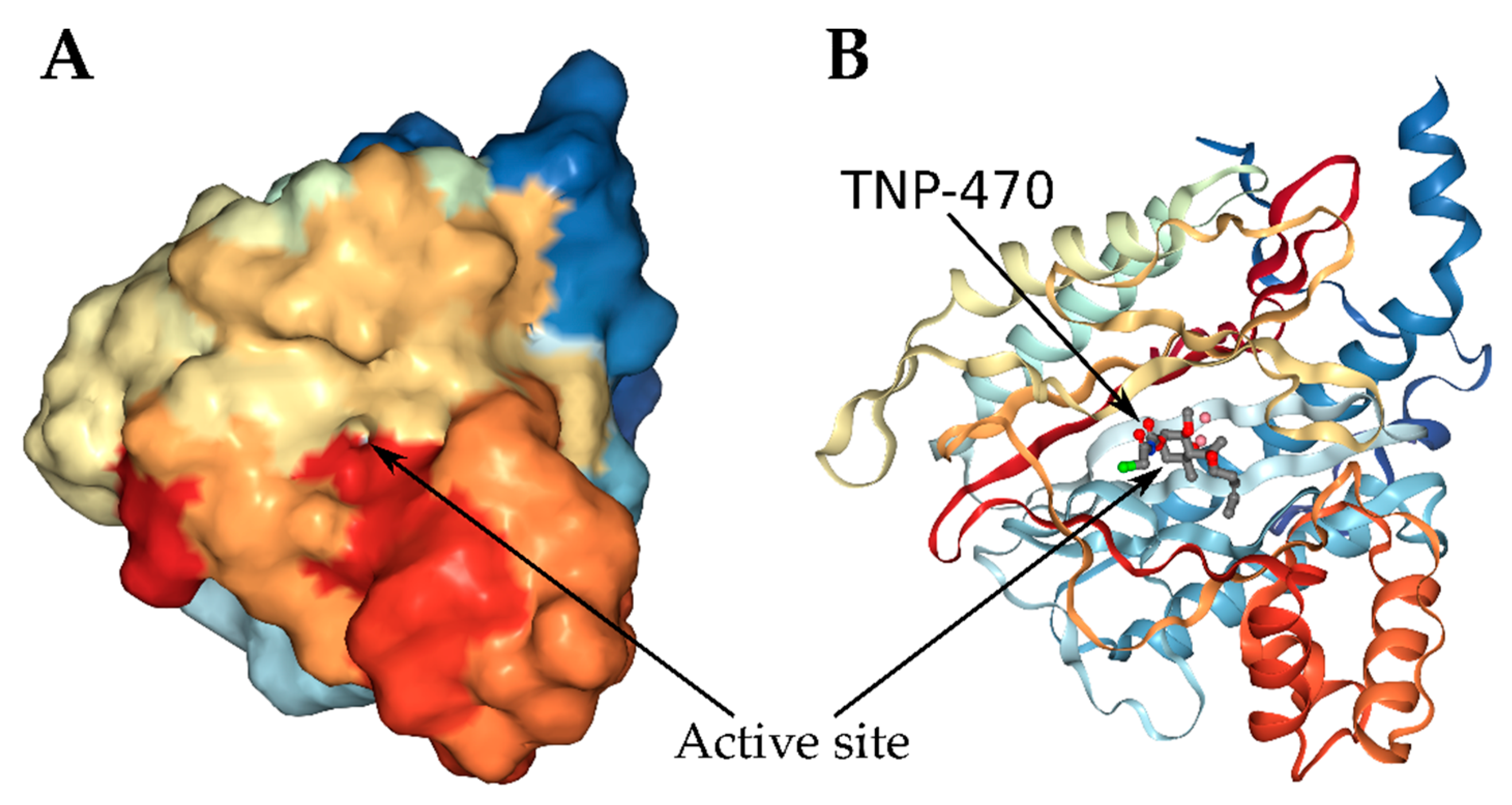
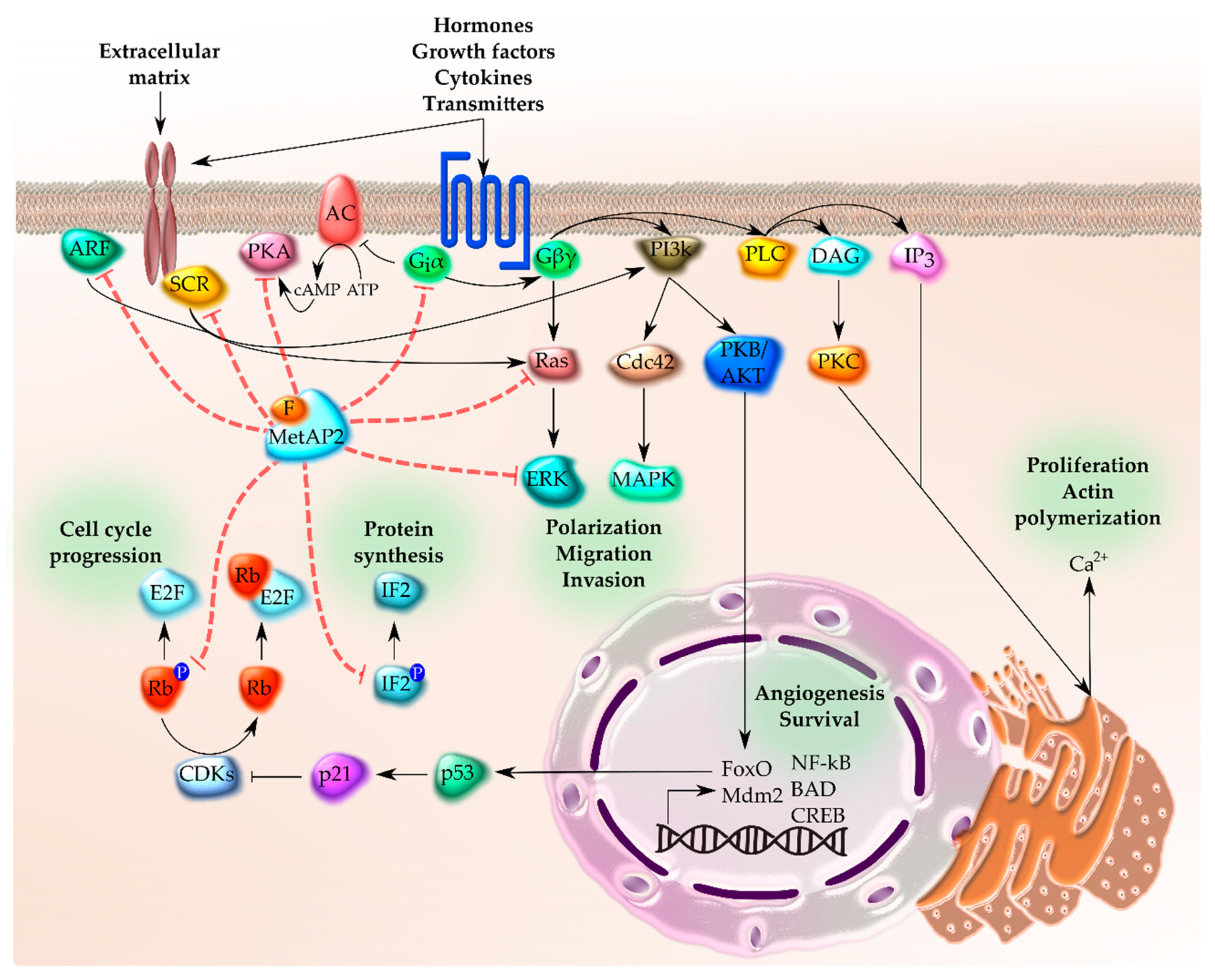
4.1. Methionine Aminopeptidases as Cellular Targets of the Mycotoxin
4.2. Effects of Fumagillin Activity on Host Cells
4.3. Fumagillin and Other Similar Toxins
5. Fumagillin Applications
5.1. Angiogenesis and Antitumor Activities
5.2. Fumagillin Antibiotic Uses
5.2.1. Treatment of Microsporidiosis
5.2.2. Treatment of Other Parasitosis
5.3. Other Applications
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Latgé, J.P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999, 12, 310–350. [Google Scholar] [CrossRef] [PubMed]
- Tekaia, F.; Latgé, J.P. Aspergillus fumigatus: Saprophyte or pathogen? Curr. Opin. Microbiol. 2005, 8, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, T.R.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin. Microbiol. Rev. 2009, 22, 447–465. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Sugui, J.A. Aspergillus fumigatus—What makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef]
- Knox, B.P.; Blachowicz, A.; Palmer, J.M.; Romsdahl, J.; Huttenlocher, A.; Wang, C.C.; Keller, N.P.; Venkateswaran, K. Characterization of Aspergillus fumigatus isolates from air and surfaces of the international Space Station. mSphere 2016, 1, e00227-16. [Google Scholar] [CrossRef]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2017, 10, 296–322. [Google Scholar] [CrossRef]
- Blachowicz, A.; Chiang, A.J.; Romsdahl, J.; Kalkum, M.; Wang, C.C.C.; Venkateswaran, K. Proteomic characterization of Aspergillus fumigatus isolated from air and surfaces of the International Space Station. Fungal Genet. Biol. 2019, 124, 39–46. [Google Scholar] [CrossRef]
- Alshareef, F.; Robson, G.D. Prevalence, persistence, and phenotypic variation of Aspergillus fumigatus in the outdoor environment in Manchester, UK, over a 2-year period. Med. Mycol. 2014, 52, 367–375. [Google Scholar] [CrossRef]
- Amarsaikhan, N.; O’Dea, E.M.; Tsoggerel, A.; Owegi, H.; Gillenwater, J.; Templeton, S.P. Isolate-dependent growth, virulence, and cell wall composition in the human pathogen Aspergillus fumigatus. PLoS ONE 2014, 9, 100430. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, G.M.; Keller, N.P. Crossover fungal pathogens: The biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet. Biol. 2013, 61, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Raffa, N.; Keller, N.P. A call to arms: Mustering secondary metabolites for success and survival of an opportunistic pathogen. PLoS Pathog. 2019, 15, 1007606. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.W.; Bentley, R. What’s in a name?—Microbial secondary metabolism. Adv. Appl. Microbiol. 1989, 34, 1–28. [Google Scholar] [CrossRef]
- Arias, M.; Santiago, L.; Vidal-García, M.; Redrado, S.; Lanuza, P.; Comas, L.; Domingo, M.P.; Rezusta, A.; Gálvez, E.M. Preparations for invasion: Modulation of host lung immunity during pulmonary aspergillosis by gliotoxin and other fungal secondary metabolites. Front. Immunol. 2018, 9, 2549. [Google Scholar] [CrossRef] [PubMed]
- Kamei, K.; Watanabe, A. Aspergillus mycotoxins and their effect on the host. Med. Mycol. 2005, 43, S95–S99. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Keller, N.P.; Bok, J.W.; Chung, D.; Perrin, R.M.; Shwab, E.K. LaeA, a global regulator of Aspergillus toxins. Med. Mycol. 2006, 44, 83–85. [Google Scholar] [CrossRef]
- Raffa, N.; Osherov, N.; Keller, N.P. Copper utilization, regulation, and acquisition by Aspergillus fumigatus. Int. J. Mol. Sci. 2019, 20, 1980. [Google Scholar] [CrossRef]
- Romsdahl, J.; Wang, C.C.C. Recent advances in the genome mining of Aspergillus secondary metabolites (covering 2012–2018). Med. Chem. Commun. 2019, 10, 840–866. [Google Scholar] [CrossRef]
- Shankar, J.; Tiwari, S.; Shishodia, S.K.; Gangwar, M.; Hoda, S.; Thakur, R.; Vijayaraghavan, P. Molecular insights into development and virulence determinants of Aspergilli: A proteomic perspective. Front. Cell. Infect. Microbiol. 2018, 8, 180. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Rank, C.; Nielsen, K.F.; Larsen, T.O. Metabolomics of Aspergillus fumigatus. Med. Mycol. 2009, 47, 53–71. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Bignell, E.; Cairns, T.C.; Throckmorton, K.; Nierman, W.C.; Keller, N.P. Secondary metabolite arsenal of an opportunistic pathogenic fungus. Philos Trans R. Soc. B 2016, 371, 20160023. [Google Scholar] [CrossRef]
- Lind, A.L.; Wisecaver, J.H.; Lameiras, C.; Wiemann, P.; Palmer, J.M.; Keller, N.P.; Rodrigues, F.; Goldman, G.H.; Rokas, A. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol. 2017, 15. [Google Scholar] [CrossRef]
- Lind, A.L.; Lim, F.Y.; Soukup, A.A.; Keller, N.P.; Rokas, A. An LaeA- and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Hanson, F.R.; Eble, T.E. An antiphage agent isolated from Aspergillus sp. J. Bacteriol. 1949, 58, 527–529. [Google Scholar]
- Perrin, R.M.; Fedorova, N.D.; Bok, J.W.; Cramer, R.A.; Wortman, J.R.; Kim, H.S.; Nierman, W.C.; Keller, N.P. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007, 3, 50. [Google Scholar] [CrossRef]
- Wiemann, P.; Guo, C.; Palmer, J.M.; Sekonyela, R.; Wang, C.C.C.; Keller, N.P. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl. Acad. Sci. USA 2013, 110, 17065–17070. [Google Scholar] [CrossRef]
- Sin, N.; Meng, L.; Wang, M.Q.; Wen, J.J.; Bornmann, W.G.; Crews, C.M. The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc. Natl. Acad. Sci. USA 1997, 94, 6099–6103. [Google Scholar] [CrossRef]
- Mauriz, J.L.; Martín-Renedo, J.; García-Palomo, A.; Tuñón, M.J.; González-Gallego, J. Methionine aminopeptidases as potential targets for treatment of gastrointestinal cancers and other tumours. Curr. Drug Targets 2010, 11, 1439–1457. [Google Scholar] [CrossRef]
- Vetro, J.A.; Dummitt, B.; Chang, Y.H. Methionine-aminopeptidase: Emerging role in angiogenesis. In Aminopeptidases in Biology and Disease; Hooper, N.M., Lendeckel, U., Eds.; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2004; pp. 17–44. [Google Scholar]
- Arico-Muendel, C.; Centrella, P.A.; Contonio, B.D.; Morgan, B.A.; O’Donovan, G.; Paradise, C.L.; Skinner, S.R.; Sluboski, B.; Svendsen, J.L.; White, K.F.; et al. Antiparasitic activities of novel, orally available fumagillin analogs. Bioorganic Med. Chem. Lett. 2009, 19, 5128–5131. [Google Scholar] [CrossRef]
- Novohradska, S.; Ferling, I.; Hillmann, F. Exploring Virulence Determinants of Filamentous Fungal Pathogens through Interactions with Soil Amoebae. Front. Cell. Infect. Microbiol. 2017, 7, 497. [Google Scholar] [CrossRef]
- Casadevall, A.; Fu, M.S.; Guimaraes, A.J.; Albuquerque, P. The ‘Amoeboid Predator-Fungal Animal Virulence’ Hypothesis. J. Fungi 2019, 5, 10. [Google Scholar] [CrossRef]
- Molina, J.M.; Tourneur, M.; Sarfati, C.; Chevret, S.; de Gouvello, A.; Gobert, J.G.; Balkan, S.; Derouin, F. Fumagillin treatment of intestinal microsporidiosis. N. Engl. J. Med. 2002, 346, 1963–1969. [Google Scholar] [CrossRef]
- van den Heever, J.P.; Thompson, T.S.; Curtis, J.M.; Ibrahim, A.; Pernal, S.F. Fumagillin: An overview of recent scientific advances and their significance for apiculture. J. Agric. Food Chem. 2014, 62, 2728–2737. [Google Scholar] [CrossRef]
- van den Heever, J.P.; Thompson, T.S.; Otto, S.J.G.; Curtis, J.M.; Ibrahim, A.; Pernal, S.F. Evaluation of Fumagilin-B and other potential alternative chemotherapies against Nosema ceranae-infected honeybees (Apis mellifera) in cage trial assays. Apidologie 2016, 47, 617–630. [Google Scholar] [CrossRef]
- Fallon, J.P.; Reeves, E.P.; Kavanagh, K. Inhibition of neutrophil function following exposure to the Aspergillus fumigatus toxin fumagillin. J. Med. Microbiol. 2010, 59, 625–633. [Google Scholar] [CrossRef]
- Zbidah, M.; Lupescu, A.; Jilani, K.; Lang, F. Stimulation of suicidal erythrocyte death by fumagillin. Basic Clin. Pharmacol. Toxicol. 2013, 112, 346–351. [Google Scholar] [CrossRef]
- Guruceaga, X.; Ezpeleta, G.; Mayayo, E.; Sueiro-Olivares, M.; Abad-Diaz-De-Cerio, A.; Aguirre Urizar, J.M.; Liu, H.G.; Wiemann, P.; Bok, J.W.; Filler, S.G.; et al. A possible role for fumagillin in cellular damage during host infection by Aspergillus fumigatus. Virulence 2018, 9, 1548–1561. [Google Scholar] [CrossRef]
- MarvinSketch, (version 17.22.0, calculation module developed by ChemAxon, 2017).
- DrugBank. Available online: https://www.drugbank.ca/about (accessed on 6 November 2019).
- Log D Predictor. Available online: https://disco.chemaxon.com/apps/demos/logd/ (accessed on 15 October 2019).
- Garrett, E.R.; Eble, T.E. Studies on the stability of fumagillin. I. Photolytic degradation in alcohol solution. J. Am. Pharm. Assoc. Sci. Ed. 1954, 43, 385–390. [Google Scholar] [CrossRef]
- Eble, T.E.; Garrett, E.R. Studies on the stability of fumagillin. II. Photolytic degradation of crystalline fumagillin. J. Am. Pharm. Assoc. Sci. Ed. 1954, 43, 536–538. [Google Scholar] [CrossRef]
- Garrett, E.R. Studies on the stability of fumagillin. III. Thermal degradation in the presence and absence of air. J. Am. Pharm. Assoc. Sci. Ed. 1954, 43, 539–543. [Google Scholar] [CrossRef]
- Brackett, J.M.; Arguello, M.D.; Schaar, J.C. Determination of fumagillin by high-performance liquid-chromatography. J. Agric. Food Chem. 1988, 36, 762–764. [Google Scholar] [CrossRef]
- Kochansky, J.; Nasr, M. Laboratory studies on the photostability of fumagillin, the active ingredient of Fumidil B. Apidologie 2004, 35, 301–310. [Google Scholar] [CrossRef]
- Dmitrovic, J.; Durden, D.A. Analysis of fumagillin in honey by LC-MS/MS. J. AOAC Int. 2013, 96, 687–695. [Google Scholar] [CrossRef]
- Assil, H.I.; Sporns, P. ELISA and HPLC methods for analysis of fumagillin and its decomposition products in honey. J. Agric. Food Chem. 1991, 39, 2206–2213. [Google Scholar] [CrossRef]
- Higes, M.; Nozal, M.J.; Alvaro, A.; Barrios, L.; Meana, A.; Martin-Hernandez, R.; Bernal, J.L.; Bernal, J. The stability and effectiveness of fumagillin in controlling Nosema ceranae (Microsporidia) infection in honey bees (Apis mellifera) under laboratory and field conditions. Apidologie 2011, 42, 364–377. [Google Scholar] [CrossRef]
- Van den Heever, J.P.; Thompson, T.S.; Curtis, J.M.; Pernal, S.F. Stability of dicyclohexylamine and fumagillin in honey. Food Chem. 2015, 179, 152–158. [Google Scholar] [CrossRef]
- Nozal, M.J.; Bernal, J.L.; Martin, M.T.; Bernal, J.; Alvaro, A.; Martin, R.; Higes, M. Trace analysis of fumagillin in honey by liquid chromatography-diode array-electrospray ionization mass spectrometry. J. Chromatogr. A 2008, 1190, 224–231. [Google Scholar] [CrossRef]
- Van den Heever, J.P.; Thompson, T.S.; Curtis, J.M.; Pernal, S.F. Determination of dicyclohexylamine and fumagillin in Honey by LC-MS/MS. Food Anal. Meth. 2015, 8, 767–777. [Google Scholar] [CrossRef]
- Lopez, M.I.; Pettis, J.S.; Smith, I.B.; Chu, P.S. Multiclass determination and confirmation of antibiotic residues in honey using LC-MS/MS. J. Agric. Food Chem. 2008, 56, 1553–1559. [Google Scholar] [CrossRef] [PubMed]
- Ivešić, M.; Krivohlavek, A.; Žuntar, I.; Tolić, S.; Šikić, S.; Musić, V.; Pavlić, I.; Bursik, A.; Galić, N. Monitoring of selected pharmaceuticals in surface waters of Croatia. Environ. Sci. Pollut. Res. 2017, 24, 23389–23400. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Chua, X.; Ling, Y.; Huang, J.; Chang, J. Multi-mycotoxin analysis in dairy products by liquid chromatography coupled to quadrupole orbitrap mass spectrometry. J. Chromatogr. A 2014, 1345, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Kanda, M.; Sasamoto, T.; Takeba, K.; Hayashi, H.; Kusano, T.; Matsushima, Y.; Nakajima, T.; Kanai, S.; Takano, I. Rapid Determination of Fumagillin Residues in Honey by Liquid Chromatography-Tandem Mass Spectrometry Using the QuEChERS Method. J. AOAC Int. 2011, 94, 878–885. [Google Scholar]
- Guyonnet, J.; Richard, M.; Hellings, P. Determination of fumagillin in muscle-tissue of rainbow-trout using automated ion-pairing liquid-chromatography. J. Chromatogr. B-Biomed. Appl. 1995, 666, 354–359. [Google Scholar] [CrossRef]
- Fekete, J.; Romvari, Z.; Szepesi, I.; Morovjan, G. Liquid-chromatographic determination of the antibiotic fumagillin in fish meat samples. J. Chromatogr. A 1995, 712, 378–381. [Google Scholar] [CrossRef]
- Fekete, J.; Romvari, Z.; Gebefugi, I.; Kettrup, A. Comparative study on determination of fumagillin in fish by normal and reversed phase chromatography. Chromatographia 1998, 48, 48–52. [Google Scholar] [CrossRef]
- Lin, H.C.; Chooi, Y.H.; Dhingra, S.; Xu, W.; Calvo, A.M.; Tang, Y.J. The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of β-trans-bergamotene. J. Am. Chem. Soc. 2013, 135, 4616–4619. [Google Scholar] [CrossRef]
- Lin, H.C.; Tsunematsu, Y.; Dhingra, S.; Xu, W.; Fukutomi, M.; Chooi, Y.H.; Cane, D.E.; Calvo, A.M.; Watanabe, K.; Tang, Y. Generation of complexity in fungal terpene biosynthesis, discovery of a multifunctional cytochrome P450 in the fumagillin pathway. J. Am. Chem. Soc. 2014, 136, 4426–4436. [Google Scholar] [CrossRef]
- Dhingra, S.; Lind, A.L.; Lin, H.C.; Tang, Y.; Rokas, A.; Calvo, A.M. The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS ONE 2013, 8, 77147. [Google Scholar] [CrossRef]
- NCBI (National Center for Biotechnology Information). Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 6 November 2019).
- AspGD (Aspergillus Genome Database). Available online: http://www.aspgd.org/ (accessed on 6 November 2019).
- UniProtKB (UniProt Knowledgebase). Available online: https://www.uniprot.org/ (accessed on 6 November 2019).
- Gravelat, F.N.; Doedt, T.; Chiang, L.Y.; Liu, H.; Filler, S.G.; Patterson, T.F.; Sheppard, D.C. In Vivo analysis of Aspergillus fumigatus developmental gene expression determined by real-time reverse transcription-PCR. Infect. Immun. 2008, 76, 3632–3639. [Google Scholar] [CrossRef]
- Vödisch, M.; Scherlach, K.; Winkler, R.; Hertweck, C.; Braun, H.P.; Roth, M.; Haas, H.; Werner, E.R.; Brakhage, A.A.; Kniemeyer, O. Analysis of the Aspergillus fumigatus proteome reveals metabolic changes and the activation of the pseurotin A biosynthesis gene cluster in response to hypoxia. J. Proteome Res. 2011, 10, 2508–2524. [Google Scholar] [CrossRef]
- Mascotti, M.L.; Ayub, M.J.; Dudek, H.; Sanz, M.K.; Fraaije, M.W. Cloning, overexpression and biocatalytic exploration of a novel Baeyer-Villiger monooxygenase from Aspergillus fumigatus Af293. AMB Express 2013, 3, 33. [Google Scholar] [CrossRef]
- Tsunematsu, Y.; Fukutomi, M.; Saruwatari, T.; Noguchi, H.; Hotta, K.; Tang, Y.; Watanabe, K. Elucidation of pseurotin biosynthetic pathway points to trans-acting C-methyltransferase: Generation of chemical diversity. Angew. Chem. Int. Ed. 2014, 53, 8475–8479. [Google Scholar] [CrossRef]
- Bruns, S.; Seidler, M.; Albrecht, D.; Salvenmoser, S.; Remme, N.; Hertweck, C.; Brakhage, A.A.; Kniemeyer, O.; Müller, F.M. Functional genomic profiling of Aspergillus fumigatus biofilm reveals enhanced production of the mycotoxin gliotoxin. Proteomics 2010, 10, 3097–3107. [Google Scholar] [CrossRef]
- Lind, A.L.; Smith, T.D.; Saterlee, T.; Calvo, A.M.; Rokas, A. Regulation of secondary metabolism by the Velvet complex is temperature-responsive in Aspergillus. G3 Bethesda 2016, 6, 4023–4033. [Google Scholar] [CrossRef]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 2004, 3, 527–535. [Google Scholar] [CrossRef]
- Bok, J.W.; Balajee, S.A.; Marr, K.A.; Andes, D.; Nielsen, K.F.; Frisvad, J.C.; Keller, N.P. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell 2005, 4, 1574–1582. [Google Scholar] [CrossRef]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef]
- Amaike, S.; Keller, N.P. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot Cell 2009, 8, 1051–1060. [Google Scholar] [CrossRef]
- Gehrke, A.; Heinekamp, T.; Jacobsen, I.D.; Brakhage, A.A. Heptahelical receptors GprC and GprD of Aspergillus fumigatus are essential regulators of colony growth, hyphal morphogenesis, and virulence. Appl. Environ. Microbiol. 2010, 76, 3989–3998. [Google Scholar] [CrossRef]
- Kim, S.S.; Kim, Y.H.; Shin, K.S. The developmental regulators, FlbB and FlbE, are involved in the virulence of Aspergillus fumigatus. J. Microbiol. Biotechnol. 2013, 23, 766–770. [Google Scholar] [CrossRef]
- Johns, A.; Scharf, D.H.; Gsaller, F.; Schmidt, H.; Heinekamp, T.; Straßburger, M.; Oliver, J.D.; Birch, M.; Beckmann, N.; Dobb, K.S.; et al. A nonredundant phosphopantetheinyl transferase, PptA, is a novel antifungal target that directs secondary metabolite, siderophore, and lysine biosynthesis in Aspergillus fumigatus and is critical for pathogenicity. MBio 2017, 8. [Google Scholar] [CrossRef]
- Manfiolli, A.O.; de Castro, P.A.; Dos Reis, T.F.; Dolan, S.; Doyle, S.; Jones, G.; Riaño Pachón, D.M.; Ulaş, M.; Noble, L.M.; Mattern, D.J.; et al. Aspergillus fumigatus protein phosphatase PpzA is involved in iron assimilation, secondary metabolite production, and virulence. Cell. Microbiol. 2017, 19. [Google Scholar] [CrossRef]
- Wu, M.Y.; Mead, M.E.; Lee, M.K.; Ostrem Loss, E.M.; Kim, S.C.; Rokas, A.; Yu, J.H. Systematic dissection of the evolutionarily conserved WetA developmental regulator across a genus of filamentous fungi. mBio 2018, 9. [Google Scholar] [CrossRef]
- Lind, A.L.; Wisecaver, J.H.; Smith, T.D.; Feng, X.; Calvo, A.M.; Rokas, A. Examining the evolution of the regulatory circuit controlling secondary metabolism and development in the fungal genus Aspergillus. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef]
- Yu, Y.; Blachowicz, A.; Will, C.; Szewczyk, E.; Glenn, S.; Gensberger-Reigl, S.; Nowrousian, M.; Wang, C.C.C.; Krappmann, S. Mating-type factor-specific regulation of the fumagillin/pseurotin secondary metabolite supercluster in Aspergillus fumigatus. Mol. Microbiol. 2018, 110, 1045–1065. [Google Scholar] [CrossRef]
- Conrad, T.; Kniemeyer, O.; Henkel, S.G.; Krüger, T.; Mattern, D.J.; Valiante, V.; Guthke, R.; Jacobsen, I.D.; Brakhage, A.A.; Vlaic, S.; et al. Module-detection approaches for the integration of multilevel omics data highlight the comprehensive response of Aspergillus fumigatus to caspofungin. BMC Syst. Biol. 2018, 1, 88. [Google Scholar] [CrossRef]
- Eshwika, A.; Kelly, J.; Fallon, J.P.; Kavanagh, K. Exposure of Aspergillus fumigatus to caspofungin results in the release, and de novo biosynthesis, of gliotoxin. Med. Mycol. 2013, 51, 121–127. [Google Scholar] [CrossRef]
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; König, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015, 6, 299. [Google Scholar] [CrossRef]
- Arfin, S.M.; Kendall, R.L.; Hall, L.; Weaver, L.H.; Stewart, A.E.; Matthews, B.W.; Bradshaw, R.A. Eukaryotic methionyl aminopeptidases: Two classes of cobalt-dependent enzymes. Proc. Natl. Acad. Sci. USA 1995, 92, 7714–7718. [Google Scholar] [CrossRef]
- Li, X.; Chang, Y.H. Evidence that the human homologue of a rat initiation factor-2 associated protein (p67) is a methionine aminopeptidase. Biochem. Biophys. Res. Commun. 1996, 227, 152–159. [Google Scholar] [CrossRef]
- Meinnel, T.; Mechulam, Y.; Blanquet, S. Methionine as translation start signal: A review of the enzymes of the pathway in Escherichia coli. Biochimie 1993, 75, 1061–1075. [Google Scholar] [CrossRef]
- Boissel, J.P.; Kasper, T.J.; Shah, S.C.; Malone, J.I.; Bunn, H.F. Amino-terminal processing of proteins: Hemoglobin South Florida, a variant with retention of initiator methionine and N(α)-acetylation. Proc. Natl. Acad. Sci. USA 1985, 82, 8448–8452. [Google Scholar] [CrossRef]
- Chang, Y.H.; Teichert, U.; Smith, J.A. Molecular cloning, sequencing, deletion, and overexpression of a methionine aminopeptidase gene from Saccharomyces cerevisiae. J. Biol. Chem. 1992, 267, 8007–8011. [Google Scholar]
- Chen, S.; Vetro, J.A.; Chang, Y.H. The specificity in vivo of two distinct methionine aminopeptidases in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2002, 398, 87–93. [Google Scholar] [CrossRef]
- Walker, K.W.; Bradshaw, R.A. Yeast Methionine Aminopeptidase, I. Alteration of substrate specificity by site-directed mutagenesis. J. Biol. Chem. 1999, 274, 13403–13409. [Google Scholar] [CrossRef]
- Catalano, A.; Romano, M.; Robuffo, I.; Strizzi, L.; Procopio, A. Methionine aminopeptidase-2 regulates human mesothelioma cell survival: Role of Bcl-2 expression and telomerase activity. Am. J. Pathol. 2001, 159, 721–731. [Google Scholar] [CrossRef]
- Lowther, W.T.; Matthews, B.W. Structure and function of the methionine aminopeptidases. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000, 1477, 157–167. [Google Scholar] [CrossRef]
- Endo, H.; Takenaga, K.; Kanno, T.; Satoh, H.; Mori, S. Methionine aminopeptidase 2 is a new target for the metastasis-associated protein, S100A4. J. Biol. Chem. 2002, 277, 26396–26402. [Google Scholar] [CrossRef]
- Bradshaw, R.A.; Brickey, W.W.; Walker, K.W. N-terminal processing: The methionine aminopeptidase and N(α)-acetyl transferase families. Trends Biochem. Sci. 1998, 23, 263–267. [Google Scholar] [CrossRef]
- Ray, M.K.; Datta, B.; Chakraborty, A.; Chattopadhyay, A.; Meza-Keuthen, S.; Gupta, N.K. The eukaryotic initiation factor 2-associated 67-kDa polypeptide (p67) plays a critical role in regulation of protein synthesis initiation in animal cells. Proc. Natl. Acad. Sci. USA 1992, 89, 539–543. [Google Scholar] [CrossRef]
- Datta, B.; Ray, M.K.; Chakrabarti, D.; Wylie, D.E.; Gupta, N.K. Glycosylation of eukaryotic peptide chain initiation factor 2 (eIF-2)-associated 67-kDa polypeptide (p67) and its possible role in the inhibition of eIF-2 kinase-catalyzed phosphorylation of the eIF-2 α-subunit. J. Biol. Chem. 1989, 264, 20620–20624. [Google Scholar]
- Atanassova, A.; Sugita, M.; Sugiura, M.; Pajpanova, T.; Ivanov, I. Molecular cloning, expression and characterization of three distinctive genes encoding methionine aminopeptidases in cyanobacterium Synechocystis sp. strain PCC6803. Arch. Microbio. 2003, 180, 185–193. [Google Scholar] [CrossRef]
- Giglione, C.; Serero, A.; Pierre, M.; Boisson, B.; Meinnel, T. Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J. 2000, 19, 5916–5929. [Google Scholar] [CrossRef]
- Leszczyniecka, M.; Bhatia, U.; Cueto, M.; Nirmala, N.R.; Towbin, H.; Vattay, A.; Wang, B.; Zabludoff, S.; Phillips, P.E. MAP1D, a novel methionine aminopeptidase family member is overexpressed in colon cancer. Oncogene 2006, 25, 3471–3478. [Google Scholar] [CrossRef][Green Version]
- Ingber, D.; Fujita, T.; Kishimoto, S.; Kanamaru, T.; Brem, H.; Folkman, J. Synthetic analogues of fumagillin that inhibit angiogenesis and suppress tumour growth. Nature 1990, 348, 555–557. [Google Scholar] [CrossRef]
- Liu, S.; Widom, J.; Kemp, C.W.; Crews, C.M.; Clardy, J. Structure of Human Methionine Aminopeptidase-2 complexed with Fumagillin. Science 1998, 282, 1324–1328. [Google Scholar] [CrossRef]
- Rose, A.S.; Bradley, A.R.; Valasatava, Y.; Duarte, J.M.; Prlić, A.; Rose, P.W. NGL viewer: Web-based molecular graphics for large complexes. Bioinformatics 2018, 34, 3755–3758. [Google Scholar] [CrossRef]
- RCSB PDB (RCSB Protein Data Bank). Available online: http://www.rcsb.org/3d-view/1B6A (accessed on 21 November 2019).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Datta, R.; Choudhury, P.; Bhattacharya, M.; Soto Leon, F.; Zhou, Y.; Datta, B. Protection of translation initiation factor eIF2 phosphorylation correlates with eIF2-associated glycoprotein p67 levels and requires the lysine-rich domain I of p67. Biochimie 2001, 83, 919–931. [Google Scholar] [CrossRef]
- Buscà, R.; Pouysségur, J.; Lenormand, P. ERK1 and ERK2 map kinases: Specific roles or functional redundancy? Front. Cell. Dev. Biol. 2016, 4, 1–23. [Google Scholar] [CrossRef]
- Datta, B.; Majumdar, A.; Datta, R.; Balusu, R. Treatment of cells with the angiogenic inhibitor fumagillin results in increased stability of eukaryotic initiation factor 2-associated glycoprotein, p67, and reduced phosphorylation of extracellular signal-regulated kinases. Biochemistry 2004, 43, 14821–14831. [Google Scholar] [CrossRef]
- Datta, R.; Choudhury, P.; Ghosh, A.; Datta, B. A glycosylation site, 60SGTS63, of p67 is required for its ability to regulate the phosphorylation and activity of eukaryotic initiation factor 2α. Biochemistry 2003, 42, 5453–5460. [Google Scholar] [CrossRef]
- Bernier, S.G.; Taghizadeh, N.; Thompson, C.D.; Westlin, W.F.; Hannig, G. Methionine aminopeptidases type I and type II are essential to control cell proliferation. J. Cell. Biochem. 2005, 95, 1191–1203. [Google Scholar] [CrossRef]
- Sawanyawisuth, K.; Wongkham, C.; Pairojkul, C.; Saeseow, O.T.; Riggins, G.J.; Araki, N.; Wongkham, S. Methionine aminopeptidase 2 over-expressed in cholangiocarcinoma: Potential for drug target. Acta Oncol. 2007, 46, 378–385. [Google Scholar] [CrossRef]
- Kanno, T.; Endo, H.; Takeuchi, K.; Moristhita, Y.; Fukayama, M.; Mori, S. High expression of methionine aminopeptidase type 2 in germinal center B cells and their neoplastic counterparts. Lab. Investig. 2002, 82, 893–901. [Google Scholar] [CrossRef]
- Chang, S.Y.P.; McGary, E.C.; Chang, S. Methionine aminopeptidase gene of Escherichia coli is essential for cell growth. J. Bacteriol. 1989, 171, 4071–4072. [Google Scholar] [CrossRef][Green Version]
- Miller, C.G.; Strauch, K.L.; Kukral, A.M.; Miller, J.L.; Wingfield, P.T.; Mazzei, G.J.; Werlen, R.C.; Graber, P.; Movva, N.R. N-terminal methionine-specific peptidase in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 1987, 84, 2718–2722. [Google Scholar] [CrossRef]
- Li, X.; Chang, Y.H. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl. Acad. Sci. USA 1995, 92, 12357–12361. [Google Scholar] [CrossRef]
- Keller, N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11, 671–677. [Google Scholar] [CrossRef]
- Buss, J.E.; Mumby, S.M.; Casey, P.J.; Gilman, A.G.; Sefton, B.M. Myristoylated alpha subunits of guanine nucleotide-binding regulatory proteins. Proc. Natl. Acad. Sci. USA 1987, 84, 7493–7497. [Google Scholar] [CrossRef]
- Yang, Z.; Wensel, T.G. N-Myristoylation of the rod outer segment G protein, transducin, in cultured retinas. J. Biol. Chem. 1992, 267, 23197–23201. [Google Scholar]
- Neves, S.R.; Ram, P.T.; Iyengar, R. G protein pathways. Science 2002, 296, 1636–1639. [Google Scholar] [CrossRef]
- Van Keulen, S.C.; Rothlisberger, U. Effect of N-terminal myristoylation on the active conformation of Gαi1-GTP. Biochemistry 2017, 56, 271–280. [Google Scholar] [CrossRef]
- Gresset, A.; Sondek, J.; Harden, T.K. The phospholipase C isozymes and their regulation. Subcellular Biochemistry. In Phosphoinositides I: Enzymes of Synthesis and Degradation; Balla, T., Wymann, M., York, J., Eds.; Springer: Dordrecht, The Netherlands, 2012; Volume 58, pp. 61–94. [Google Scholar]
- Xu, J.; Wang, F.; Van Keymeulen, A.; Herzmark, P.; Straight, A.; Kelly, K.; Takuwa, Y.; Sugimoto, N.; Mitchison, T.; Bourne, H.R. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell 2003, 114, 201–214. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB signaling: Navigating the network. Cell 2018, 169, 381–405. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Src protein–tyrosine kinase structure and regulation. Biochem. Biophys. Res. Commun. 2004, 324, 1155–1164. [Google Scholar] [CrossRef]
- Tillo, S.E.; Xiong, W.H.; Thakahashi, M.; Miao, S.; Andrade, A.L.; Fortin, D.A.; Yang, G.; Qin, M.; Smoody, B.F.; Stork, P.J.S.; et al. Liberated PKA catalytic subunits associate with the membrane via myristoylation to preferentially phosphorylate membrane substrates. Cell Rep. 2017, 19, 617–629. [Google Scholar] [CrossRef]
- Behnia, R.; Munro, S. Organelle identity and the signposts for membrane traffic. Nature 2005, 438, 597–604. [Google Scholar] [CrossRef]
- Aitken, A.; Cohen, P.; Santikarn, S.; Williams, D.H.; Calder, A.G.; Smith, A.; Kleet, C.B. Identification of the NHZ-terminal blocking group of calcineurin B as myristic acid. FEBS Lett. 1982, 150, 314–318. [Google Scholar] [CrossRef]
- D’Souza-Schorey, C.; Chavrier, P. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006, 7, 347–358. [Google Scholar] [CrossRef]
- Mahajan-Thakur, S.; Bien-Möller, S.; Marx, S.; Schroeder, H.; Rauch, B.H. Sphingosine 1-phosphate (S1P) signaling in glioblastoma multiforme—A systematic review. Int. J. Mol. Sci. 2017, 18, 2448. [Google Scholar] [CrossRef]
- Erpel, T.; Courtneidge, S.A. Src family protein tyrosine kinases and cellular signal transduction pathways. Curr. Opin. Cell Biol. 1995, 7, 176–182. [Google Scholar] [CrossRef]
- Warder, S.E.; Tucker, L.A.; Mcloughlin, S.M.; Strelitzer, T.J.; Meuth, J.L.; Zhang, Q.; Sheppard, G.S.; Richardson, P.L.; Lesniewski, R.; Davidsen, S.K.; et al. Discovery, identification, and characterization of candidate pharmacodynamic markers of methionine aminopeptidase-2 inhibition. J. Proteome Res. 2008, 7, 4807–4820. [Google Scholar] [CrossRef]
- Frottin, F.; Bienvenut, W.V.; Bignon, J.; Jacquet, E.; Vaca Jacome, A.S.; Van Dorsselaer, A.; Cianferani, S.; Carapito, C.; Meinnel, T.; Giglione, C.; et al. MetAP1 and MetAP2 drive cell selectivity for a potent anti-cancer agent in synergy, by controlling glutathione redox state. Oncotarget 2016, 7, 63306–63323. [Google Scholar] [CrossRef]
- Okrój, M.; Kamysz, W.; Slominska, E.M.; Mysliwski, A.; Bigda, J. A novel mechanism of action of the fumagillin analog, TNP-470, in the B16F10 murine melanoma cell line. Anticancer Drugs 2005, 16, 817–823. [Google Scholar] [CrossRef]
- Liu, Y.; Kahn, R.A.; Prestegard, J.H. Structure and Membrane Interaction of Myristoylated ARF1. Structure 2009, 17, 79–87. [Google Scholar] [CrossRef]
- Abad, A.; Fernández-Molina, J.V.; Bikandi, J.; Ramírez, A.; Margareto, J.; Sendino, J.; Hernando, F.L.; Pontón, J.; Garaizar, J.; Rementeria, A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 2010, 27, 155–182. [Google Scholar] [CrossRef] [PubMed]
- Sigg, H.P.; Weber, H.P. Isolierung und strukturaufklarung von ovalicin. Helv. Chim. Acta 1968, 51, 1395–1408. [Google Scholar] [CrossRef]
- Griffith, E.C.; Su, Z.; Turk, B.E.; Chen, S.; Chang, Y.H.; Wu, Z.; Biemann, K.; Liu, J.O. Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem. Biol. 1997, 4, 461–471. [Google Scholar] [CrossRef]
- Mangmool, S.; Kurose, H. Gi/o protein-dependent and -independent actions of pertussis toxin (PTX). Toxins 2011, 3, 884–899. [Google Scholar] [CrossRef]
- Locht, C.; Coutte, L.; Mielcarek, N. The ins and outs of pertussis toxin. FEBS J. 2011, 278, 4668–4682. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.; Holmgren, J. Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell. Mol. Life Sci. 2008, 65, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.J. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon 2001, 39, 1793–1803. [Google Scholar] [CrossRef]
- El-Kenawi, A.E.; El-Remessy, A.B. Angiogenesis inhibitors in cancer therapy: Mechanistic perspective on classification and treatment rationales. Br. J. Pharmacol. 2013, 170, 712–729. [Google Scholar] [CrossRef]
- Kusaka, M.; Sudo, K.; Fujita, T.; Marui, S.; Itoh, F.; Ingber, D.; Folkman, J. Potent anti-angiogenic action of AGM-1470: Comparison to the fumagillin parent. Biochem. Biophys. Res. Commun. 1991, 174, 1070–1076. [Google Scholar] [CrossRef]
- Chun, E.; Han, C.K.; Yoon, J.H.; Sim, T.B.; Kim, Y.K.; Lee, K.Y. Novel inhibitors targeted to methionine aminopeptidase 2 (MetAP2) strongly inhibit the growth of cancers in xenografted nude model. Int. J. Cancer 2005, 114, 124–130. [Google Scholar] [CrossRef]
- Kidoikhammouan, S.; Seubwai, W.; Silsirivanit, A.; Wongkham, S.; Sawanyawisuth, K.; Wongkham, C. Blocking of methionine aminopeptidase-2 by TNP-470 induces apoptosis and increases chemosensitivity of cholangiocarcinoma. J. Cancer Res. Ther. 2019, 15, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Niwano, M.; Arii, S.; Mori, A.; Ishigami, S.; Harada, T.; Mise, M.; Furutani, M.; Fujioka, M.; Imamura, M. Inhibition of tumor growth and microvascular angiogenesis by the potent angiogenesis inhibitor, TNP-470, in rats. Surg. Today 1998, 28, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Mauriz, J.L.; Gonzalez, P.; Durán, M.C.; Molpeceres, V.; Culebras, J.M.; Gonzalez-Gallego, J. Cell-cycle inhibition by TNP-470 in an in vivo model of hepatocarcinoma is mediated by a p53 and p21WAF1/CIP1 mechanism. Transl. Res. 2007, 149, 46–53. [Google Scholar] [CrossRef]
- Bhargava, P.; Marshall, J.L.; Rizvi, N.; Dahut, W.; Yoe, J.; Figuera, M.; Phipps, K.; Ong, V.S.; Kato, A.; Hawkins, M.J. A Phase I and pharmacokinetic study of TNP-470 administered weekly to patients with advanced cancer. Clin. Cancer Res. 1999, 5, 1989–1995. [Google Scholar]
- Audemard, A.; Le Bellec, M.L.; Carluer, L.; Dargère, S.; Verdon, R.; Castrale, C.; Lobbedez, T.; Hurault de Ligny, B. Fumagillin-induced aseptic meningoencephalitis in a kidney transplant recipient with microsporidiosis. Transpl. Infect. Dis. 2012, 14, 147–149. [Google Scholar] [CrossRef]
- Molina, J.M.; Goguel, J.; Sarfati, C.; Chastang, C.; Desportes-Livage, I.; Michiels, J.F.; Maslo, C.; Katlama, C.; Cotte, L.; Leport, C.; et al. Potential efficiency of fumagillin in intestinal microsporidiosis due to Enterocytozoon bieneusi in patients with HIV Infection: Results of a drug screening study. AIDS 1997, 11, 1603–1610. [Google Scholar] [CrossRef]
- Hou, L.; Mori, D.; Takase, Y.; Meihua, P.; Kai, K.; Tokunaga, O. Fumagillin inhibits colorectal cancer growth and metastasis in mice: In vivo and in vitro study of anti-angiogenesis. Pathol. Int. 2009, 59, 448–461. [Google Scholar] [CrossRef]
- Tanaka, S.; Arii, S. Current status and perspective of antiangiogenic therapy for cancer: Hepatocellular carcinoma. Int. J. Clin. Oncol. 2006, 11, 82–89. [Google Scholar] [CrossRef]
- Ogawa, H.; Sato, Y.; Kondo, M.; Takahashi, N.; Oshima, T.; Sasaki, F.; Une, Y.; Nishihira, J.; Todo, S. Combined treatment with TNP-470 and 5-fluorouracil effectively inhibits growth of murine colon cancer cells in vitro and liver metastasis in vivo. Oncol. Rep. 2000, 7, 467–472. [Google Scholar] [CrossRef]
- Wang, J.; Sheppard, G.S.; Lou, P.; Kawai, M.; BaMaung, N.; Erickson, S.A.; Tucker-Garcia, L.; Park, C.; Bouska, J.; Wang, Y.C.; et al. Tumor suppression by a rationally designed reversible inhibitor of methionine aminopeptidase-2. Cancer Res. 2003, 63, 7861–7869. [Google Scholar]
- Hotz, H.G.; Reber, J.A.; Hotz, B.; Sanghavi, P.C.; Yu, T.; Foitzik, T.; Buhr, H.J.; Hines, O.J. Angiogenesis inhibitor TNP-470 reduces human pancreatic cancer growth. J. Gastrointest. Surg. 2001, 5, 131–138. [Google Scholar] [CrossRef]
- Kawarada, Y.; Ishikura, H.; Kishimoto, T.; Saito, K.; Takahashi, T.; Kato, H.; Yoshiki, T. Inhibitory effects of the antiangiogenic agent TNP-470 on establishment and growth of hematogenous metastasis of human pancreatic carcinoma in SCID beige mice in vivo. Pancreas 1997, 15, 251–257. [Google Scholar] [CrossRef]
- Shishido, T.; Yasoshima, T.; Denno, R.; Mukaiya, M.; Sato, N.; Hirata, K. Inhibition of liver metastasis of human pancreatic carcinoma by angiogenesis inhibitor TNP-470 in combination with cisplatin. Jpn. J. Cancer Res. 1998, 89, 963–969. [Google Scholar] [CrossRef]
- Kato, H.; Ishikura, H.; Kawarada, Y.; Furuya, M.; Kondo, S.; Kato, H.; Yoshiki, T. Anti-angiogenic treatment for peritoneal dissemination of pancreas adenocarcinoma: A study using TNP-470. Jpn. J. Cancer Res. 2001, 92, 67–73. [Google Scholar] [CrossRef]
- Miyazaki, J.; Tsuzuki, Y.; Matsuzaki, K.; Hokari, R.; Okada, Y.; Kawaguchi, A.; Nagao, S.; Itoh, K.; Miura, S. Combination therapy with tumor-lysate pulsed dendritic cells and antiangiogenic drug TNP-470 for mouse pancreatic cancer. Int. J. Cancer 2005, 117, 499–505. [Google Scholar] [CrossRef]
- Ho, D.H.; Wong, R.H. TNP-470 skews DC differentiation to Th1-stimulatory phenotypes and can serve as a novel adjuvant in a cancer vaccine. Blood Adv. 2018, 2, 1664–1679. [Google Scholar] [CrossRef]
- Yamaoka, M.; Yamamoto, T.; Masaki, T.; Ikeyama, S.; Sudo, K.; Fujita, T. Angiogenesis inhibitor TNP-470 (AGM-1470) potently inhibits the tumor growth of hormone-independent human breast and prostate carcinoma cell lines. Cancer Res. 2003, 53, 5233–5236. [Google Scholar]
- Tucker, L.A.; Zhang, Q.; Sheppard, G.S.; Jiang, F.; McKeegan, E.; Lesniewski, R.; Davidsen, S.K.; Bell, R.L.; Wang, J. Ectopic expression of methionine aminopeptidase-2 causes cell transformation and stimulates proliferation. Oncogene 2008, 27, 3967–3976. [Google Scholar] [CrossRef]
- Logothetis, C.J.; Wu, K.K.; Finn, L.D.; Figg, W.; Ghaddar, H.; Gutterman, J.U. Phase I trial of the angiogenesis inhibitor TNP-470 for progressive androgen-independent prostate cancer. Clin. Cancer Res. 2001, 7, 1198–1203. [Google Scholar]
- Bo, H.; Ghazizadeh, M.; Shimizu, H.; Kurihara, Y.; Egawa, S.; Moriyama, Y.; Tajiri, T.; Kawanami, O. Effect of ionizing irradiation on human esophageal cancer cell lines by cDNA microarray gene expression analysis. J. Nippon Med. Sch. 2004, 71, 172–180. [Google Scholar] [CrossRef]
- Conteas, C.N.; Berlin, O.G.; Ash, L.R.; Pruthi, J.S. Therapy for human gastrointestinal microsporidiosis. Am. J. Trop. Med. Hyg. 2000, 63, 121–127. [Google Scholar] [CrossRef]
- Champion, L.; Durrbach, A.; Lang, P.; Delahousse, M.; Chauvet, C.; Sarfati, C.; Glotz, D.; Molina, J.M. Fumagillin for treatment of intestinal microsporidiosis in renal transplant recipients. Am. J. Transpl. 2010, 10, 1925–1930. [Google Scholar] [CrossRef]
- Bukreyeva, I.; Angoulvant, A.; Bendib, I.; Gagnard, J.C.; Bourhis, J.H.; Dargère, S.; Bonhomme, J.; Thellier, M.; Gachot, B.; Wyplosz, B. Enterocytozoon bieneusi microsporidiosis in stem cell transplant recipients treated with fumagillin. Emerg. Infect. Dis. 2017, 23, 1039–1041. [Google Scholar] [CrossRef]
- Bailey, L.; Ball, B.V. Honey Bee Pathology, 2nd ed.; Academic Press: London, UK, 1991; pp. 1–298. [Google Scholar]
- Fries, I. Nosema apis—A parasite in the honey bee colony. Bee World 1993, 74, 5–19. [Google Scholar] [CrossRef]
- Bailey, L. Effect of fumagillin upon Nosema apis (Zander). Nature 1953, 171, 212–213. [Google Scholar] [CrossRef]
- Katznelson, H.; Jamieson, C.A. Control of Nosema disease of honeybees with fumagillin. Science 1952, 115, 70–71. [Google Scholar] [CrossRef]
- Hartwig, A.; Przełȩcka, A. Nucleic acids in the intestine of Apis mellifera infected with Nosema apis and treated with fumagillin DCH: Cytochemical and autoradiographic studies. J. Invertebr. Pathol. 1971, 18, 331–336. [Google Scholar] [CrossRef]
- Webster, T.C. Fumagillin affects Nosema apis and honey bees (Hymenopterai Apidae). J. Econ. Entomol. 1994, 87, 601–604. [Google Scholar] [CrossRef]
- Williams, G.R.; Sampson, M.A.; Shutler, D.; Rogers, R.E.L. Does fumagillin control the recently detected invasive parasite Nosema ceranae in Western honey bees (Apis mellifera)? J. Invertebr. Pathol. 2008, 99, 342–344. [Google Scholar] [CrossRef]
- Huang, W.F.; Solter, L.F.; Yau, P.M.; Imai, B.S. Nosema ceranae escapes fumagillin control in honey bees. PLoS Pathol. 2013, 9. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.Q.; Li, H.M.; Zhang, Q.H.; Gao, Y.; Li, X.J. Antibiotic residues in honey: A review on analytical methods by liquid chromatography tandem mass spectrometry. Trends Anal. Chem. 2019, 110, 344–356. [Google Scholar] [CrossRef]
- Zhang, P.; Nicholson, D.E.; Bujnicki, J.M.; Su, X.; Brendle, J.J.; Ferdig, M.; Kyle, D.E.; Milhous, W.K.; Chiang, P.K. Angiogenesis inhibitors specific for methionine aminopeptidase 2 as drugs for malaria and leishmaniasis. J. Biomed. Sci. 2002, 9, 34–40. [Google Scholar] [CrossRef]
- Chen, X.; Xie, S.; Bhat, S.; Kumar, N.; Shapiro, T.A.; Liu, J.O. Fumagillin and fumarranol interact with P. falciparum methionine aminopeptidase 2 and inhibit malaria parasite growth in vitro and in vivo. Chem. Biol 2009, 16, 193–202. [Google Scholar] [CrossRef]
- Hillmann, F.; Novohradská, S.; Mattern, D.J.; Forberger, T.; Heinekamp, T.; Westermann, M.; Winckler, T.; Brakhage, A.A. Virulence determinants of the human pathogenic fungus Aspergillus fumigatus protect against soil amoeba predation. Environ. Microbiol. 2015, 17, 2858–2869. [Google Scholar] [CrossRef]
- Watanabe, N.; Nishihara, Y.; Yamaguchi, T.; Koito, A.; Miyoshi, H.; Kakeya, H.; Osada, H. Fumagillin suppresses HIV-1 infection of macrophages through the inhibition of Vpr activity. FEBS Lett. 2006, 580, 2598–2602. [Google Scholar] [CrossRef]
- Cao, Y. Angiogenesis modulates adipogenesis and obesity. J. Clin. Investig. 2007, 117, 2362–2368. [Google Scholar] [CrossRef]
- Lijnen, H.R.; Frederix, L.; Van Hoef, B. Fumagillin reduces adipose tissue formation in murine models of nutritionally induced obesity. Obesity 2010, 18, 2241–2246. [Google Scholar] [CrossRef]
- Scroyen, I.; Christiaens, V.; Lijnen, H.R. Effect of fumagillin on adipocyte differentiation and adipogenesis. Biochim. Biophys. Acta 2010, 1800, 425–429. [Google Scholar] [CrossRef]
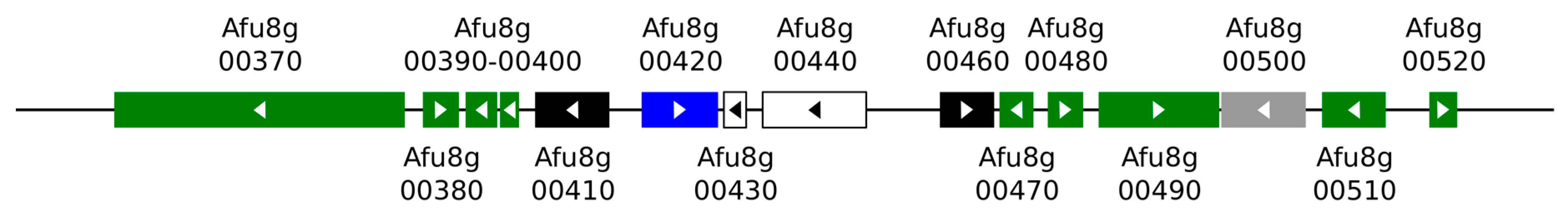
| Gene Name b | Other Gene Names c | Protein Abbreviated (Complete) Name d | UniProtKB d | Deletion Mutant | Ref. | |
|---|---|---|---|---|---|---|
| Afu8g00370 | fma-PKS | af370 fmaB | Fma-PKS (Fumagillin dodecapentaenoate synthase/Fumagillin biosynthesis polyketide synthase) | Q4WAY3 | Fumagillin production abolished Fumagillol production | [28,62] |
| Afu8g00380 | fma-AT | af380 fmaC | Fma-AT (Polyketide transferase af380/Fumagillin biosynthesis acyltransferase) | Q4WAY4 | Fumagillin production abolished Fumagillol production | [62] |
| Afu8g00390 /00400 | Afu8g00390 | af390-400 fmaD | Fma-MT (O-methyltransferase af390-400/Fumagillin biosynthesis methyltransferase) | A0A067Z9B6 | Fumagillin production abolished Demethyl-fumagillin production | [28,63] |
| Afu8g00410 | metAP | fpaII | MAP2-1/MetAP2-1 (Methionine aminopeptidase 2-1/Peptidase M) | Q4WAY7 | [68] | |
| Afu8g00420 | fumR | fapR | FumR (C6 finger transcription factor fumR/Fumagillin gene cluster regulator) | Q4WAY8 | Fumagillin and pseurotin gene clusters silenced | [28,64] |
| Afu8g00430 | Afu8g00430 | (EthD domain-containing protein) | Q4WAY9 | [62,69] | ||
| Afu8g00440 | Afu8g00440 | psoF | PsoF (Baeyer-Villiger monooxygenase/Dual-functional monooxygenase/methyltransferase psoF/Pseurotin biosynthesis protein F) | Q4WAZ0 | Pseurotin production abolished | [28,62,69,70,71] |
| Afu8g00460 | Afu8g00460 | fpaI | (Methionine aminopeptidase) | Q4WAZ1 | [28,62] | |
| Afu8g00470 | Afu8g00470 | af470 fmaE | Fma-ABM (Monooxygenase af470/Fumagillin biosynthesis antibiotic biosynthesis monooxygenase superfamily monooxygenase) | Q4WAZ2 | Fumagillin production abolished Prefumagillin accumulation | [28,62,63] |
| Afu8g00480 | Afu8g00480 | af480 fmaF | Fma-C6H (Dioxygenase af480/Fumagillin biosynthesis cluster C-6 hydroxylase) | Q4WAZ3 | Fumagillin production abolished 6-demethoxy-fumagillin accumulation | [28,62,63] |
| Afu8g00490 | Afu8g00490 | af490 | Fma-KR (Stereoselective keto-reductase af490/Fumagillin biosynthesis cluster keto-reductase) | Q4WAZ4 | Fumagillin production strongly decreased | [62,63,72] |
| Afu8g00500 | Afu8g00500 | (Acetate-CoA ligase, putative) | Q4WAZ5 | [62,72] | ||
| Afu8g00510 | Afu8g00510 | af510 fmaG | Fma-P450 (Multifunctional cytochrome P450 monooxygenase af510/ Fumagillin biosynthesis cluster P450 monooxygenase) | Q4WAZ6 | Fumagillin production abolished, β-trans-bergamotene accumulation | [28,62,63] |
| Afu8g00520 | fma-TC | af520 fmaA | Fma-TC (Fumagillin beta-trans-bergamotene synthase af520/Fumagillin biosynthesis terpene cyclase) | M4VQY9 | Fumagillin production abolished | [28,62] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guruceaga, X.; Perez-Cuesta, U.; Abad-Diaz de Cerio, A.; Gonzalez, O.; Alonso, R.M.; Hernando, F.L.; Ramirez-Garcia, A.; Rementeria, A. Fumagillin, a Mycotoxin of Aspergillus fumigatus: Biosynthesis, Biological Activities, Detection, and Applications. Toxins 2020, 12, 7. https://doi.org/10.3390/toxins12010007
Guruceaga X, Perez-Cuesta U, Abad-Diaz de Cerio A, Gonzalez O, Alonso RM, Hernando FL, Ramirez-Garcia A, Rementeria A. Fumagillin, a Mycotoxin of Aspergillus fumigatus: Biosynthesis, Biological Activities, Detection, and Applications. Toxins. 2020; 12(1):7. https://doi.org/10.3390/toxins12010007
Chicago/Turabian StyleGuruceaga, Xabier, Uxue Perez-Cuesta, Ana Abad-Diaz de Cerio, Oskar Gonzalez, Rosa M. Alonso, Fernando Luis Hernando, Andoni Ramirez-Garcia, and Aitor Rementeria. 2020. "Fumagillin, a Mycotoxin of Aspergillus fumigatus: Biosynthesis, Biological Activities, Detection, and Applications" Toxins 12, no. 1: 7. https://doi.org/10.3390/toxins12010007
APA StyleGuruceaga, X., Perez-Cuesta, U., Abad-Diaz de Cerio, A., Gonzalez, O., Alonso, R. M., Hernando, F. L., Ramirez-Garcia, A., & Rementeria, A. (2020). Fumagillin, a Mycotoxin of Aspergillus fumigatus: Biosynthesis, Biological Activities, Detection, and Applications. Toxins, 12(1), 7. https://doi.org/10.3390/toxins12010007






