Identification and Evaluations of Novel Insecticidal Proteins from Plants of the Class Polypodiopsida for Crop Protection against Key Lepidopteran Pests
Abstract
1. Introduction
2. Results
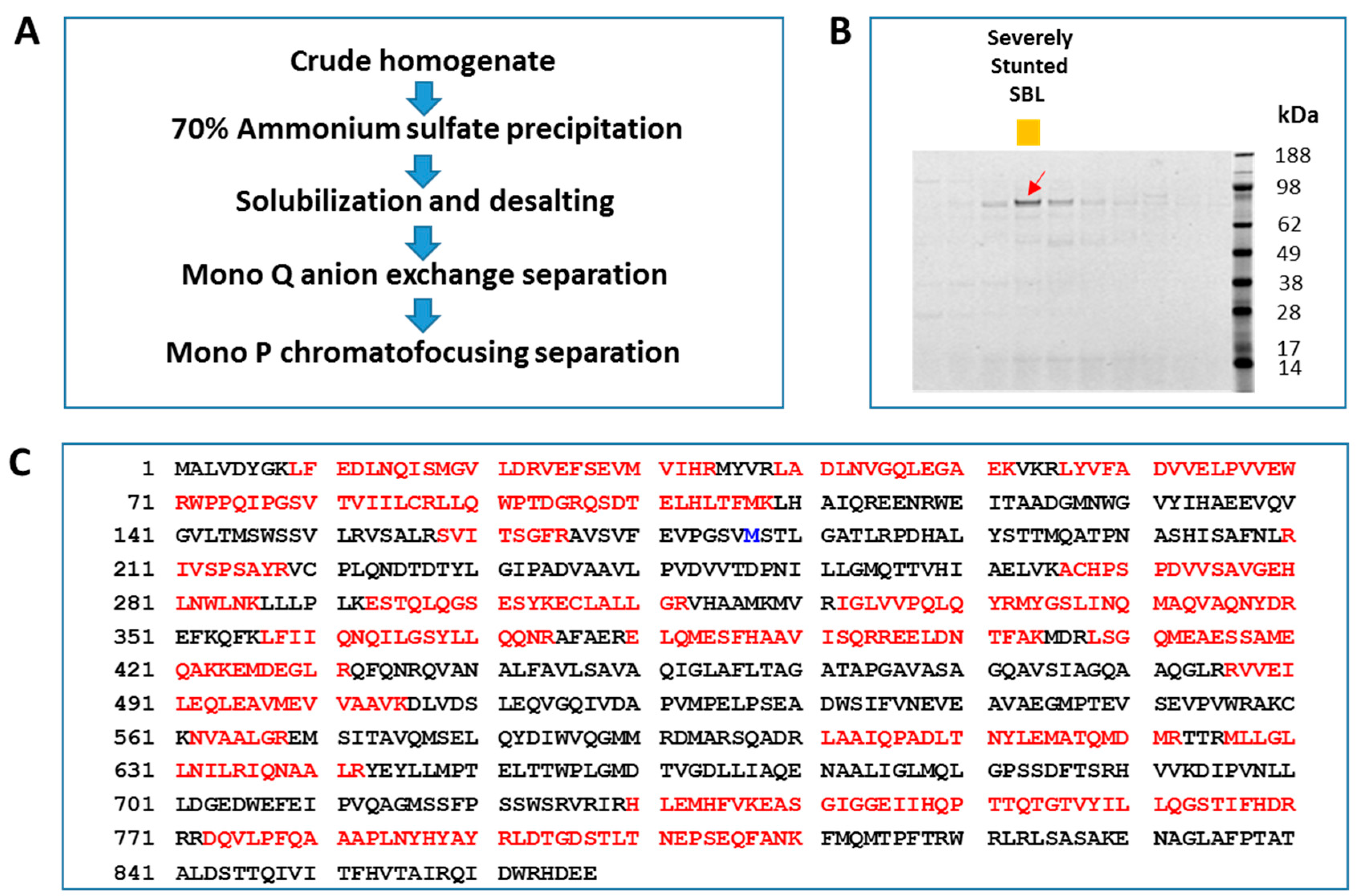
2.1. Plant Diversity Screening, Protein Purification, and Identification
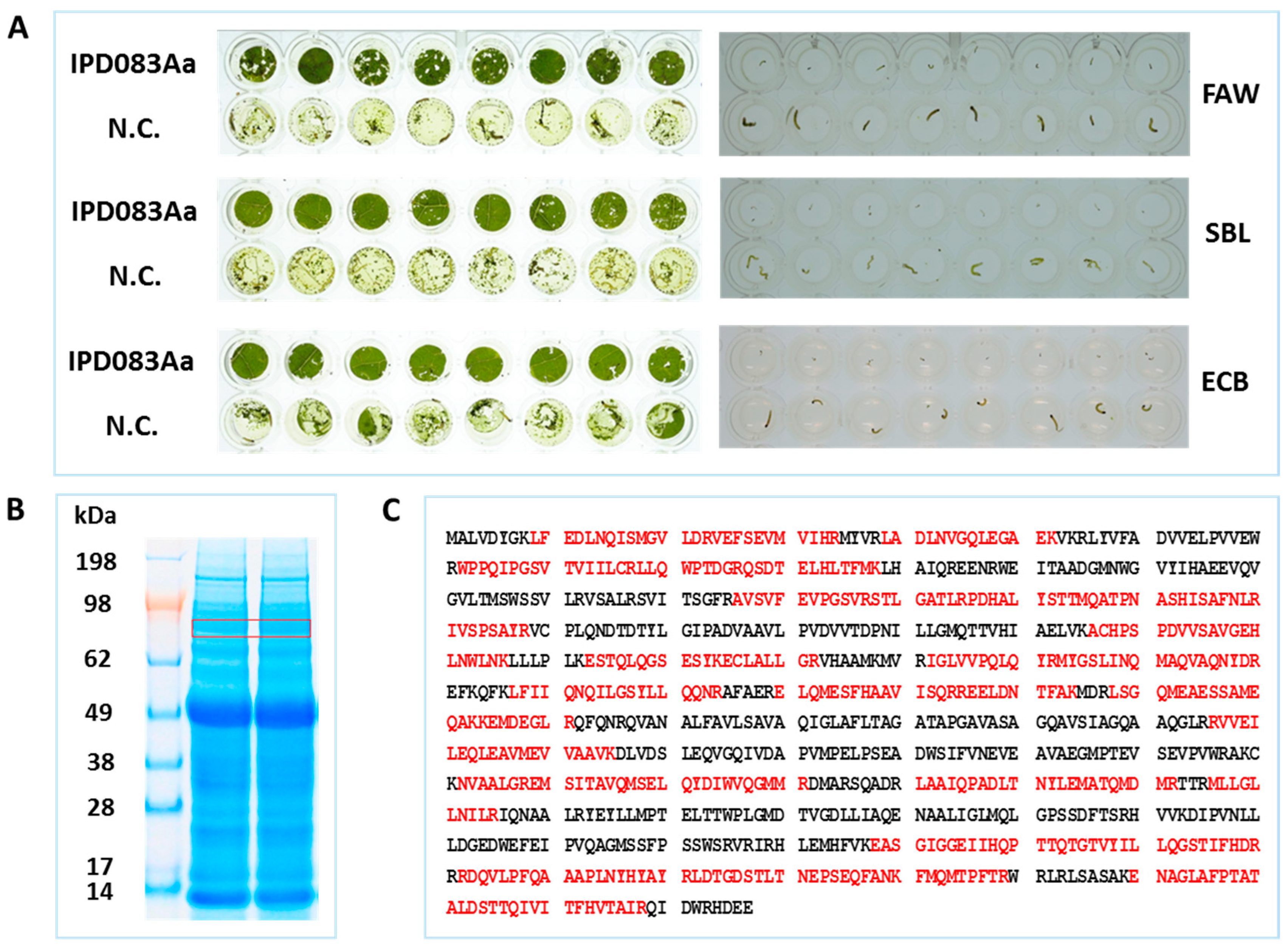
2.2. Insecticidal Activity Confirmation
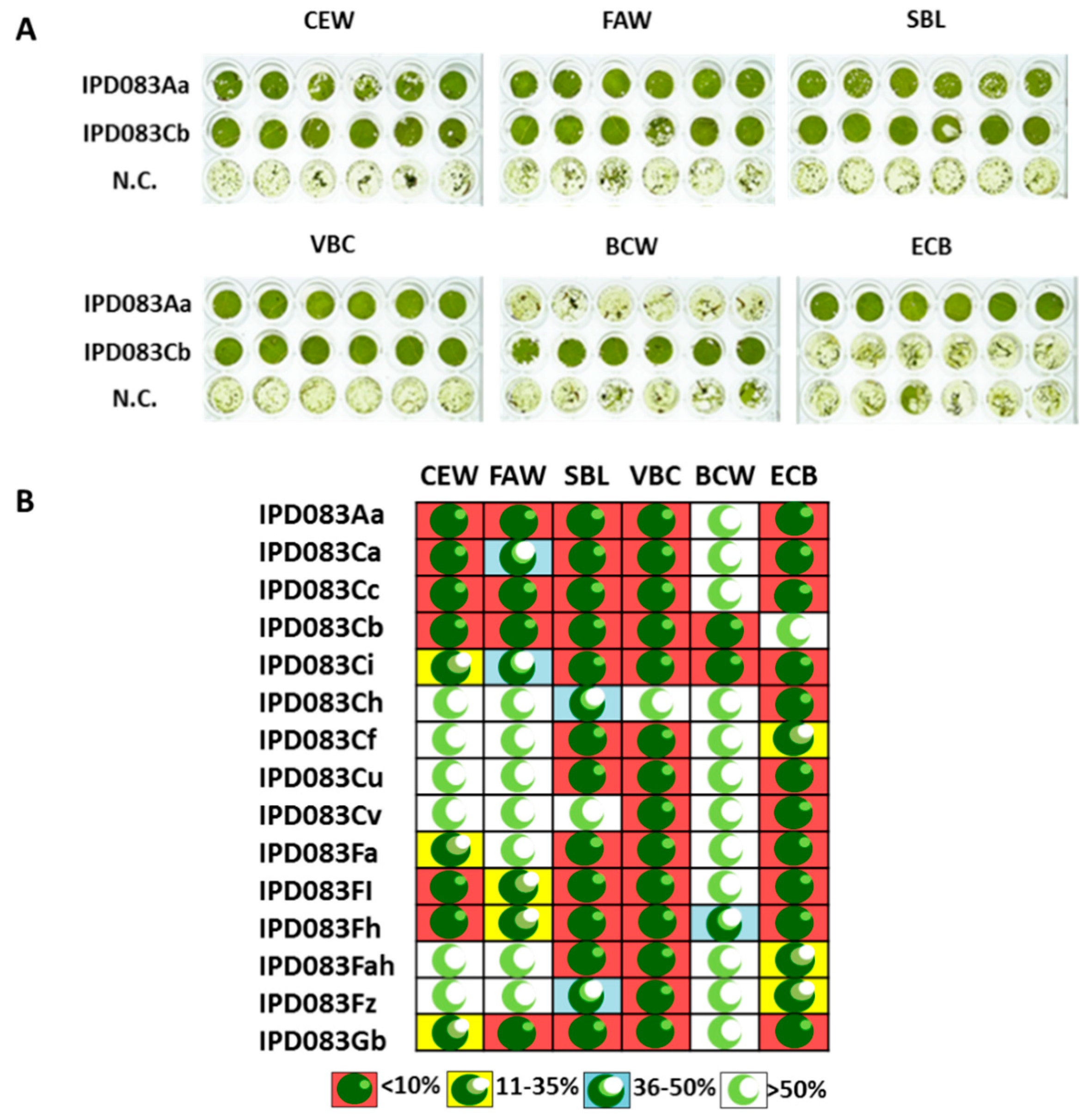
2.3. Homolog Identification, Insecticidal Activity, and Spectrum Assessment
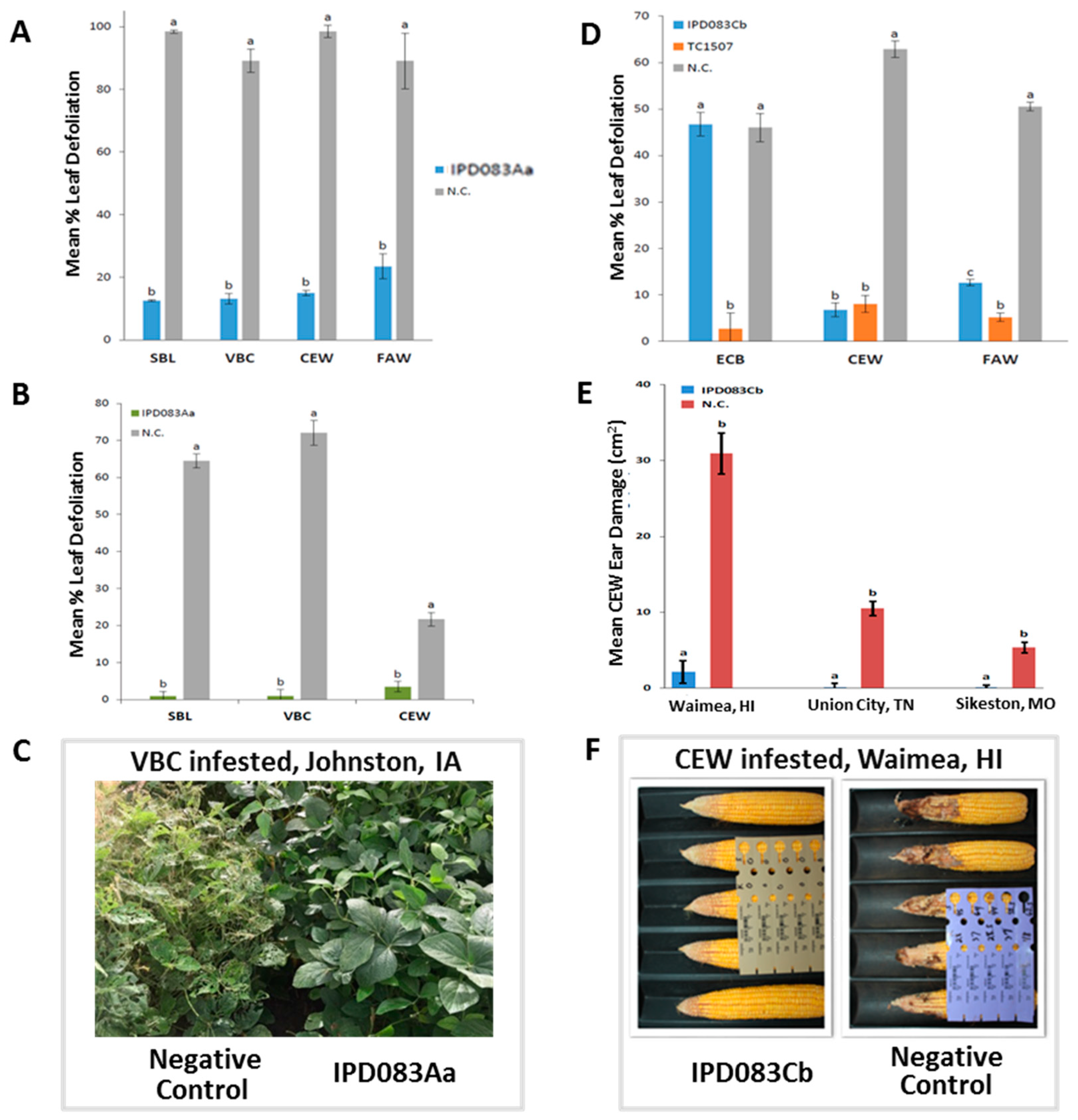
2.4. Evaluation of Insecticidal Activity of IPD083Aa in Soybean Plants
2.5. Evaluation of Insecticidal Activity of IPD083Cb in Corn Plants
2.6. Recombinant Protein Characterization, Activity, and Spectrum Evaluation
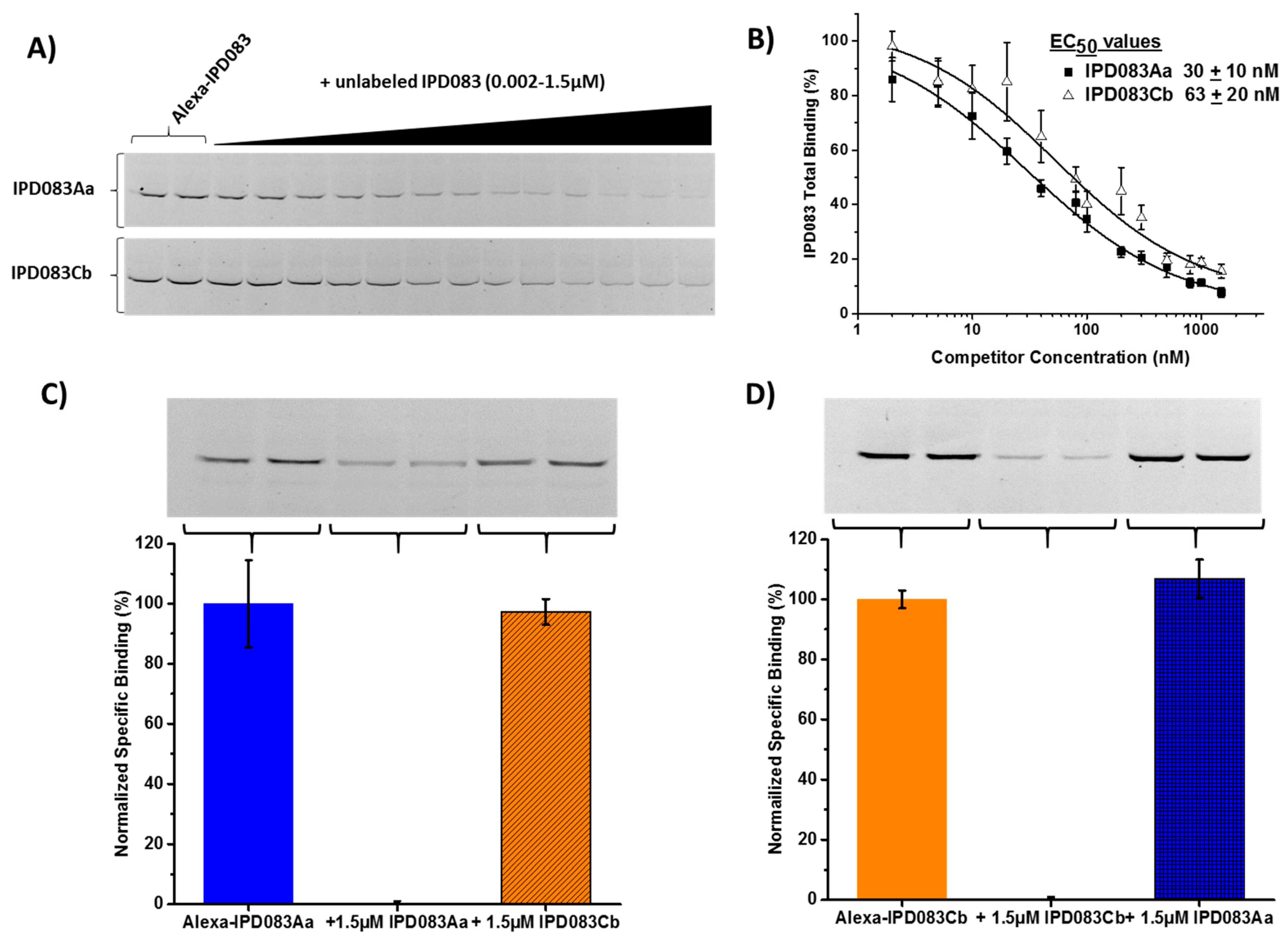
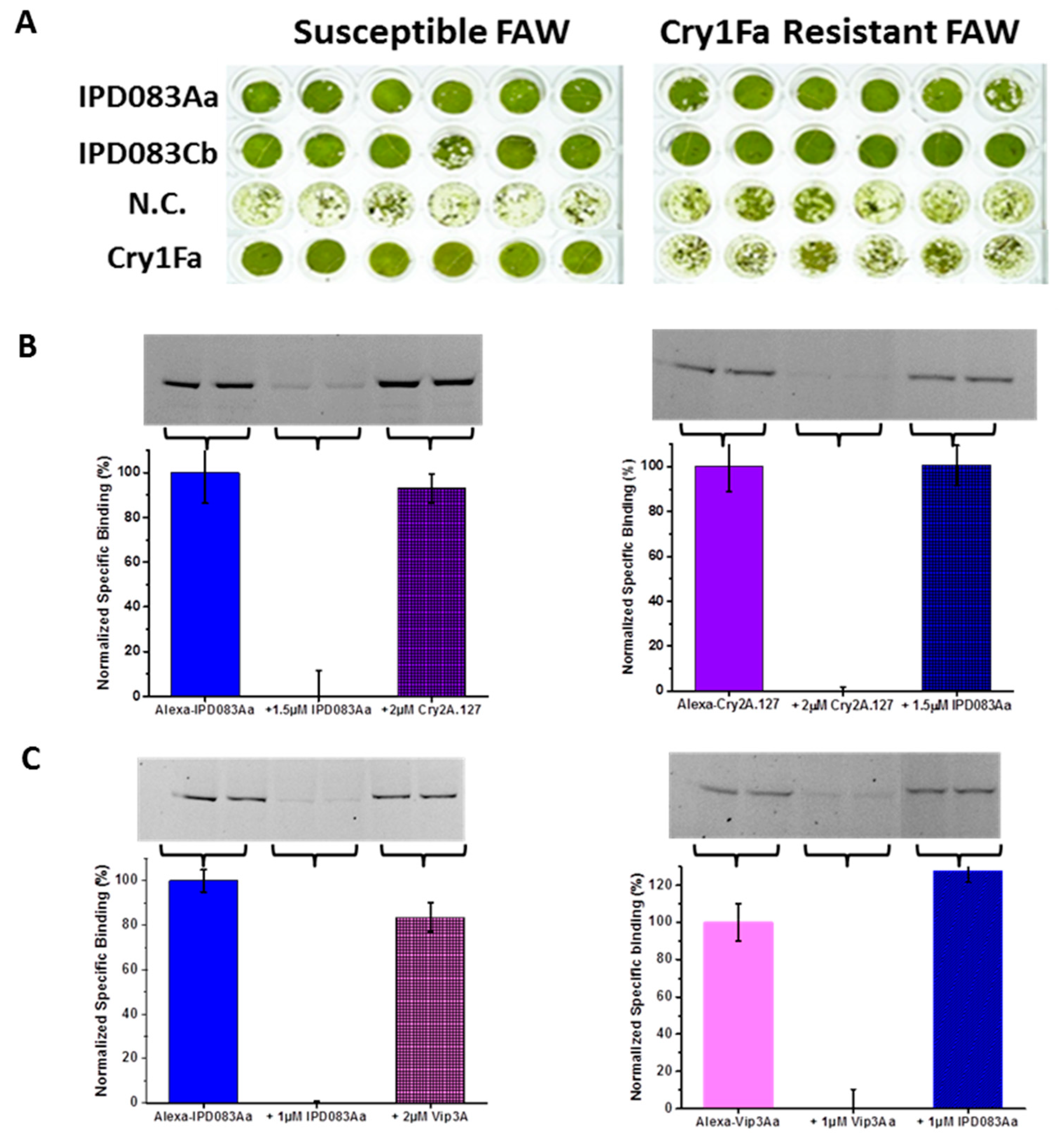
2.7. Evaluation of the Interaction Between IPD083 Proteins and CEW or FAW Midgut Tissues
2.8. Evaluation of the Target Sites of IPD083 Proteins Against Major Classes of Lepidopteran Active Bt Proteins
3. Discussion
4. Materials and Methods
4.1. Artificial Diet Insect Bioassays for Plant Diversity Screening and Protein Purification
4.2. Generation of Plant Transcriptome Sequence Libraries
4.3. Plant Sample Preparation, Protein Purification, and Identification
4.4. Functional Verification of IPD083Aa and its Homologs in the Common Bean Transient Expression System
4.5. Vector Construction, Transformation, and Greenhouse Efficacy Evaluation for Soybean
4.6. Field Evaluation of Transgenic Soybean Plants Expressing the IPD083Aa Protein
4.7. Vector Construction, Transformation, and Greenhouse Efficacy Evaluation for Corn
4.8. Field Evaluation of Transgenic Corn Plants Expressing the IPD083Cb Protein
4.9. Test for FAW Cross-Resistance to IPD083Aa and IPD083Cb Caused by Selection With Cry1Fa
4.10. Recombinant Protein Production and Functional Verification of IPD083Aa and IPD083Cb
4.11. Insect Brush Border Membrane Vesicle (BBMV) Binding of IPD083 Proteins and Competition Binding Studies
4.12. Recombinant Bt Protein Production for Binding Studies
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data and Materials Availability
References
- Onstad, D.W.; Crain, P.R. Major economic issues in integrated pest management. In Economics of Integrated Pest Management of Insects; Onstad, D.W., Crain, P.R., Eds.; CABI: Wallingford, UK, In Press.
- Oerke, E.C. Estimated crop losses due to pathogens, animal pests and weeds. In Crop Production and Crop Protection: Estimated Losses in Major Food and Cash Crops; Oerke, E.C., Dehne, H.W., Schonebeck, F., Weber, A., Eds.; Elsevier: Amsterdam, The Netherlands, 1994; pp. 77–88. [Google Scholar]
- Higley, L.C.; Boethel, D.J. Handbook of Soybean Insect Pests; Entomological Society of America: Lanham, MD, USA, 1994. [Google Scholar]
- Steffey, K.L.; Gray, M.E.; Andow, D.A.; Rice, M. Handbook of Corn Insects; Entomological Society of America: Lanham, MD, USA, 1999. [Google Scholar]
- National Academies of Sciences Engineering and Medicine. Genetically Engineered Crops: Experiences and Prospects; The National Academies Press: Washington, DC, USA, 2016; p. 606. [Google Scholar] [CrossRef]
- Nelson, M.E.; Alves, A.P. Plant incorporated protectants and insect resistance. In Insect Resistance Management: Biology, Economics, and Prediction, 2nd ed.; Onstad, D.W., Ed.; Academic Press: London, UK, 2014; pp. 99–147. [Google Scholar]
- Storer, N.P.; Thompson, G.D.; Head, G.P. Application of pyramided traits against Lepidoptera in insect resistance management for Bt crops. GM Crops Food 2012, 3, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Carriere, Y.; Fabrick, J.A.; Tabashnik, B.E. Can pyramids and seed mixtures delay resistance to Bt crops? Trends Biotechnol. 2016, 34, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E. Tips for battling billion-dollar beetles. Science 2016, 354, 552–553. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Carriere, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 2017, 35, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Mota-Sanchez, D.; Whalon, M.E.; Hollingworth, R.M.; Carriere, Y. Defining terms for proactive management of resistance to Bt crops and pesticides. J. Econ. Entomol. 2014, 107, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Pogue, M.G. A world revision of the genus Spodoptera Guenée (Lepidoptera: Noctuidae); American Entomological Society Philadelphia: Philadelphia, PA, USA, 2002. [Google Scholar]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamo, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in west and central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [PubMed]
- Sharanabasappa; Kalleshwaraswamy, C.M.; Asokan, R.; Swamy, H.M.M.; Maruthi, M.S.; Pavithra, H.B.; Hegde, K.; Navi, S.; Prabhu, S.T.; Goergen, G. First report of the fall armyworm, Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae), an alien invasive pest on maize in India. Pest Management Horticultural Ecosyst. 2018, 24, 23–29. [Google Scholar]
- FAO. Briefing Note on FAO Actions on Fall Armyworm; Food and Agriculture Organization of the United Nations: Rome, Italy, 5 March 2019; pp. 1–6. [Google Scholar]
- Storer, N.P.; Babcock, J.M.; Schlenz, M.; Meade, T.; Thompson, G.D.; Bing, J.W.; Huckaba, R.M. Discovery and characterization of field resistance to Bt maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. J. Econ. Entomol. 2010, 103, 1031–1038. [Google Scholar] [CrossRef]
- Gatehouse, J.A. Prospects for using proteinase inhibitors to protect transgenic plants against attack by herbivorous insects. Curr. Protein Pept. Sci. 2011, 12, 409–416. [Google Scholar] [CrossRef]
- Macedo, M.; Oliveira, C.; Oliveira, C. Insecticidal activity of plant lectins and potential application in crop protection. Molecules 2015, 20, 2014. [Google Scholar] [CrossRef]
- Sampson, K.; Zaitseva, J.; Stauffer, M.; Vande Berg, B.; Guo, R.; Tomso, D.; McNulty, B.; Desai, N.; Balasubramanian, D. Discovery of a novel insecticidal protein from Chromobacterium piscinae, with activity against western corn rootworm, Diabrotica virgifera virgifera. J. Invertebr. Pathol. 2017, 142, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.Z.; O’Rear, J.; Schellenberger, U.; Rosen, B.A.; Park, Y.J.; McDonald, M.J.; Zhu, G.; Xie, W.; Kassa, A.; Procyk, L.; et al. A selective insecticidal protein from Pseudomonas mosselii for corn rootworm control. Plant Biotechnol. J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yalpani, N.; Altier, D.; Barry, J.; Kassa, A.; Nowatzki, T.M.; Sethi, A.; Zhao, J.Z.; Diehn, S.; Crane, V.; Sandahl, G.; et al. An Alcaligenes strain emulates Bacillus thuringiensis producing a binary protein that kills corn rootworm through a mechanism similar to Cry34Ab1/Cry35Ab1. Sci. Rep. 2017, 7, 3063. [Google Scholar] [CrossRef] [PubMed]
- Schellenberger, U.; Oral, J.; Rosen, B.A.; Wei, J.Z.; Zhu, G.; Xie, W.; McDonald, M.J.; Cerf, D.C.; Diehn, S.H.; Crane, V.C.; et al. A selective insecticidal protein from Pseudomonas for controlling corn rootworms. Science 2016, 354, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Upadhyay, S.K.; Mishra, M.; Saurabh, S.; Singh, R.; Singh, H.; Thakur, N.; Rai, P.; Pandey, P.; Hans, A.L.; et al. Expression of an insecticidal fern protein in cotton protects against whitefly. Nat. Biotechnol. 2016, 34, 1046–1051. [Google Scholar] [CrossRef] [PubMed]
- Christenhusz, M.J.M.; Chase, M.W. Trends and concepts in fern classification. Ann. Botany 2014, 113, 571–594. [Google Scholar] [CrossRef]
- Hirosawa, M.; Hoshida, M.; Ishikawa, M.; Toya, T. MASCOT: Multiple alignment system for protein sequences based on three-way dynamic programming. Comput. Appl. Biosci. 1993, 9, 161–167. [Google Scholar] [CrossRef]
- Kapila, J.; Rycke, R.D.; Montagu, M.V.; Angenon, G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997, 122, 101–108. [Google Scholar] [CrossRef]
- Schob, H.; Kunz, C.; Meins, F., Jr. Silencing of transgenes introduced into leaves by agroinfiltration: A simple, rapid method for investigating sequence requirements for gene silencing. Mol. Gen. Genet. 1997, 256, 581–585. [Google Scholar]
- Dey, N.; Maiti, I.B. Structure and promoter/leader deletion analysis of mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Mol. Biol. 1999, 40, 771–782. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Roesler, K.; Shen, B.; Bermudez, E.; Li, C.; Hunt, J.; Damude, H.G.; Ripp, K.G.; Everard, J.D.; Booth, J.R.; Castaneda, L.; et al. An improved variant of soybean type 1 diacylglycerol acyltransferase increases the oil content and cecreases the soluble carbohydrate content of soybeans. Plant Physiol. 2016, 171, 878–893. [Google Scholar] [CrossRef] [PubMed]
- Grefen, C.; Donald, N.; Hashimoto, K.; Kudla, J.; Schumacher, K.; Blatt, M.R. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010, 64, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.; Lenderts, B.; Glassman, K.; Berezowska-Kaniewska, M.; Christensen, H.; Asmus, T.; Zhen, S.; Chu, U.; Cho, M.J.; Zhao, Z.Y. Optimized Agrobacterium-mediated sorghum transformation protocol and molecular data of transgenic sorghum plants. In Vitro Cell Dev. Biol. Plant 2014, 50, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.J.; Wu, E.; Kwan, J.; Yu, M.; Banh, J.; Linn, W.; Anand, A.; Li, Z.; TeRonde, S.; Register, J.C., 3rd; et al. Agrobacterium-mediated high-frequency transformation of an elite commercial maize (Zea mays L.) inbred line. Plant Cell Rep. 2014, 33, 1767–1777. [Google Scholar] [CrossRef]
- Christensen, A.H.; Quail, P.H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Wolt, J.D. A mixture toxicity approach for environmental risk assessment of multiple insect resistance genes. Environ. Toxicol. Chem. 2011, 30, 763–772. [Google Scholar] [CrossRef]
- Jurat-Fuentes, J.L.; Crickmore, N. Specificity determinants for Cry insecticidal proteins: Insights from their mode of action. J. Invertebr. Pathol. 2016. [Google Scholar] [CrossRef]
- Ferre, J.; Van Rie, J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 2002, 47, 501–533. [Google Scholar] [CrossRef]
- Velez, A.M.; Spencer, T.A.; Alves, A.P.; Moellenbeck, D.; Meagher, R.L.; Chirakkal, H.; Siegfried, B.D. Inheritance of Cry1F resistance, cross-resistance and frequency of resistant alleles in Spodoptera frugiperda (Lepidoptera: Noctuidae). Bull. Entomol. Res. 2013, 103, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Rodriguez, C.S.; Hernandez-Martinez, P.; Van Rie, J.; Escriche, B.; Ferre, J. Shared midgut binding sites for Cry1A.105, Cry1Aa, Cry1Ab, Cry1Ac and Cry1Fa proteins from Bacillus thuringiensis in two important corn pests, Ostrinia nubilalis and Spodoptera frugiperda. PLoS ONE 2013, 8, e68164. [Google Scholar] [CrossRef] [PubMed]
- Sanchis, V. From microbial sprays to insect-resistant transgenic plants: History of the biospesticide Bacillus thuringiensis. A review. Agron. Sustain. Dev. 2011, 31, 217–231. [Google Scholar] [CrossRef]
- Ciosi, M.; Miller, N.J.; Kim, K.S.; Giordano, R.; Estoup, A.; Guillemaud, T. Invasion of Europe by the western corn rootworm, Diabrotica virgifera virgifera: Multiple transatlantic introductions with various reductions of genetic diversity. Mol. Ecol. 2008, 17, 3614–3627. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cornejo, J.; Wechsler, S.J.; Livingston, M.; Mitchell, L. Genetically Engineered Crops in the United States; U.S. Department of Agriculture: Washington, DC, USA, 2014.
- Mohan, S.; Ma, P.W.; Williams, W.P.; Luthe, D.S. A naturally occurring plant cysteine protease possesses remarkable toxicity against insect pests and synergizes Bacillus thuringiensis toxin. PLoS ONE 2008, 3, e1786. [Google Scholar] [CrossRef] [PubMed]
- Dodt, M.; Roehr, J.T.; Ahmed, R.; Dieterich, C. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 2012, 1, 895–905. [Google Scholar] [CrossRef]
- Le, H.S.; Schulz, M.H.; McCauley, B.M.; Hinman, V.F.; Bar-Joseph, Z. Probabilistic error correction for RNA sequencing. Nucleic Acids Res. 2013, 41, e109. [Google Scholar] [CrossRef] [PubMed]
- Crusoe, M.R.; Alameldin, H.F.; Awad, S.; Boucher, E.; Caldwell, A.; Cartwright, R.; Charbonneau, A.; Constantinides, B.; Edvenson, G.; Fay, S.; et al. The khmer software package: Enabling efficient nucleotide sequence analysis. F1000Res. 2015, 4, 900. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Lv, M.J.; Zhu, X.G.; Chin, F.Y. IDBA-tran: A more robust de novo de Bruijn graph assembler for transcriptomes with uneven expression levels. Bioinformatics 2013, 29, i326–i334. [Google Scholar] [CrossRef]
- Schulz, M.H.; Zerbino, D.R.; Vingron, M.; Birney, E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012, 28, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Huang, W.; He, G.; Gu, S.; Li, S.; et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Callis, J.; Carpenter, T.; Sun, C.W.; Vierstra, R.D. Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 1995, 139, 921–939. [Google Scholar] [PubMed]
- Bubner, B.; Baldwin, I.T. Use of real-time PCR for determining copy number and zygosity in transgenic plants. Plant Cell Rep. 2004, 23, 263–271. [Google Scholar] [CrossRef]
- Fehr, W.R.; Caviness, C.E.; Burmood, D.T.; Pennington, J.S. Stage of Development Descriptions for Soybeans, Glycine Max (L.) Merrill1. Crop Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Ortega, M.A.; All, J.N.; Boerma, H.R.; Parrott, W.A. Pyramids of QTLs enhance host-plant resistance and Bt-mediated resistance to leaf-chewing insects in soybean. Theor. Appl. Genet. 2016, 129, 703–715. [Google Scholar] [CrossRef]
- An, G.; Mitra, A.; Choi, H.K.; Costa, M.A.; An, K.; Thornburg, R.W.; Ryan, C.A. Functional analysis of the 3’ control region of the potato wound-inducible proteinase inhibitor II gene. Plant Cell 1989, 1, 115–122. [Google Scholar]
- Komari, T. Transformation of cultured cells of Chenopodium quinoa by binary vectors that carry a fragment of DNA from the virulence region of pTiBo542. Plant Cell Rep. 1990, 9, 303–306. [Google Scholar] [CrossRef]
- Komari, T.; Hiei, Y.; Saito, Y.; Murai, N.; Kumashiro, T. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 1996, 10, 165–174. [Google Scholar] [CrossRef]
- Beirne, J.; Truchan, H.; Rao, L. Development and qualification of a size exclusion chromatography coupled with multiangle light scattering method for molecular weight determination of unfractionated heparin. Anal. Bioanal. Chem. 2011, 399, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Wolfersberger, M.; Luethy, P.; Maurer, A.; Parenti, P.; Sacchi, F.V.; Giordana, B.; Hanozet, G.M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Bioch. Physiol. 1987, 86A, 301–308. [Google Scholar] [CrossRef]
- Bermudez, E.; Emig, R.; McBride, K.; Yamamoto, T. Bacillus thuringiensis crystal polypeptides, polynucleotides, and compositions thereof. U.S. Patent 7,943,734, 17 May 2011. Seq ID No. 134. [Google Scholar]
- Kain, W.; Song, X.; Janmaat, A.F.; Zhao, J.Z.; Myers, J.; Shelton, A.M.; Wang, P. Resistance of Trichoplusia ni populations selected by Bacillus thuringiensis sprays to cotton plants expressing pyramided Bacillus thuringiensis toxins Cry1Ac and Cry2Ab. Appl. Environ. Microbiol. 2015, 81, 1884–1890. [Google Scholar] [CrossRef] [PubMed]
- Cerf, D.; Cong, R.; Freeman, M.; McBride, K.E.; Yamamoto, T. Bacillus thuringiensis crystal polypeptides, polynucleotides, and compositions thereof. U.S. Patent 8,530,411, 10 September 2013. Seq ID No. 22. [Google Scholar]
- Willcoxon, M.I.; Dennis, J.R.; Lau, S.; Xie, W.; You, Y.; Leng, S.; Fong, R.C.; Tamamoto, T. A high-throughput, in-vitro assay for Bacillus thuringiensis insecticidal proteins. J. Biotechnol. 2016, 217, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Crespo, A.L.; Spencer, T.A.; Tan, S.Y.; Siegfried, B.D. Fitness costs of Cry1Ab resistance in a field-derived strain of Ostrinia nubilalis (Lepidoptera: Crambidae). J. Econ. Entomol. 2010, 103, 1386–1393. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Protein Sequence ID to IPD083Aa (%) | GenBank Accession Number | Organism |
|---|---|---|---|
| IPD083Aa | 100 | KY588369 | Adiantum pedatum L. |
| IPD083Ca | 70 | KY588372 | Adiantum trapeziforme |
| IPD083Cc | 70 | KY588373 | Adiantum trapeziforme |
| IPD083Cb | 71 | KY588370 | Adiantum trapeziforme |
| IPD083Ci | 71 | KY588371 | Rumohra adiantiformis |
| IPD083Ch | 74 | KY588375 | Polystichum tsus-simense |
| IPD083Cf | 77 | KY588374 | Lygodium flexuosum |
| IPD083Cu | 72 | KY588376 | Dryopteris intermedia |
| IPD083Cv | 74 | KY588377 | Dryopteris intermedia |
| IPD083Fa | 48 | KY588378 | Polypodium musifolium |
| IPD083Fl | 48 | KY588379 | Davallia tyermanii |
| IPD083Fh | 46 | KY588380 | Phyllitis scolopendium |
| IPD083Fah | 49 | KY588383 | Coniogramme venusta |
| IPD083Fz | 45 | KY588382 | Osmunda regalis |
| IPD083Gb | 38 | KY588381 | Adiantum raddianum |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Schepers, E.; Lum, A.; Rice, J.; Yalpani, N.; Gerber, R.; Jiménez-Juárez, N.; Haile, F.; Pascual, A.; Barry, J.; et al. Identification and Evaluations of Novel Insecticidal Proteins from Plants of the Class Polypodiopsida for Crop Protection against Key Lepidopteran Pests. Toxins 2019, 11, 383. https://doi.org/10.3390/toxins11070383
Liu L, Schepers E, Lum A, Rice J, Yalpani N, Gerber R, Jiménez-Juárez N, Haile F, Pascual A, Barry J, et al. Identification and Evaluations of Novel Insecticidal Proteins from Plants of the Class Polypodiopsida for Crop Protection against Key Lepidopteran Pests. Toxins. 2019; 11(7):383. https://doi.org/10.3390/toxins11070383
Chicago/Turabian StyleLiu, Lu, Eric Schepers, Amy Lum, Janet Rice, Nasser Yalpani, Ryan Gerber, Nuria Jiménez-Juárez, Fikru Haile, Alejandra Pascual, Jennifer Barry, and et al. 2019. "Identification and Evaluations of Novel Insecticidal Proteins from Plants of the Class Polypodiopsida for Crop Protection against Key Lepidopteran Pests" Toxins 11, no. 7: 383. https://doi.org/10.3390/toxins11070383
APA StyleLiu, L., Schepers, E., Lum, A., Rice, J., Yalpani, N., Gerber, R., Jiménez-Juárez, N., Haile, F., Pascual, A., Barry, J., Qi, X., Kassa, A., Heckert, M. J., Xie, W., Ding, C., Oral, J., Nguyen, M., Le, J., Procyk, L., ... Wu, G. (2019). Identification and Evaluations of Novel Insecticidal Proteins from Plants of the Class Polypodiopsida for Crop Protection against Key Lepidopteran Pests. Toxins, 11(7), 383. https://doi.org/10.3390/toxins11070383






