Polyphasic Assessment of Aflatoxin Production Potential in Selected Aspergilli
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Selected Aspergilli
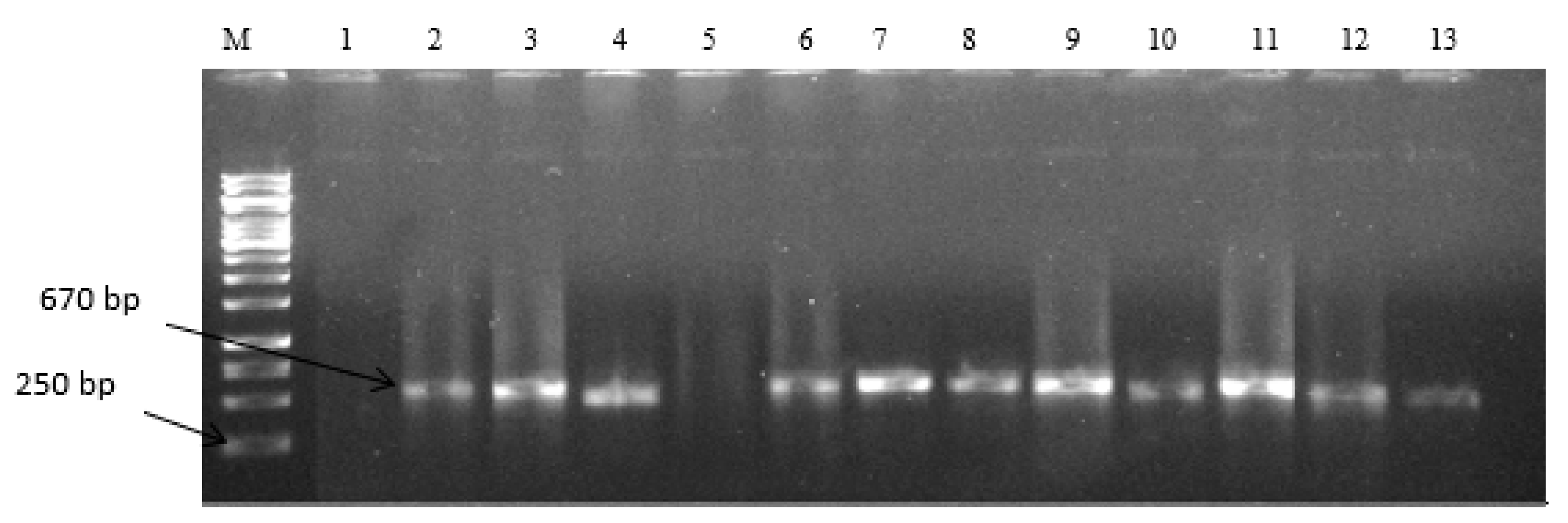
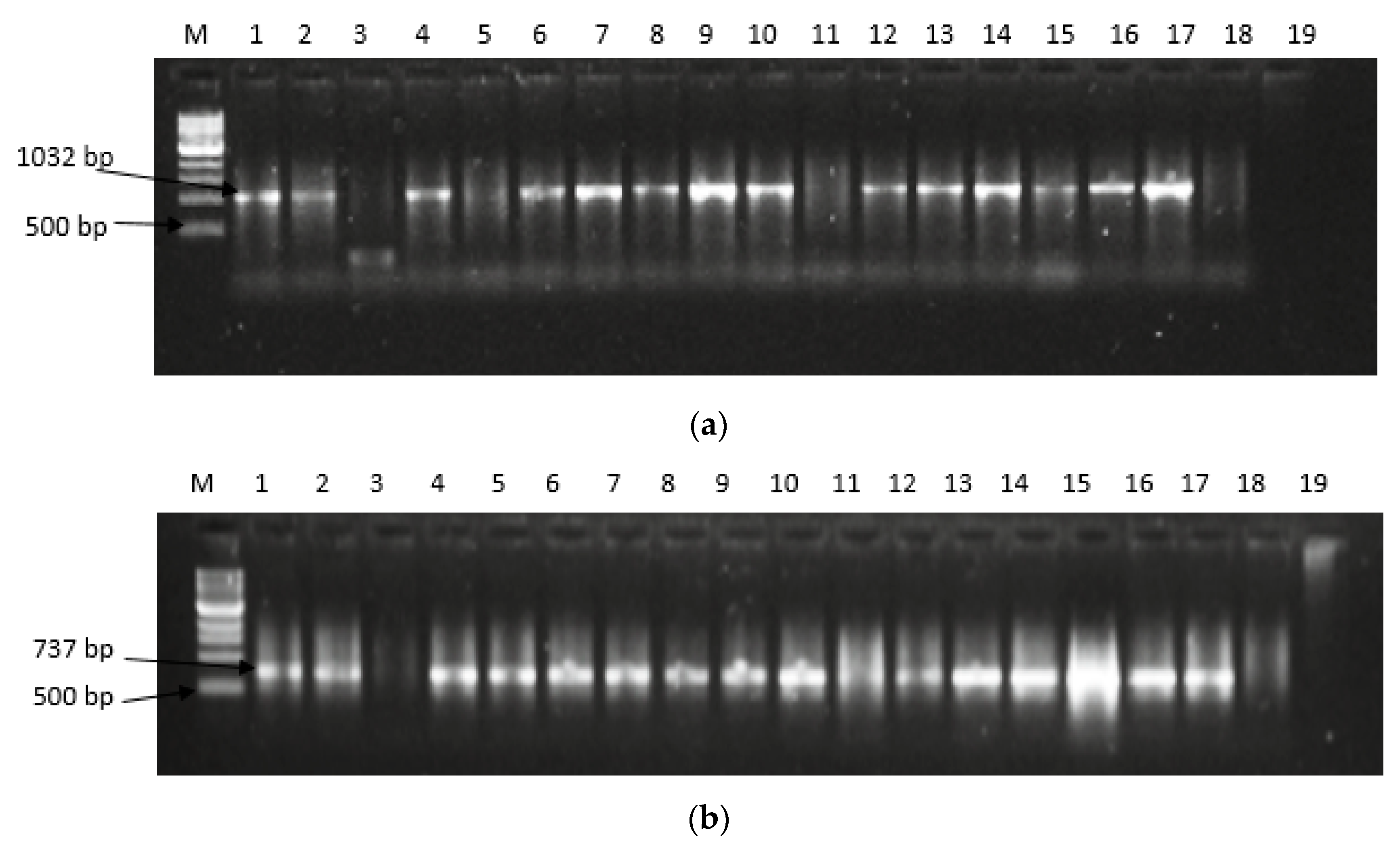
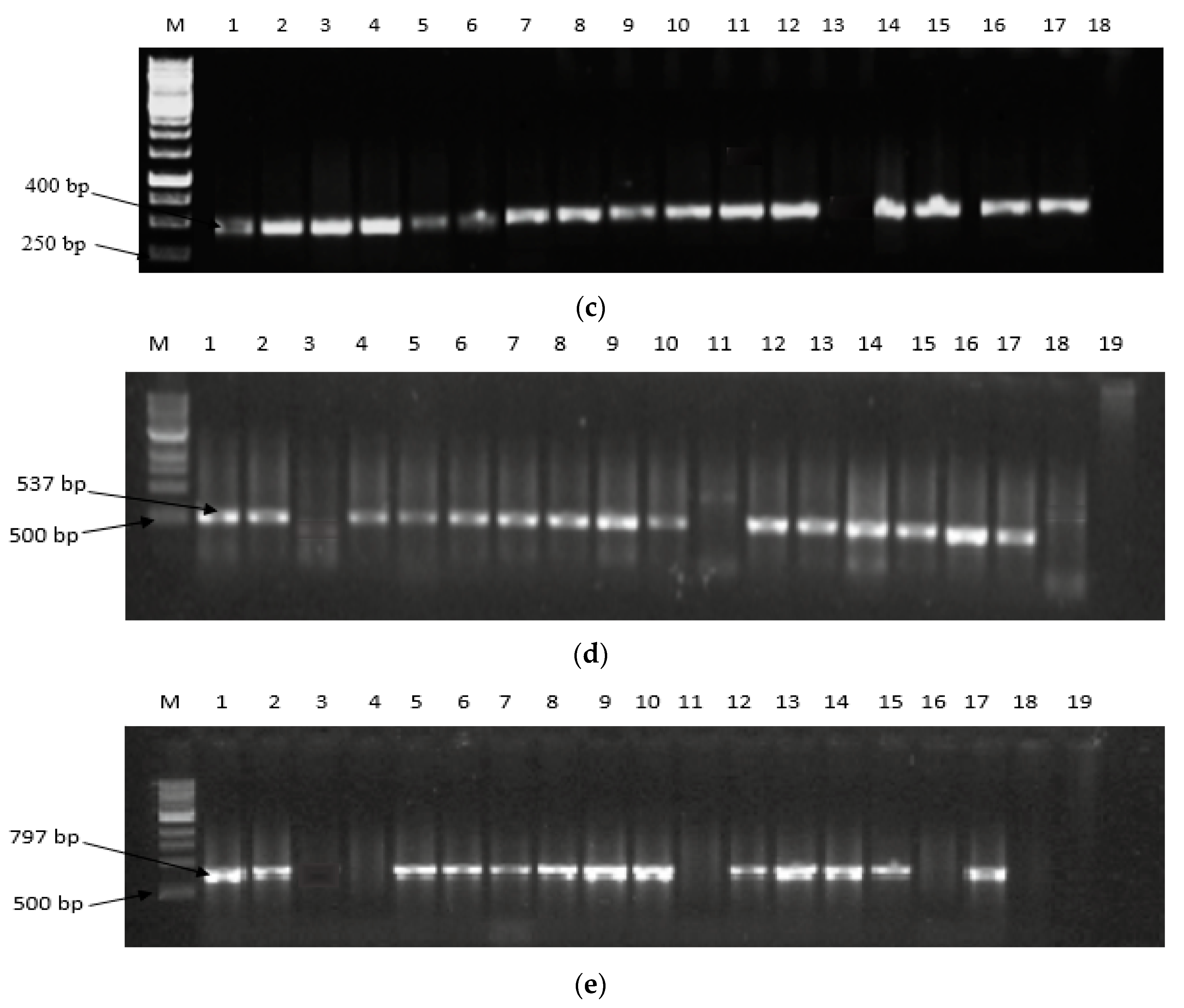
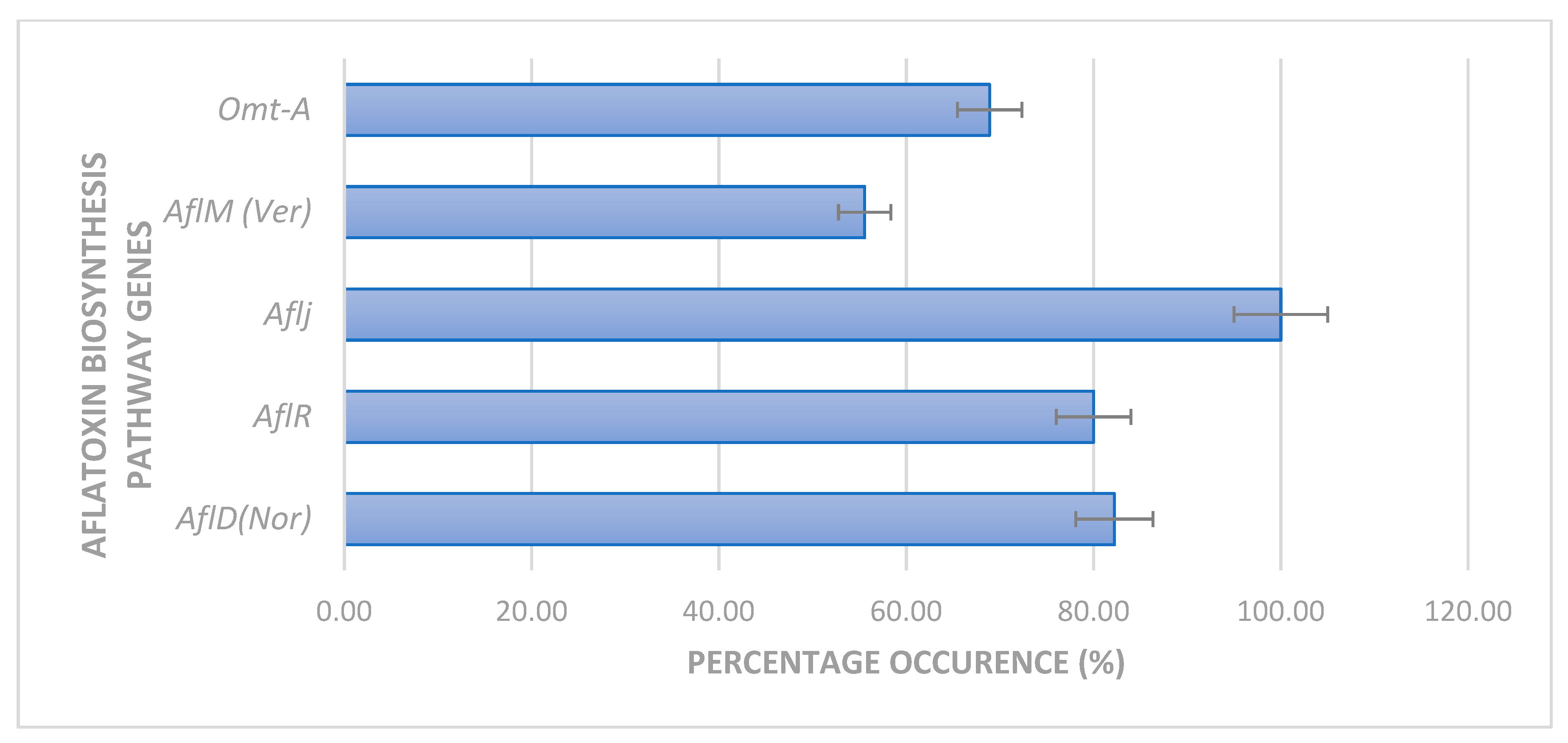
2.2. Expression of Aflatoxin Biosynthesis Pathway Genes
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Materials
5.2. Culturing of Fungi
5.3. Molecular Identification of Presumptive Aspergilli
PCR Amplification of ITS1 and ITS4 of Presumptive Aspergillus Isolates
5.4. Expression of Aflatoxin Biosynthesis Pathway Genes in Filamentous Aspergilli
5.5. Evaluation of Filamentous Aspergillus spp. for Aflatoxin Production
5.5.1. β-Cyclodextrin Neutral Red Desiccated Coconut Agar (β-CNRDCA) Assay
5.5.2. Yeast Extract Sucrose (Ammonium Vapor) Test Assay
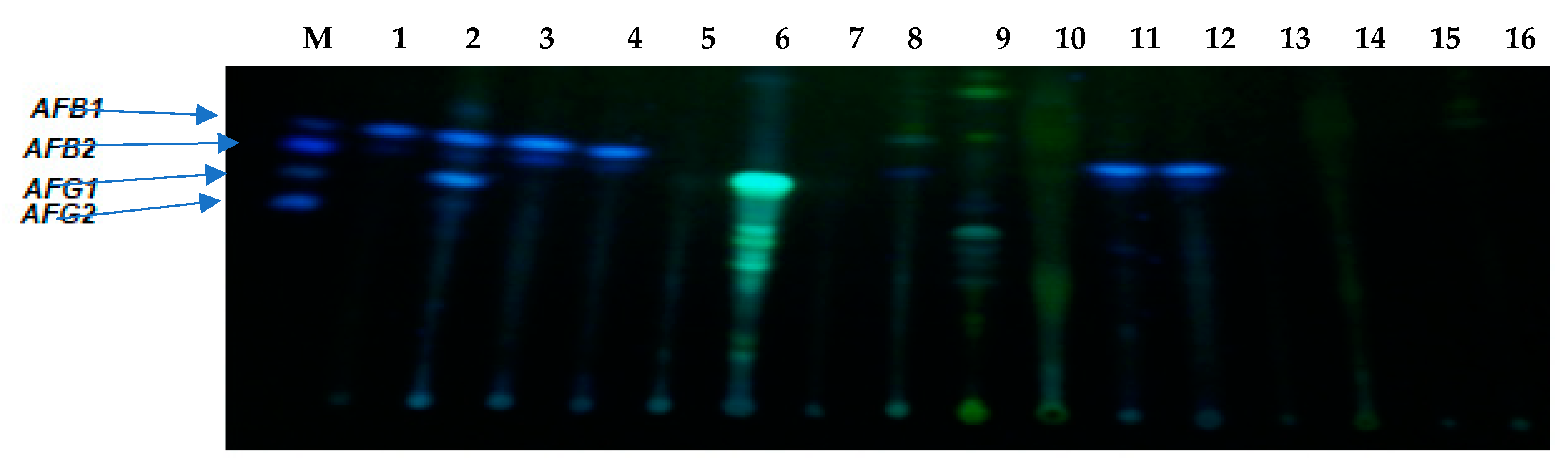
5.6. Aflatoxin Production by Aspergilli in Wheat Flour Using Thin-Layer Chromatography (TLC) Assay
5.7. Quantification of Aflatoxin by High-Performance Liquid Chromatography
5.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ominski, K.H.; Marquardi, R.R.; Sinha, R.N.; Abramson, D. Ecological aspects of growth and mycotoxin production by storage fungi. In Mycotoxins in Grains: Compounds Other than Aflatoxins; Miller, J.D., Trenholm, H.L., Eds.; Eagan Press: St. Paul, MN, USA, 1994; pp. 287–314. [Google Scholar]
- Bhat, R.; Rai, R.V.; Karim, A.A. Mycotoxins in food and feed: Present status and future concerns. Comp. Rev. Food Sci. Food Saf. 2010, 9, 57–81. [Google Scholar] [CrossRef]
- Charoenpornsook, K.; Kavisarasai, P. Mycotoxins in animal feedstuffs of Thailand. KMITL Sci. Technol. J. 2006, 6, 25–28. [Google Scholar]
- Francesca, B.; Chiara, M. Mycotoxins in Food, Food Industrial Processes—Methods and Equipment; InTechOpen: Rijeka, Croatia, 2012; Volume 10, pp. 169–199. [Google Scholar]
- Bryden, W.L. Mycotoxins in the food chain: Human health implications. Asia Pac. J. Clin. Nutr. 2007, 16, 95–101. [Google Scholar] [PubMed]
- Castegnaro, M.; Pfohl-Leszkowicz, A. Les mycotoxines: Contaminants omniprésents dans l’alimentation animale et humaine, dans La sécurité alimentaire du consommateur. In Tec & Doc; Lavoisier: Paris, France, 2002; pp. 127–179. [Google Scholar]
- Food and Agriculture Organisation (FAO). Post Harvest Losses in Quality of Food Grains; Food and Agriculture Organisation: Rome, Italy, 1983; Volume 29, p. 103. [Google Scholar]
- Brera, C.B.; De Santis, D.F.; Miraglia, M. Mycotoxins. In Food Contaminants and Residue Analysis; Pico, Y., Ed.; Elsevier: Oxford, UK, 2008; pp. 363–419. [Google Scholar]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef]
- CAST. Mycotoxins: Risks in Plant, Animal and Human Systems; Task Force Report N 139; CAST: Ames, IA, USA, 2003. [Google Scholar]
- Niessen, L. PCR-based diagnosis and quantification of mycotoxin producing fungi. Int. J. Food Microbiol. 2007, 119, 38–46. [Google Scholar] [CrossRef]
- Bankole, S.; Schollenberger, M.; Drochner, W. Mycotoxins in food systems in Sub Saharan Africa: A review. Mycotoxin Res. 2006, 22, 163–169. [Google Scholar] [CrossRef]
- Nonaka, Y.; Saitob, K.; Hanioka, N.; Narimatsu, S.; Kataoka, H. Determination of aflatoxins in food samples by automated on-line in-tube solid-phase microextraction coupled with liquid chromatography–mass spectrometry. J. Chromatogr. 2009, 1216, 4416–4422. [Google Scholar] [CrossRef]
- EC. Commission Regulation (EC)No. 2174/2003 of 12 December 2003 Amending Regulation (EC) No. 466/2001 as Regards Aflatoxins. Off. J. Eur. Union 2003, 12, 326. [Google Scholar]
- Codex Alimentarius Commission. Joint FAO/WHO Food Standards Program, Codex Committee on Food Additives and Contaminants; CODEX: Hague, The Netherlands, 2001. [Google Scholar]
- Ojuri, O.T.; Ezekiel, C.N.; Sulyok, M.; Ezeokoli, O.T.; Oyedele, O.A.; Ayeni, K.I.; Eskola, M.K.; Šarkanj, B.; Hajšlováf, J.; Adeleke, R.A.; et al. Assessing the mycotoxicological risk from consumption of complementary foods by infants and young children. Niger. Food Chem. Toxicol. 2018, 121, 37–50. [Google Scholar] [CrossRef]
- Gbashi, S.; Madala, N.E.; De Saeger, S.; De Boevre, M.; Adekoya, I.; Adebo, O.A.; Njobeh, P.B. The socio-economic impact of mycotoxin contamination in Africa. In Fungi and Mycotoxins—Their Occurrence, Impact on Health and the Economy as well as Pre- and Postharvest Management Strategies; Njobeh, P.B., Stepman, F., Eds.; InTech: Rijeka, Croatia, 2019. [Google Scholar]
- Van Egmond, H.P.; Jonker, M.A. Worldwide regulations on aflatoxins—The situation in 2002. J. Toxicol. Toxin Rev. 2004, 23, 273–293. [Google Scholar] [CrossRef]
- Rodrigues, P.; Santos, C.; Venâncio, A.; Lima, N. Species identification of Aspergillus section Flavi isolates from Portuguese almonds using phenotypic, including MALDI-TOF ICMS, and molecular approaches. J. Appl. Microbiol. 2011, 111, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Nleya, N.; Adetunji, M.; Mwanza, M. Current status of mycotoxin contamination of food commodities in Zimbabwe. Toxins 2018, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Adetunji, M.C.; Ngoma, L.; Atanda, O.O.; Mwanza, M. A polyphasic method for the identification of aflatoxigenic Aspergilla from cashew nuts. World J. Microbiol. Biotechnol. 2019, 35, 8. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.W.; Ito, Y.; Horn, B.W.; Goto, T. Aspergillus bombycis, a new aflatoxigenic species and genetic variation in its sibling species, A. nomius. Mycologia 2001, 93, 689–703. [Google Scholar] [CrossRef]
- Mircea, C.; Poiata, A.; Tuchilus, C.; Agoroae, L.; Butnaru, E.; Stanescu, U. Aflatoxigenic fungi isolated from medicinal herbs. Toxicol. Lett. 2008, 180, 32–246. [Google Scholar] [CrossRef]
- Saleemullah, A.I.; Khalil, I.A.; Shah, H. Aflatoxin content of stored and artificially innoculated cereals and nuts. Food Chem. 2006, 98, 699–703. [Google Scholar] [CrossRef]
- Rodrigues, P.; Venâncio, A.; Kozakiewicz, Z.; Lima, N. A polyphasic approach to the identification of aflatoxigenic and non-aflatoxigenic strains of Aspergillus Section Flavi isolated from Portuguese almonds. Internat. J. Food Microbiol. 2009, 129, 187–193. [Google Scholar] [CrossRef]
- Food and Agricultural Organization (FAO); World Health Organization (WHO). Standards programme. In Codex Alimentarius Commission, 3rd ed.; Alinorm: Rome, Italy, 1992; p. 93/12. [Google Scholar]
- EU. Setting Maximum Levels for Certain Contaminants in Foodstuffs. Off. J. Eur. Union 2006, 364, 5–24. [Google Scholar]
- EC. Directives 2002/32/EC of the European Parliament and of the Council on Undesirable Substances in Animal Feed; European, C., Ed.; EC: Brussels, Belgium, 2002; pp. 1–27. [Google Scholar]
- Moss, M.O. Risk assessment for aflatoxins in foodstuffs. Int. Biodeterior. Biodegrad. 2002, 50, 137–142. [Google Scholar] [CrossRef]
- Makun, H.A.; Dutton, M.F.; Njobeh, P.B.; Gbodi, T.A.; Haruna, O.G. Aflatoxin Contamination in Foods and Feeds: A Special Focus on Africa, in Trends in Vital Food and Control Engineering; Eissa, P.A.A., Ed.; InTech Open: Rijeka, Croatia, 2012; Volume 10, pp. 187–290. [Google Scholar]
- Makun, H.A.; Dutton, M.F.; Njobeh, P.B.; Mwanza, M.; Kabiru, A.Y. Natural multi- mycotoxin occurrence in rice from Niger State, Nigeria. Mycotoxin Res. 2011, 27, 97–104. [Google Scholar] [CrossRef]
- Lee, C.Z.; Liou, G.Y.; Yuan, G.F. Comparison of the aflR gene sequences of strains in Aspergillus section Flavi. Microbiology 2006, 152, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Georgianna, D.R.; Hawkridge, A.M.; Muddiman, D.C.; Payne, G.A. Temperature-dependent regulation of proteins in Aspergillus flavus: Whole organism stable isotope labeling by amino acids. J. Proteome Res. 2008, 7, 2973–2979. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, D.; Ehrlich, K.C.; Cleveland, T.E. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 2003, 61, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Renqing, Z. The Function, Accumulation and Localization of the Nor-1 Protein Involved in Aflatoxin Biosynthesis; the Function of the Flu? Gene Associated with Sporulatton in Aspergillus Parasiticus. In Department of Food Science and Human Nutrition and Multidisciplinary Graduate Program in Environmental Toxicology; Michigan State University: East Lansing, MI, USA, 1997; p. 246. [Google Scholar]
- Ryan, L.A.; Dal Bello, F.; Arendt, E.K.; Koehler, P. Detection and quantitation of 2,5-diketopiperazines in wheat sourdough and bread. J. Agric. Food Chem. 2009, 57, 9563–9568. [Google Scholar] [CrossRef]
- Bhatnagar, D.; Cary, J.W.; Ehrlich, K.; Yu, J.; Cleveland, T.E. Understanding the genetics of regulation of aflatoxin production and Aspergillus flavus development. Mycopathologia 2006, 162, 155–166. [Google Scholar] [CrossRef]
- Chiou, C.H.; Miller, M.; Wilson, D.L.; Trail, F.; Linz, J.E. Chromosomal location plays a role in regulation of aflatoxin gene expression in Aspergillus parasiticus. Appl. Environ. Microbiol. 2002, 68, 306–315. [Google Scholar] [CrossRef]
- Yu, J.; Chang, P.-K.; Bhatnagar, D.; Cleveland, T.E. Cloning of sugar utilization gene cluster in Aspergillus parasiticus. Biochim. Biophys. Acta 2000, 1493, 211–214. [Google Scholar] [CrossRef]
- Atanda, O.; Ogunrinu, M.; Olorunfemi, F. A neutral red desiccated coconut agar for rapid detection of aflatoxigenic fungi and visual determination of aflatoxins. World Mycotoxin J. 2011, 4, 147–155. [Google Scholar] [CrossRef]
- Ezekiel, C.N.; Udom, I.E.; Frisvad, J.C.; Adetunji, M.C.; Houbraken, J.; Fapohunda, S.O.; Samson, R.A.; Atanda, O.O.; Agi-Otto, M.C.; Onashile, O.A. Assessment of aflatoxigenic Aspergillus and other fungi in millet and sesame from Plateau State, Nigeria. Mycology 2014, 5, 16–22. [Google Scholar] [CrossRef]
- Degola, F.; Dall’Asta, C.; Restivo, F. Development of a simple and high-throughput method for detecting aflatoxins production in culture media. Lett. Appl. Microbiol. 2012, 55, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.K.; Zablotowicz, R.M.; Weaver, R.M.; Horn, B.W.; Xie, W.; Shier, W.T. Comparison of Cultural and Analytical Methods for Determination of Aflatoxin Production by Missisissipi Delta Aspergillus isolates. Can. J. Microbiol. 2005, 50, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Sudini, H.; Srilakshmi, P.; Vijay, K.K.K.; Njoroge, S.M.; Osiru, M.; Anitha, S.; Waliyar, F. Detection of aflatoxigenic Aspergillus strains by cultural and molecular methods: A critical review African. J. Microbiol. Res. 2015, 9, 484–491. [Google Scholar]
- Cary, J.; Ehrlich, K.; Beltz, S.; Harris-Coward, P.; Klich, M. Characterization of the Aspergillus ochraceoroseus aflatoxin/sterigmatocystin biosynthetic gene cluster. Mycologia 2009, 101, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Klich, M.; Yu, J.; Chang, P.-K.; Mullaney, E.; Bhatnagar, D.; Cleveland, T. Hybridization of genes involved in aflatoxin biosynthesis to DNA of aflatoxigenic and non-aflatoxigenic Aspergilli. Appl. Microbiol. Biotechnol. 1995, 44, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Wagacha, J.M.; Muthomi, J.W. Mycotoxin problem in Africa: Current status, implications to food safety and health and possible management strategies. Int. J. Food Microbiol. 2008, 124, 1–12. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer (IARC). Monographs on the Evaluation of Carcinogenic Risk to Humans. In Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins; IARC, Ed.; World Health Organisation: Lyon, France, 1993; p. 489. [Google Scholar]
- Mngadi, P.T.; Govinden, R.; Odhav, B. Co-occurring mycotoxins in animal feeds. Afri. J. Biotechnol. 2008, 7, 2239–2243. [Google Scholar]
- Kangethe, E.K.; Langa, K.A. Aflatoxin B1 and M1 contamination of animal feeds and milk from urban centers in Kenya. Afr. Health Sci. 2009, 9, 218–226. [Google Scholar]
- Elzupir, A.O.; Younis, M.H.; Himmat, F.M.; Elhussein, A.M. Determination of Aflatoxins in Animal Feed in Khartoum State, Sudan. J. Anim. Vet. Adv. 2009, 8, 1000–1003. [Google Scholar]
- Mwanza, M.; Ndou, R.V.; Dzoma, B.; Nyirenda, M.; Bakunzi, F. Canine aflatoxicosis outbreak in South Africa; A possible multi-mycotoxins aetiology Art 133. J. South Afr. Vet. Assoc. 2013, 84, 5. [Google Scholar]
- Zinedine, A.; Juan, C.; Soriano, J.M.; Moltó, J.C.; Idrissi, L.; Mañes, J. Limited survey for the occurrence of aflatoxins in cereals and poultry feeds from Rabat, Morocco. Internat. J. Food Microbiol. 2007, 115, 124–127. [Google Scholar] [CrossRef]
- Adebayo-Tayo, B.C.; Ettah, A.E. Microbiological quality and aflatoxin B1 level in poultry and livestock feeds. Niger. J. Microbiol. 2010, 24, 2145–2152. [Google Scholar]
- Magnani, M.; Fernandes, T.; Prete, C.E.C.; Homechim, M.; Ono, E.Y.S.; Vilas-Boas, L.A.; Sartori, D.; Furlaneto, M.C.; Fungaro, M.H.P. Molecular identification of Aspergillus spp. isolated from coffee beans. Sci. Agric. 2005, 62, 45–49. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemists (AOAC). Official Methods of Analysis, 18th ed.; Association of Official Analytical Chemists, Ed.; AOAC: Washington, DC, USA, 2007. [Google Scholar]
- Hassane, A.M.A.; El-Shanawany, A.A.; Abo-Dahab, N.F.; Abdel-Hadi, A.M.; Abdul-Raouf, U.M.; Mwanza, M. Influence of Different Moisture Contents and Temperature on Growth and Production of Aflatoxin B1 by a Toxigenic Aspergillus flavus Isolate in Wheat Flour. J. Ecol. Heal. Environ. 2017, 5, 77–83. [Google Scholar] [CrossRef]
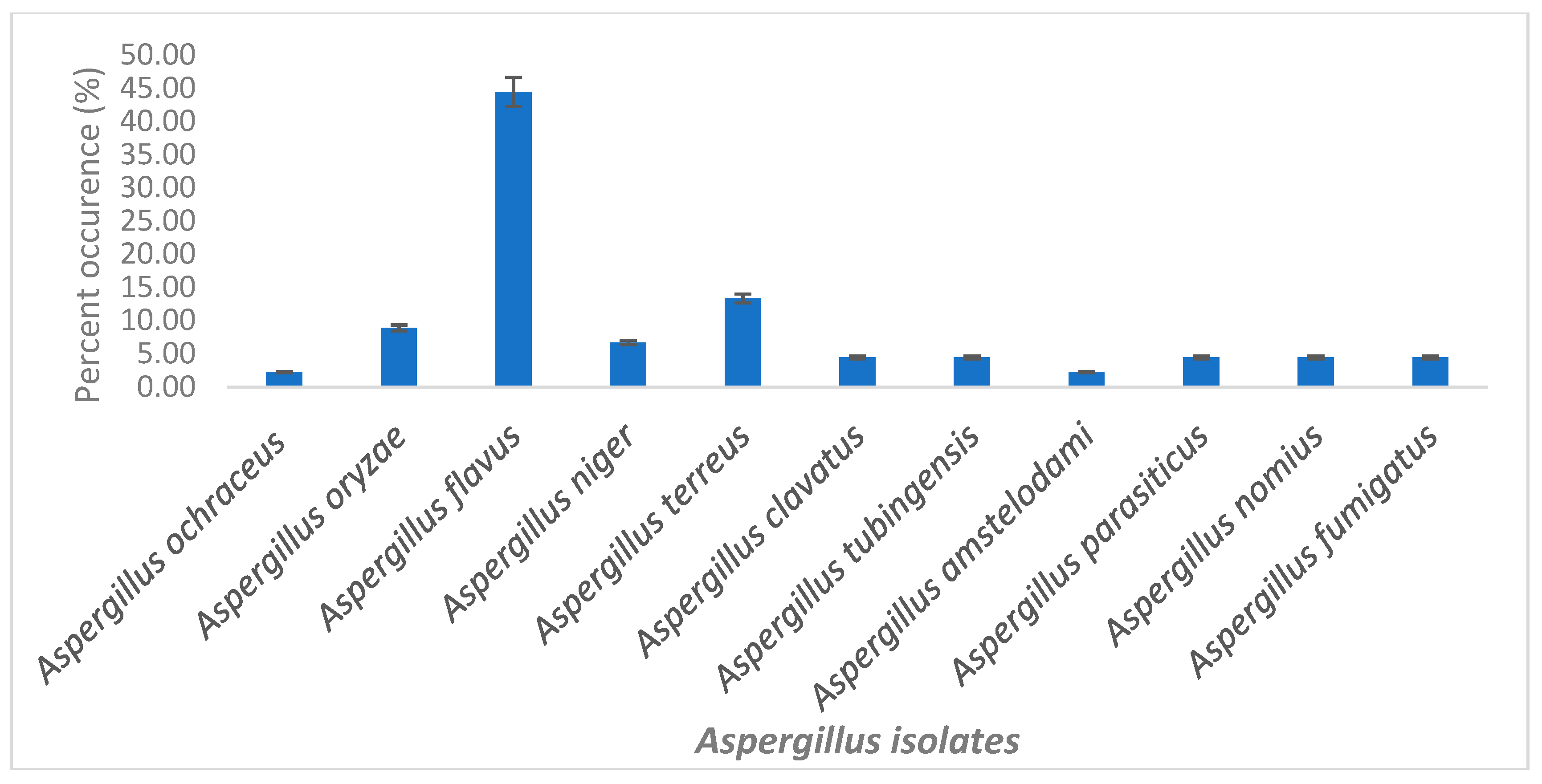
| SN | ID | Isolates Name | Assession No | Media | Colour and Morphology |
|---|---|---|---|---|---|
| 1 | FG1 | Aspergillus oryzae | MG647833 | MEA | Line Green |
| 2 | FG2 | Aspergillus flavus | MG659619 | MEA | Line green |
| 3 | FG3 | Aspergillus niger | MG647838 | MEA | Black |
| 4 | FG4 | Aspergillus flavus | MG659628 | MEA | Green |
| 5 | FG5 | Aspergillus niger | MG647849 | MEA | Black |
| 6 | FG6 | Aspergillus terreus | MG647840 | MEA | Dark green |
| 7 | FG7 | Aspergillus flavus | MG659631 | MEA | Line green |
| 8 | FG8 | Aspergillus tubingensis | MG647844 | MEA | Green |
| 9 | FG9 | Aspergillus flavus | MH270531 | MEA | Green |
| 10 | FG10 | Aspergillus flavus | MG647845 | MEA | Green |
| 11 | FG11 | Aspergillus terreus | MG647846 | MEA | Dark green |
| 12 | FG12 | Aspergillus parasiticus | MG659626 | MEA | Green |
| 13 | FG13 | Aspergillus amstelodami | MG647851 | MEA | Line Green |
| 14 | FG14 | Aspergillus flavus | MH270612 | MEA | Green |
| 15 | FG15 | Aspergillus nomius | MG659621 | MEA | Green |
| 16 | FG16 | Aspergillus tubingensis | MG647853 | MEA | Military green |
| 17 | FG17 | Aspergillus terreus | MG647863 | MEA | dark green |
| 18 | FG18 | Aspergillus flavus | MG659635 | MEA | Green |
| 19 | FG19 | Aspergillus flavus | MH270544 | MEA | Green |
| 20 | FG20 | Aspergillus flavus | MG659673 | MEA | Green |
| 21 | FG21 | Aspergillus niger | MG647867 | MEA | Black |
| 22 | FG22 | Aspergillus flavus | MH270559 | MEA | Green |
| 23 | FG23 | Aspergillus flavus | MG659626 | MEA | Green |
| 24 | FG24 | Aspergillus flavus | MH270559 | MEA | Green |
| 25 | FG25 | Aspergillus oryzae | MH270563 | MEA | Green |
| 26 | FG26 | Aspergillus flavus | MH270574 | MEA | Green |
| 27 | FG27 | Aspergillus clavatus | MG647850 | MEA | Line Green |
| 28 | FG28 | Aspergillus fumigatus | MG647855 | MEA | Bluish green |
| 29 | FG29 | Aspergillus fumigatus | MG647869 | MEA | Bluish green |
| 30 | FG30 | Aspergillus terreus | MG647840 | MEA | Dark green |
| 31 | FG31 | Aspergillus flavus | MH270578 | MEA | Green |
| 32 | FG32 | Aspergillus flavus | MG647868 | MEA | Green |
| 33 | FG33 | Aspergillus flavus | MH270581 | MEA | Green |
| 34 | FG34 | Aspergillus oryzae | MG659690 | MEA | Military green |
| 35 | FG35 | Aspergillus clavatus | MG647856 | MEA | Green |
| 36 | FG36 | Aspergillus oryzae | MG659633 | MEA | Green |
| 37 | FG37 | Aspergillus terreus | MG647866 | MEA | Green |
| 38 | FG38 | Aspergillus terreus | MG647852 | MEA | Green |
| 39 | FG39 | Aspergillus flavus | MG647857 | MEA | Green |
| 40 | FG40 | Aspergillus flavus | MG659627 | MEA | Green |
| 41 | FG41 | Aspergillus parasiticus | MG659687 | MEA | Green |
| 42 | FG42 | Aspergillus flavus | MG659676 | MEA | Green |
| 43 | FG43 | Aspergillus nomius | MH270600 | MEA | Green |
| 44 | FG44 | Aspergillus ochraceoroseus | MH270530 | MEA | Green |
| 45 | FG45 | Aspergillus flavus | MG647871 | MEA | Green |
| 46 | Control 1 | Aspergillus flavus | ATCC 259622TM | MEA | Green |
| 47 | Control 2 | Saccharomyces cerevisiae | MEA | Creamy |
| SN | ID | Isolates Name | Assession No | aflD (Nor-A) | aflR | aflJ | aflM (ver-1) | Omt-A | YES (NH4 Vapor Test) | (β-CDNRDCA) | TLC | HPLC | Selected Isolates | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yellow Pigment | UV-Florescence | |||||||||||||
| 1 | FG1 | Aspergillus oryzae | MG647833 | POS | NEG | POS | POS | NEG | POS | POS | - | nd | nd | |
| 2 | FG2 | Aspergillus flavus | MG659619 | NEG | POS | POS | NEG | POS | POS | POS | - | nd | nd | |
| 3 | FG3 | Aspergillus niger | MG647838 | POS | POS | POS | NEG | POS | NEG | NEG | - | nd | nd | |
| 4 | FG4 | Aspergillus flavus | MG659628 | POS | POS | POS | POS | POS | POS | POS | ± | NEG | POS | ** |
| 5 | FG5 | Aspergillus niger | MG647849 | POS | POS | POS | NEG | POS | NEG | NEG | - | nd | nd | |
| 6 | FG6 | Aspergillus terreus | MG647840 | NEG | POS | POS | NEG | POS | NEG | NEG | - | nd | nd | |
| 7 | FG7 | Aspergillus flavus | MG659631 | POS | NEG | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 8 | FG8 | Aspergillus tubingensis | MG647844 | POS | NEG | POS | POS | NEG | NEG | NEG | - | nd | nd | |
| 9 | FG9 | Aspergillus flavus | MH270531 | POS | POS | POS | POS | POS | NEG | NEG | - | NEG | POS | ** |
| 10 | FG10 | Aspergillus flavus | MG647845 | POS | POS | POS | POS | POS | POS | POS | + | NEG | POS | ** |
| 11 | FG11 | Aspergillus terreus | MG647846 | NEG | POS | POS | NEG | NEG | NEG | POS | ++ | nd | nd | |
| 12 | FG12 | Aspergillus parasiticus | MG659626 | POS | POS | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 13 | FG13 | Aspergillus amstelodami | MG647851 | NEG | POS | POS | NEG | NEG | NEG | NEG | - | nd | nd | |
| 14 | FG14 | Aspergillus flavus | MH270612 | POS | POS | POS | POS | POS | POS | POS | ++++ | POS | POS | ** |
| 15 | FG15 | Aspergillus nomius | MG659621 | POS | POS | POS | POS | POS | POS | POS | ++++ | POS | POS | ** |
| 16 | FG16 | Aspergillus tubingensis | MG647853 | NEG | NEG | POS | NEG | POS | NEG | NEG | - | nd | nd | |
| 17 | FG17 | Aspergillus terreus | MG647863 | POS | POS | POS | NEG | NEG | NEG | NEG | - | nd | nd | |
| 18 | FG18 | Aspergillus flavus | MG659635 | POS | POS | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 19 | FG19 | Aspergillus flavus | MH270544 | POS | POS | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 20 | FG20 | Aspergillus flavus | MG659673 | POS | POS | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 21 | FG21 | Aspergillus niger | MG647867 | POS | POS | POS | NEG | NEG | NEG | POS | ++ | nd | nd | |
| 22 | FG22 | Aspergillus flavus | MH270559 | POS | POS | POS | POS | POS | POS | POS | ++ | POS | POS | ** |
| 23 | FG23 | Aspergillus flavus | MG659626 | POS | POS | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 24 | FG24 | Aspergillus flavus | MH270559 | POS | POS | POS | POS | POS | POS | POS | +/- | POS | POS | ** |
| 25 | FG25 | Aspergillus oryzae | MH270563 | POS | POS | POS | POS | POS | POS | POS | +/- | POS | POS | ** |
| 26 | FG26 | Aspergillus flavus | MH270574 | NEG | POS | POS | POS | NEG | POS | NEG | - | nd | nd | |
| 27 | FG27 | Aspergillus clavatus | MG647850 | POS | POS | POS | NEG | NEG | NEG | POS | - | nd | nd | |
| 28 | FG28 | Aspergillus fumigatus | MG647855 | NEG | POS | POS | NEG | POS | NEG | NEG | - | nd | nd | |
| 29 | FG29 | Aspergillus fumigatus | MG647869 | POS | POS | POS | NEG | POS | NEG | POS | - | nd | nd | |
| 30 | FG30 | Aspergillus terreus | MG647840 | POS | POS | POS | NEG | NEG | NEG | NEG | - | nd | nd | |
| 31 | FG31 | Aspergillus flavus | MH270578 | POS | POS | POS | POS | POS | POS | POS | ++ | POS | POS | ** |
| 32 | FG32 | Aspergillus flavus | MG647868 | POS | POS | POS | NEG | NEG | NEG | POS | - | nd | nd | |
| 33 | FG33 | Aspergillus flavus | MH270581 | POS | POS | POS | POS | POS | POS | POS | ++ | POS | POS | ** |
| 34 | FG34 | Aspergillus oryzae | MG659690 | POS | POS | POS | POS | POS | POS | POS | + | NEG | POS | ** |
| 35 | FG35 | Aspergillus clavatus | MG647856 | POS | POS | POS | NEG | NEG | NEG | POS | - | nd | nd | |
| 36 | FG36 | Aspergillus oryzae | MG659633 | POS | POS | POS | POS | POS | NEG | POS | + | NEG | POS | ** |
| 37 | FG37 | Aspergillus terreus | MG647866 | POS | POS | POS | NEG | POS | NEG | POS | + | nd | nd | |
| 38 | FG38 | Aspergillus terreus | MG647852 | POS | POS | POS | NEG | POS | NEG | POS | - | nd | nd | |
| 39 | FG39 | Aspergillus flavus | MG647857 | POS | POS | POS | POS | NEG | NEG | POS | - | nd | nd | |
| 40 | FG40 | Aspergillus flavus | MG659627 | POS | NEG | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 41 | FG41 | Aspergillus parasiticus | MG659687 | POS | POS | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 42 | FG42 | Aspergillus flavus | MG659676 | POS | POS | POS | POS | POS | NEG | NEG | - | POS | POS | ** |
| 43 | FG43 | Aspergillus nomius | MH270600 | POS | POS | POS | POS | POS | POS | POS | +++ | POS | POS | ** |
| 44 | FG44 | Aspergillus ochraceoroseus | MH270530 | POS | POS | POS | POS | POS | POS | POS | ++ | POS | POS | ** |
| 45 | FG45 | Aspergillus flavus | MG647871 | POS | POS | POS | POS | NEG | NEG | POS | - | nd | nd | |
| 46 | Control 1 | Aspergillus flavus ATCC | 259622TM | POS | POS | POS | POS | POS | POS | POS | +++ | POS | POS | |
| 47 | Control 2 | Saccharomyces cereviasiae | NEG | NEG | NEG | NEG | NEG | NEG | NEG | - | nd | nd | ||
| S/N | ID | Isolate Name | AFG2 (µg/g) | AFG1 (µg/g) | AFB2 (µg/g) | AFB1 (µg/g) | AFTOT (µg/g) |
|---|---|---|---|---|---|---|---|
| 1 | FG4 | Aspergillus flavus | nd | nd | 6.78 ± 0.01 r | nd | 6.78 ± 0.01 q |
| 2 | FG7 | Aspergillus flavus | 1.67 ± 0.01 p | nd | nd | 54.75 ± 3.46 f | 56.42 ± 3.45 m |
| 3 | FG9 | Aspergillus flavus | nd | nd | nd | nd | nd |
| 4 | FG10 | Aspergillus flavus | nd | 6.21 ± 0.02 c | nd | nd | 6.21 ± 0.02 q |
| 5 | FG12 | Aspergillus parasiticus | 360.06 ± 0.05 b | 745.34 ± 0.03 a | 242.52 ± 0.04 a | 34,043.71 ± 0.27 b | 35,391.63 ± 0.34 b |
| 6 | FG14 | Aspergillus flavus | nd | nd | 13.24 ± 0.01 q | 59.66 ± 0.02 e | 72.90 ± 0.01 l |
| 7 | FG15 | Aspergillus nomius | 768.52 ± 0.03 a | 396.45 ± 0.02 b | nd | 70,289.23 ± 0.67a | 71,454.21 ± 0.66 a |
| 8 | FG18 | Aspergillus flavus | 21.91 ± 0.03 i | nd | 103.15 ± 0.02 c | nd | 125.06 ± 0.03 h |
| 9 | FG19 | Aspergillus flavus | 0.98 ± 0.02 q | nd | 24.14 ± 0.01 o | nd | 25.12 ± 0.03 p |
| 10 | FG20 | Aspergillus flavus | 11.84 ± 0.04 k | nd | 66.08 ± 0.07 i | nd | 77.92 ± 0.11 j |
| 11 | FG22 | Aspergillus flavus | 3.43 ± 0.02 o | 1.88 ± 0.01 f | 50.08 ± 0.01 k | nd | 55.39 ± 0.04 m |
| 12 | FG23 | Aspergillus flavus | 72.55 ± 0.02 e | nd | 30.33 ± 0.04 m | nd | 102.88 ± 0.04 i |
| 13 | FG24 | Aspergillus flavus | 14.86 ± 0.06 j | nd | 57.77 ± 0.02 j | nd | 72.63 ± 0.06 l |
| 14 | FG25 | Aspergillus oryzae | 86.04 ± 0.04 d | nd | 95.58 ± 0.01 d | nd | 181.62 ± 0.05 f |
| 15 | FG31 | Aspergillus flavus | 253.44 ± 0.05 c | 3.19 ± 0.02 d | 73.88 ± 0.03 f | nd | 330.51 ± 0.07 e |
| 16 | FG33 | Aspergillus flavus | 38.33 ± 0.03 g | nd | 109.23 ± 0.03 a | nd | 147.57 ± 0.05 g |
| 17 | FG34 | Aspergillus oryzae | nd | nd | nd | nd | nd |
| 18 | FG36 | Aspergillus oryzae | nd | nd | nd | nd | nd |
| 19 | FG40 | Aspergillus flavus | 4.77 ± 0.02 m | nd | 71.66 ± 0.02 h | nd | 76.43 ± 0.03 k |
| 20 | FG41 | Aspergillus parasiticus | 29.44 ± 0.04 h | nd | 45.09 ± 0.01 l | 1062.56 ± 0.05 c | 1137.09 ± 0.08 c |
| 21 | FG42 | Aspergillus flavus | 4.65 ± 0.03 n | nd | 73.73 ± 0.02 g | nd | 78.39 ± 0.06 j |
| 22 | FG43 | Aspergillus nomius | 56.57 ± 0.03 f | 0.84 ± 0.01 g | 75.63 ± 0.02 e | 704.67 ± 0.02 d | 837.70 ± 0.06 d |
| 23 | FG44 | Aspergillus ochraceoroseus | 11.47 ± 0.02 l | 2.34 ± 0.02 e | 22.65 ± 0.02 p | 3.77 ± 0.02 g | 40.23 ± 0.05 n |
| 24 | Control 1 | Aspergillus flavus ATCC 46283 | 0.53 ± 0.07 s | nd | 29.35 ± 0.03 n | nd | 29.87 ± 0.05 o |
| 25 | Control 2 | Saccharomyces cerevisiae | 0.63 ± 0.03 r | nd | 0.04 ± 0.01 s | nd | 0.67 ± 0.03 |
| 26 | Standard | AFG2 (0.05) µg/mL | 13,820.28 | nd | nd | nd | nd |
| 27 | standard | AFG1 (0.05) µg/mL | nd | 4,556,586 | nd | nd | nd |
| 28 | standard | AFB2 (0.05) µg/mL | nd | nd | 1,798,698 | nd | nd |
| 29 | standard | AFB1 (0.05) µg/mL | nd | nd | nd | 3,742,170 | nd |
| 30 | standard | AFTOT (0.05) µg/mL | nd | nd | nd | nd | 11,479,481 |
| Standard | WHO/FAO limit in Feed [26] | 5 µg/kg | |||||
| European Union limit in animal feed [27] | 4 µg/kg | ||||||
| European Commission aflatoxins in animal feed [14,28] | 5–20 µg/kg | ||||||
| European Commission tolerable limit of aflatoxins in foods in Africa [28] | <5–20 µg/kg | ||||||
| European Commission tolerable limits of aflatoxins in Animal feeds [28] | <0.001–0.01 µg/kg body weight | ||||||
| European Commission aflatoxins limits for infants food [28] | 0.05–10 µg/kg | ||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akinola, S.A.; Ateba, C.N.; Mwanza, M. Polyphasic Assessment of Aflatoxin Production Potential in Selected Aspergilli. Toxins 2019, 11, 692. https://doi.org/10.3390/toxins11120692
Akinola SA, Ateba CN, Mwanza M. Polyphasic Assessment of Aflatoxin Production Potential in Selected Aspergilli. Toxins. 2019; 11(12):692. https://doi.org/10.3390/toxins11120692
Chicago/Turabian StyleAkinola, Stephen Abiola, Collins Njie Ateba, and Mulunda Mwanza. 2019. "Polyphasic Assessment of Aflatoxin Production Potential in Selected Aspergilli" Toxins 11, no. 12: 692. https://doi.org/10.3390/toxins11120692
APA StyleAkinola, S. A., Ateba, C. N., & Mwanza, M. (2019). Polyphasic Assessment of Aflatoxin Production Potential in Selected Aspergilli. Toxins, 11(12), 692. https://doi.org/10.3390/toxins11120692






