Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi
Abstract
1. Introduction
2. Results and Discussion
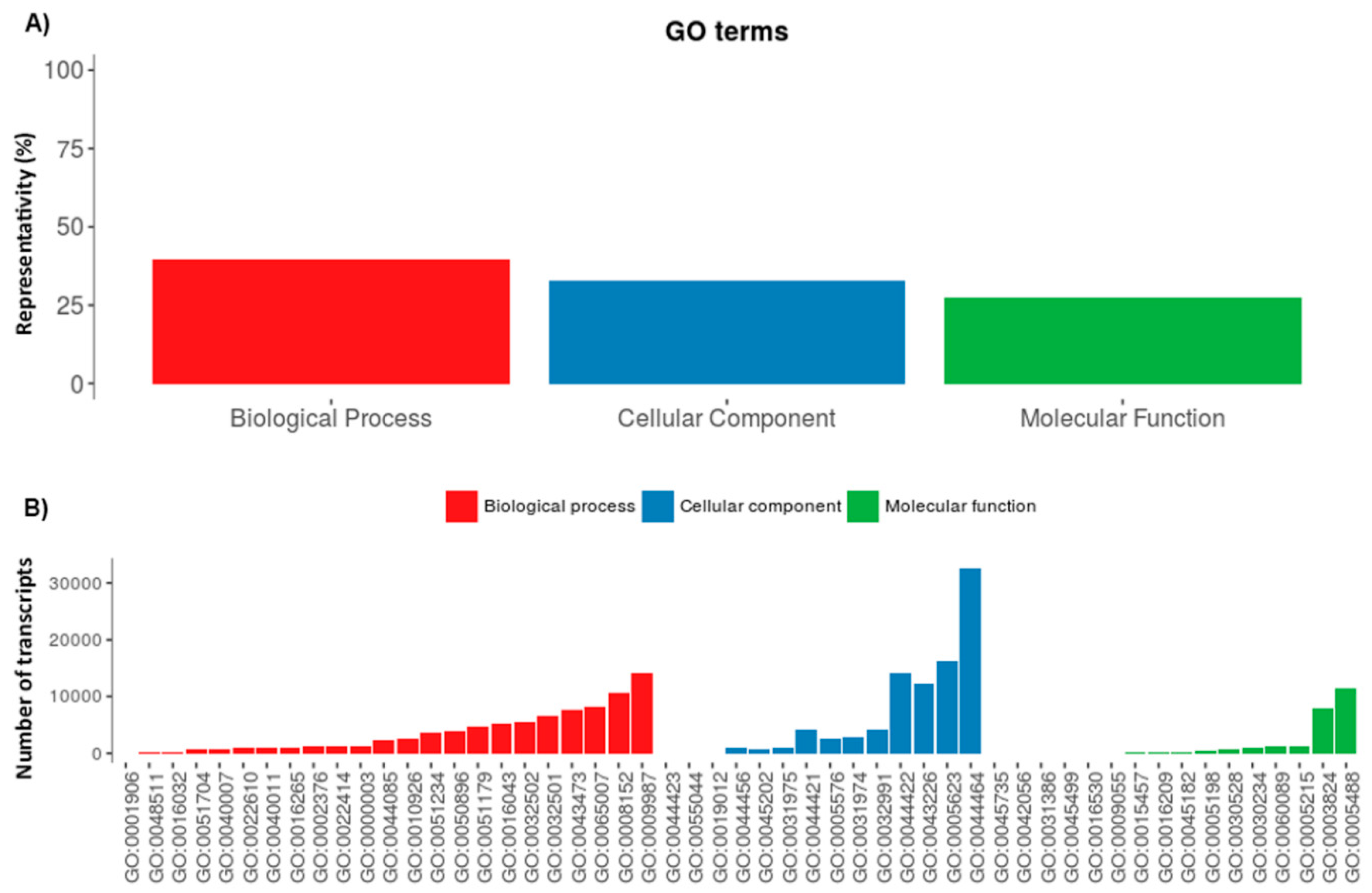
2.1. Serradigitus Gertschi Venom Gland Global Transcriptome Analysis
2.2. The Repertoire of Venom-Specific Transcripts in S. gertschi
2.2.1. Ion Channel Toxins
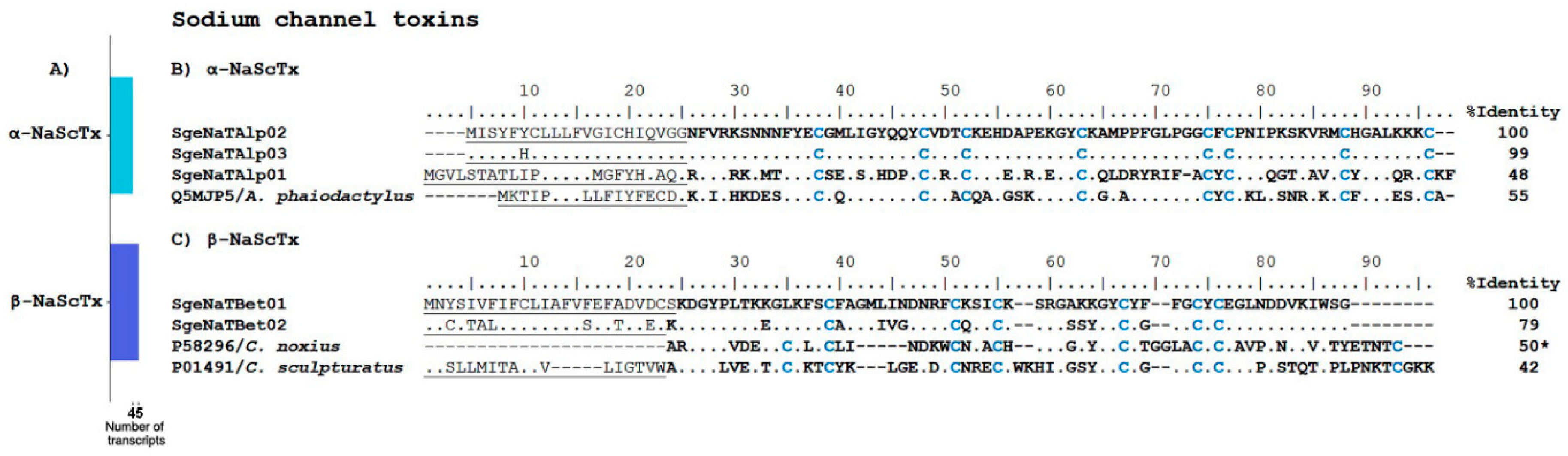
Sodium Channel Toxins
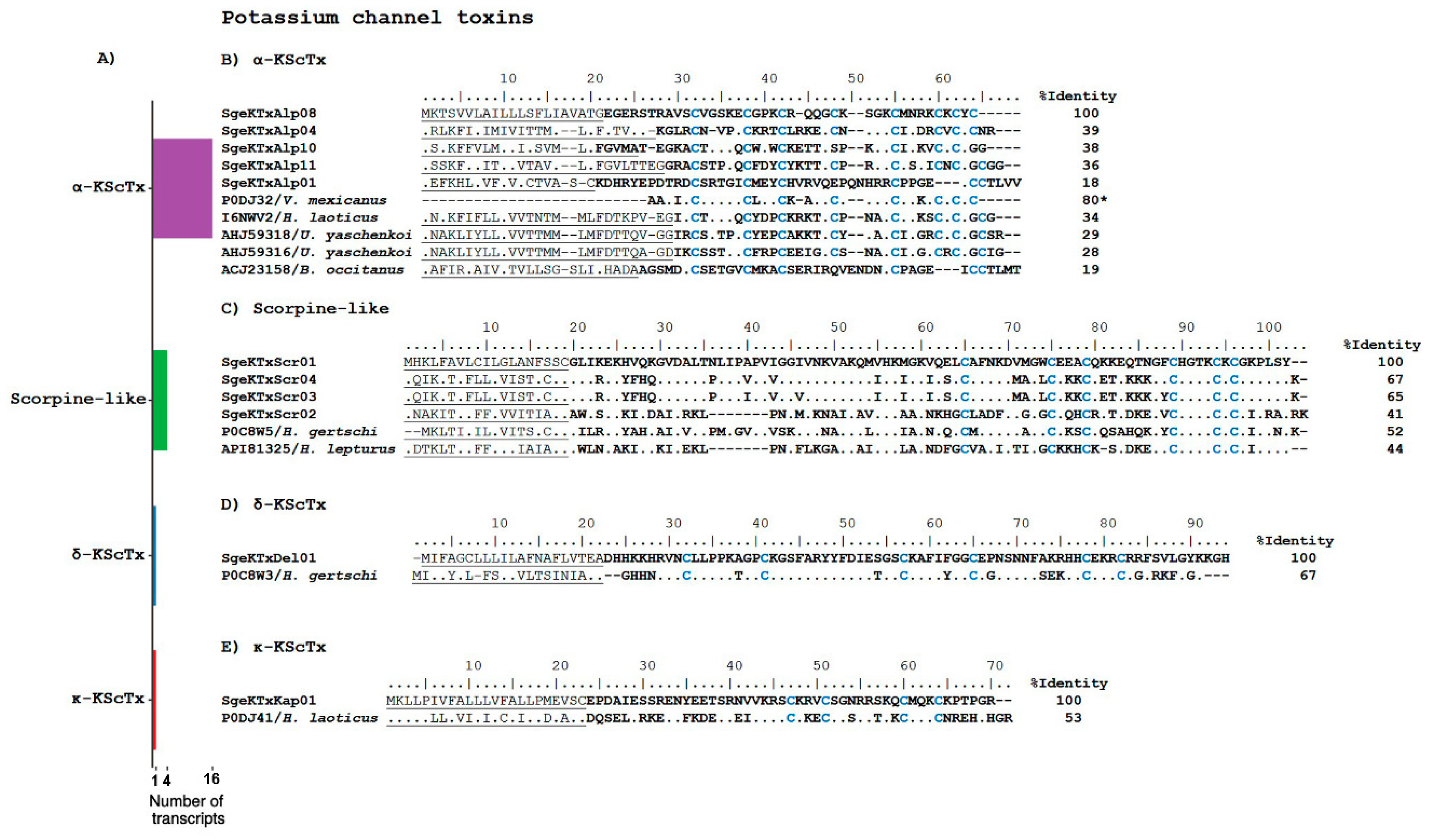
Potassium Channel Toxins
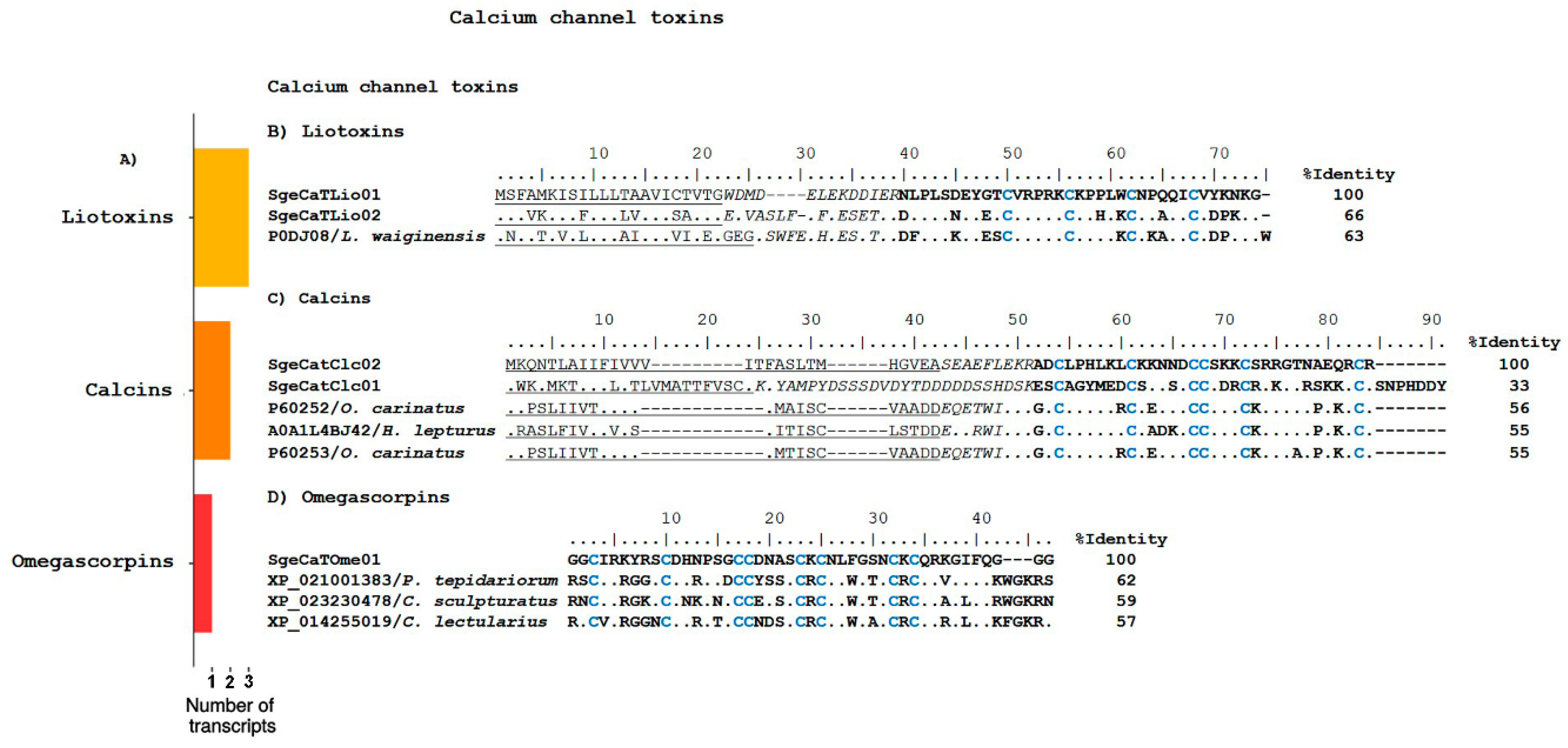
Calcium Channel Toxins
Omegascorpins
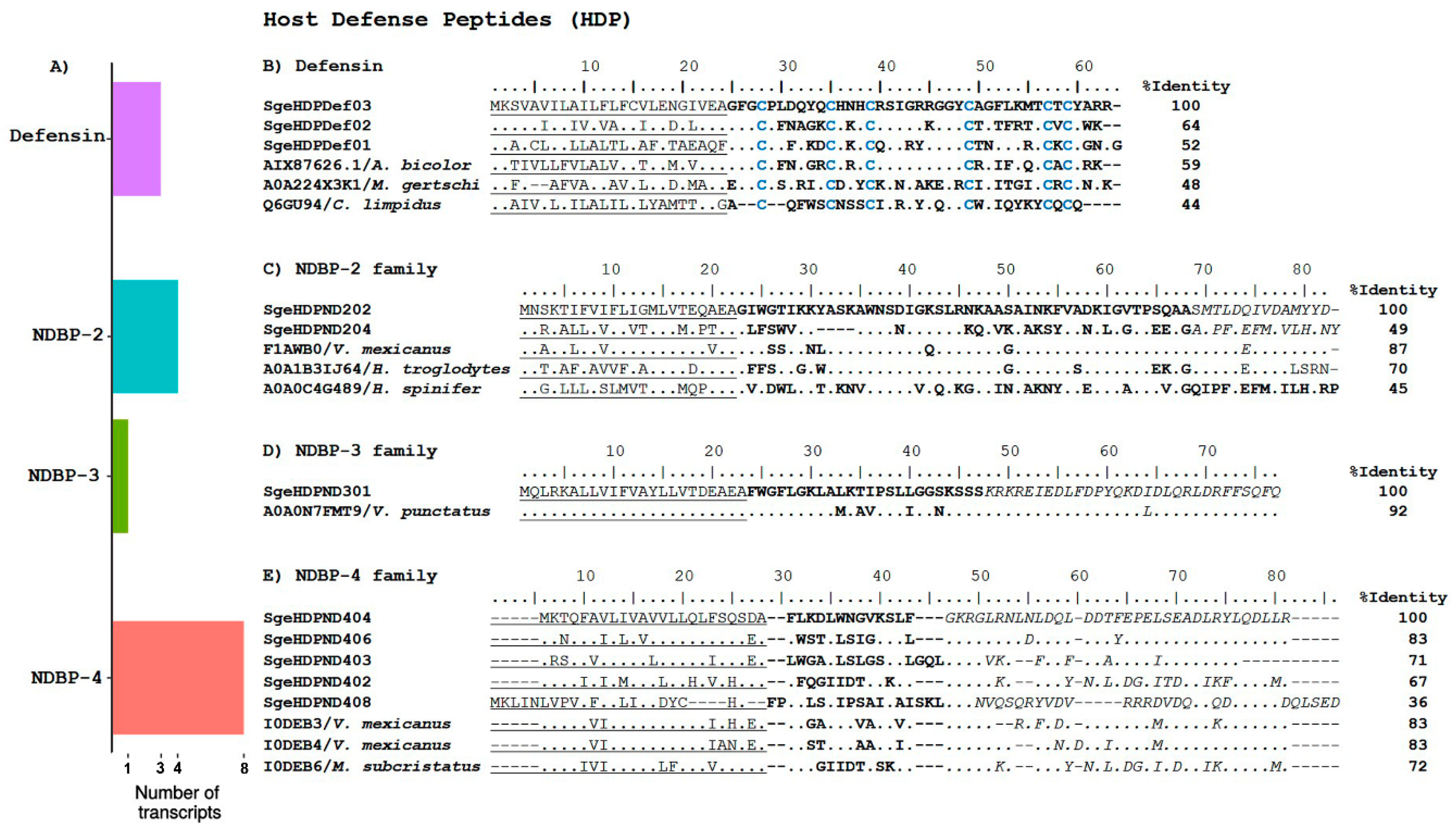
2.2.2. Host Defense Peptides
Non-Disulfide Bridged Peptides
2.2.3. Enzymes
2.2.4. Protease Inhibitors
2.2.5. Other Venom Components
La1-Like Peptides
CAP Superfamily
IGFBP Family
2.3. Proteomic Analyses
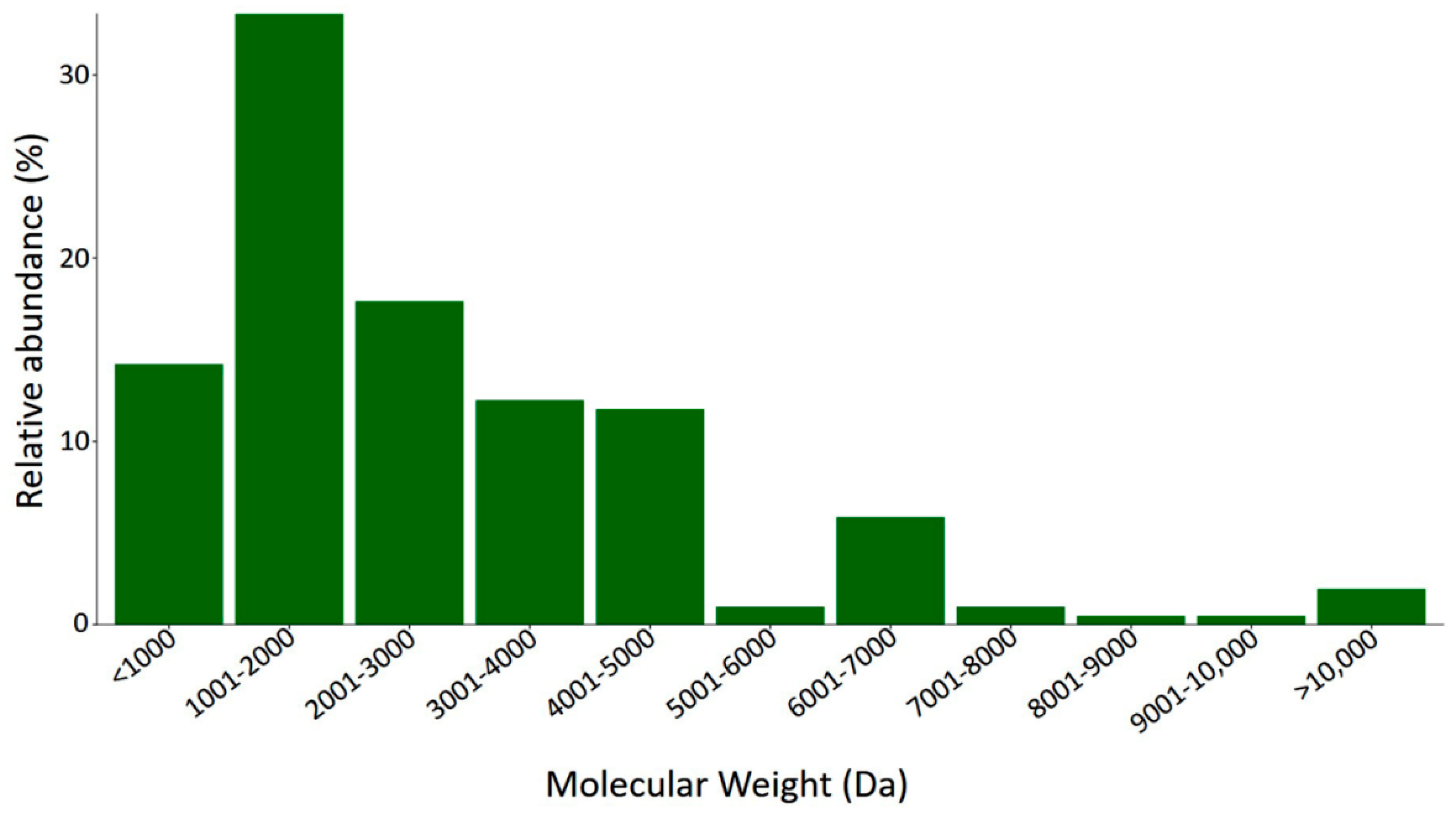
2.3.1. Mass Fingerprint Results and MS Data Analysis
2.3.2. LC-MS/MS Analysis of the Digested Venom of S. gertschi
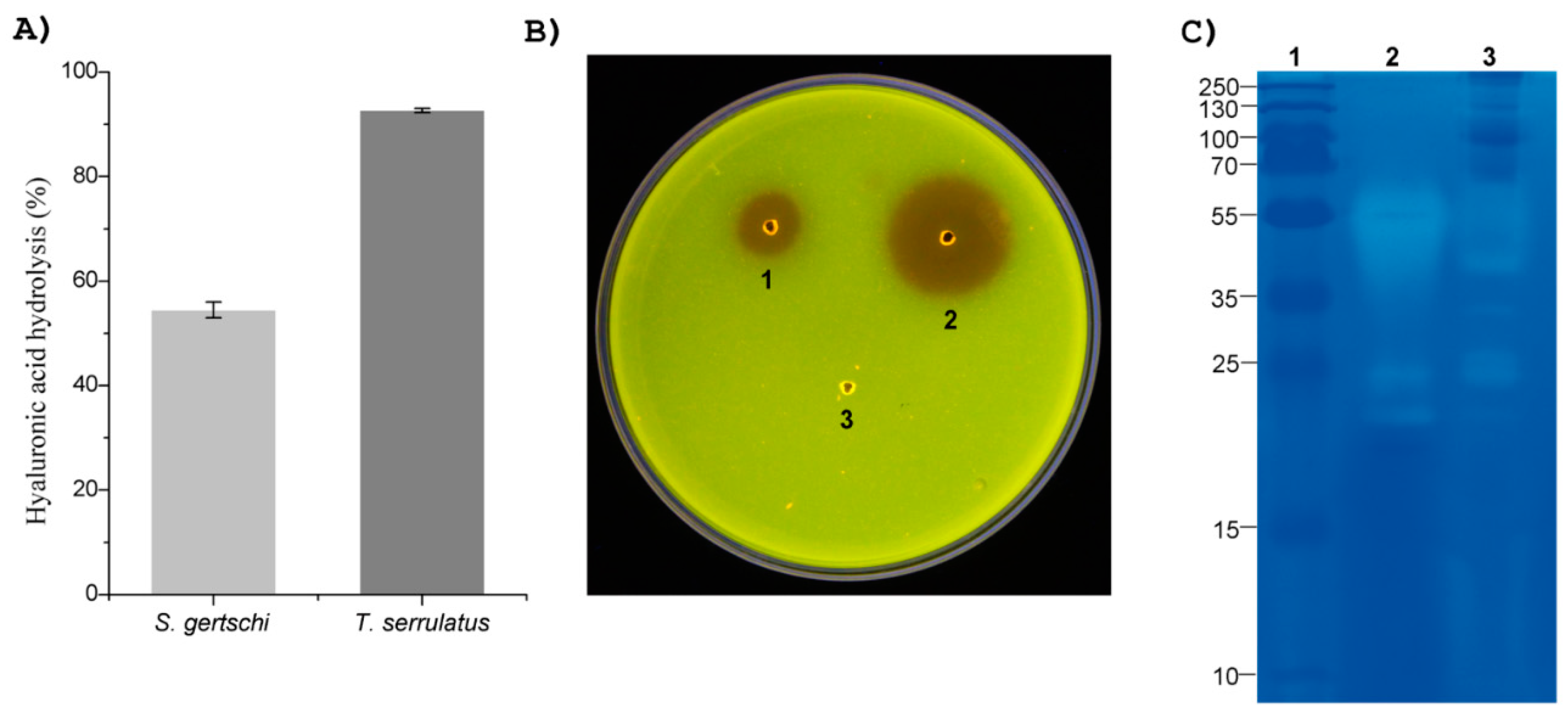
2.4. Venom Enzymatic Activities
3. Conclusions
4. Materials and Methods
4.1. Biological Material
4.2. RNA Extraction, RNA-Seq and Venom-Gland Transcriptome Assembly
4.3. Transcript Nomenclature
4.4. Bioinformatics
4.5. Molecular Mass Determination of the Venom Components
4.6. Identification of Proteome by Tryptic Digestion and LC-MS/MS Analysis
4.7. Venom Enzymatic Activities
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- González-Santillán, E.; Prendini, L. Redefinition and generic revision of the north american Vaejovid scorpion subfamily Syntropinae Kraepelin 1905, with descriptions of six new genera. Bull. Am. Museum Nat. Hist. 2013, 382, 1–71. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; Francke, O.F.; Ureta, C.; Possani, L. Scorpions from Mexico: From species diversity to venom complexity. Toxins 2015, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Prendini, L. Substratum specialization and speciation in southern African scorpions: The Effect Hypothesis revisited. Scorpions 2001, 2001, 113–138. [Google Scholar]
- Hernández-Aponte, C.A.; Silva-Sanchez, J.; Quintero-Hernández, V.; Rodríguez-Romero, A.; Balderas, C.; Possani, L.D.; Gurrola, G.B. Vejovine, a new antibiotic from the scorpion venom of Vaejovis mexicanus. Toxicon 2011, 57, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Gurrola, G.B.; Hernandez-Lopez, R.A.; Rodriguez de la Vega, R.C.; Varga, Z.; Batista, C.V.F.; Salas-Castillo, S.P.; Panyi, G.; del Rio-Portilla, F.; Possani, L.D. Structure, function, and chemical synthesis of Vaejovis mexicanus peptide 24: A novel potent blocker of Kv1.3 potassium channels of human T lymphocytes. Biochemistry 2012, 51, 4049–4061. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Gurrola-Briones, G.; Papp, F.; Rodriguez de la Vega, R.C.; Pedraza-Alva, G.; Tajhya, R.B.; Gaspar, R.; Cardenas, L.; Rosenstein, Y.; Beeton, C.; et al. Vm24, a natural immunosuppressive peptide, potently and selectively blocks Kv1.3 potassium channels of human T cells. Mol. Pharmacol. 2012, 82, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernández, V.; Ramírez-Carreto, S.; Romero-Gutiérrez, M.T.; Valdez-Velázquez, L.L.; Becerril, B.; Possani, L.D.; Ortiz, E. Transcriptome analysis of scorpion species belonging to the Vaejovis genus. PLoS ONE 2015, 10, e0117188. [Google Scholar] [CrossRef] [PubMed]
- Romero-Gutierrez, T.; Peguero-Sanchez, E.; Cevallos, M.A.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. A deeper examination of Thorellius atrox scorpion venom components with omic techonologies. Toxins 2017, 9, 399. [Google Scholar] [CrossRef] [PubMed]
- Cid-Uribe, J.I.; Santibáñez-López, C.E.; Meneses, E.P.; Batista, C.V.F.; Jiménez-Vargas, J.M.; Ortiz, E.; Possani, L.D. The diversity of venom components of the scorpion species Paravaejovis schwenkmeyeri (Scorpiones: Vaejovidae) revealed by transcriptome and proteome analyses. Toxicon 2018, 151, 47–62. [Google Scholar] [CrossRef] [PubMed]
- González-Santillán, E.; Prendini, L. Phylogeny of the north american vaejovid scorpion subfamily Syntropinae Kraepelin, 1905, based on morphology, mitochondrial and nuclear DNA. Cladistics 2015, 31, 341–405. [Google Scholar] [CrossRef]
- Williams, S.C. Scorpions of Baja California, Mexico, and adjacent islands. Occas. Pap. Calif. Acad. Sci. 1980, 135, 1–127. [Google Scholar]
- Williams, S. Scorpion bionomics. Ann. Rev. Entomol. 1987, 32, 275–295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shi, W.; Zeng, X.C.; Ge, F.; Yang, M.; Nie, Y.; Bao, A.; Wu, S.; Guoji, E. Unique diversity of the venom peptides from the scorpion Androctonus bicolor revealed by transcriptomic and proteomic analysis. J. Proteom. 2015, 128, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.; Ward, M. Venom-gland transcriptomics and venom proteomics of the black-back scorpion (Hadrurus spadix) reveal detectability challenges and an unexplored realm of animal toxin diversity. Toxicon 2017, 128, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.; Ellsworth, S.; Rokyta, D. Venom-gland transcriptomics and venom proteomics of the Hentz striped scorpion (Centruroides hentzi; Buthidae) reveal high toxin diversity in a harmless member of a lethal family. Toxicon 2018, 142, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Baradaran, M.; Jalali, A.; Soorki, M.; Jokar, M.; Galehdari, H. First transcriptome analysis of Iranian scorpion, Mesobuthus eupeus venom gland. Iran. J. Pharm. Res. 2018, in press. [Google Scholar]
- Zhong, J.; Zeng, X.C.; Zeng, X.; Nie, Y.; Zhang, L.; Wu, S.; Bao, A. Transcriptomic analysis of the venom glands from the scorpion Hadogenes troglodytes revealed unique and extremely high diversity of the venom peptides. J. Proteom. 2017, 150, 40–62. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.E.; Cid-Uribe, J.I.; Zamudio, F.Z.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Venom gland transcriptomic and venom proteomic analyses of the scorpion Megacormus gertschi Díaz-Najera, 1966 (Scorpiones: Euscorpiidae: Megacorminae). Toxicon 2017, 133, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.E.; Cid-Uribe, J.I.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Venom gland transcriptomic and proteomic analyses of the enigmatic scorpion Superstitionia donensis (Scorpiones: Superstitioniidae), with insights on the evolution of its venom components. Toxins 2016, 8, 367. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Gu, J.; Yan, Z.; Wang, M.; Ma, C.; Zhang, J.; Jiang, G.; Ge, M.; Xu, S.; Xu, Z.; et al. De novo transcriptomic analysis of the venomous glands from the scorpion Heterometrus spinifer revealed unique and extremely high diversity of the venom peptides. Toxicon 2018, 143, 1–19. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, R.C.R.; Possani, L.D. Novel paradigms on scorpion toxins that affects the activating mechanism of sodium channels. Toxicon 2007, 49, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Gurevitz, M. Mapping of scorpion toxin receptor sites at voltage-gated sodium channels. Toxicon 2012, 60, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Soleglad, M.; Fet, V. The systematics of the scorpion subfamily Uroctoninae (Scorpiones: Chactidae). Rev. Ibér. Aracnol. 2004, 10, 81–128. [Google Scholar]
- Valdez-Cruz, N.A.; Batista, C.V.F.; Zamudio, F.Z.; Bosmans, F.; Tytgat, J.; Possani, L.D. Phaiodotoxin, a novel structural class of insect-toxin isolated from the venom of the Mexican scorpion Anuroctonus phaiodactylus. Eur. J. Biochem. 2004, 271, 4753–4761. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Dominguez, M.E.; Olamendi-Portugal, T.; Garcia, U.; Garcia, C.; Arechiga, H.; Possani, L.D. Cn11, the first example of a scorpion toxin that is a true blocker of Na+ currents in crayfish neurons. J. Exp. Biol. 2002, 205, 869–876. [Google Scholar] [PubMed]
- Carcamo-Noriega, E.N.; Olamendi-Portugal, T.; Restano-Cassulini, R.; Rowe, A.; Uribe-Romero, S.J.; Becerril, B.; Possani, L.D. Intraspecific variation of Centruroides sculpturatus scorpion venom from two regions of Arizona. Arch. Biochem. Biophys. 2018, 638, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenkov, A.I.; Krylov, N.A.; Chugunov, A.O.; Grishin, E.V.; Vassilevski, A.A. Kalium: A database of potassium channel toxins from scorpion venom. Database 2016, 2016, baw056. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Vargas, J.M.; Possani, L.D.; Luna-Ramírez, K. Arthropod toxins acting on neuronal potassium channels. Neuropharmacology 2017, 127, 139–160. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Hill, J.M.; Little, M.J.; Nicholson, G.M.; King, G.F.; Alewood, P.F. Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc. Natl. Acad. Sci. USA 2011, 108, 10478–10483. [Google Scholar] [CrossRef] [PubMed]
- Torabi, E.; Asgari, S.; Khalaj, V.; Behdani, M.; Kazemi-Lomedasht, F.; Bagheri, K.P.; Shahbazzadeh, D. Corrigendum to “The first report on transcriptome analysis of the venom gland of Iranian scorpion, Hemiscorpius lepturus” [Toxicon 125 (2017) 123–130]. Toxicon 2017, 128, 60. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramirez, K.; Quintero-Hernandez, V.; Vargas-Jaimes, L.; Batista, C.V.F.; Winkel, K.D.; Possani, L.D. Characterization of the venom from the Australian scorpion Urodacus yaschenkoi: Molecular mass analysis of components, cDNA sequences and peptides with antimicrobial activity. Toxicon 2013, 63, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Possani-Postay, L.D.; Gurrola-Briones, G.; Salas-Castillo, S.P.; Batista, C.V.F.; Varga, Z.S.; Panyi, G.; Gáspár, R. Vm23 and Vm24, Two Scorpion Peptides that Block Human T-Lymphocyte Potassium Channels (Sub-Type Kv1.3) with High Selectivity and Decrease the In Vivo DTH-Response in Rats. European Patent EP2158213A1, 14 May 2007. [Google Scholar]
- Diego-Garcia, E.; Schwartz, E.F.; D’Suze, G.; Gonzalez, S.A.R.; Batista, C.V.F.; Garcia, B.I.; de la Vega, R.C.R.; Possani, L.D. Wide phylogenetic distribution of Scorpine and long-chain beta-KTx-like peptides in scorpion venoms: Identification of “orphan” components. Peptides 2007, 28, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Soorki, M.; Galehdari, H.; Baradaran, M.; Jalali, A. First venom gland transcriptomic analysis of Iranian yellow scorpion Odonthubuthus doriae. Toxicon 2016, 120, 69–77. [Google Scholar]
- Schwartz, E.F.; Diego-Garcia, E.; de la Vega, R.C.R.; Possani, L.D. Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC Genom. 2007, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, F.; Feng, J.; Yang, W.; Zeng, D.; Zhao, R.; Cao, Z.; Liu, M.; Li, W.; Jiang, L.; et al. Genomic and structural characterization of Kunitz-Type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins. PLoS ONE 2013, 8, e60201. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, R.; Peigneur, S.; Pons, T.; Alvarez, C.; González, L.; Chávez, M.A.; Tytgat, J. The kunitz-type protein ShPI-1 inhibits serine proteases and voltage-gated potassium channels. Toxins 2016, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Huys, I.; Dyason, K.; Verdonck, F.; Tytgat, J. Evolutionary trace analysis of scorpion toxins specific for K+-channels. Proteins Struct. Funct. Genet. 2004, 54, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Camargos, T.S.; Restano-Cassulini, R.; Possani, L.D.; Peigneur, S.; Tytgat, J.; Schwartz, C.A.; Alves, E.M.C.; de Freitas, S.M.; Schwartz, E.F. The new kappa-KTx 2.5 from the scorpion Opisthacanthus cayaporum. Peptides 2011, 32, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Vandendriessche, T.; Kopljar, I.; Jenkins, D.P.; Diego-Garcia, E.; Abdel-Mottaleb, Y.; Vermassen, E.; Clynen, E.; Schoofs, L.; Wulff, H.; Snyders, D.; et al. Purification, molecular cloning and functional characterization of HelaTx1 (Heterometrus laoticus): The first member of a new k-KTX subfamily. Biochem. Pharmacol. 2012, 83, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, H.H.; Kirbyf, M.S.; Lederer, W.J.; Coronado, R. Scorpion toxins targeted against the sarcoplasmic reticulum Ca(2+)-release channel of skeletal and cardiac muscle. Physiology 1992, 89, 12185–12189. [Google Scholar] [CrossRef]
- Xiao, L.; Gurrola, G.B.; Zhang, J.; Valdivia, C.R.; SanMartin, M.; Zamudio, F.Z.; Zhang, L.; Possani, L.D.; Valdivia, H.H. Structure–function relationships of peptides forming the calcin family of ryanodine receptor ligands. J. Gen. Physiol. 2016, 147, 375–394. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Vetter, I.; Lewis, R.; Peigneur, S.; Tytgat, J.; Lam, A.; Gallant, E.; Beard, N.; Alewood, P.; Dulhunty, A. Multiple actions of phi-LITX-Lw1a on ryanodine receptors reveal a functional link between scorpion DDH and ICK toxins. Proc. Natl. Acad. Sci. USA 2013, 110, 8906–8911. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Darbon, H.; Dyason, K.; Verdonck, F.; Tytgat, J. Evolutionary origin of inhibitor cystine knot peptides. FASEB J. 2003, 17, 1765–1767. [Google Scholar] [CrossRef] [PubMed]
- Shahbazzadeh, D.; Srairi-Abid, N.; Feng, W.; Ram, N.; Borchani, L.; Ronjat, M.; Akbari, A.; Pessah, I.N.; De Waard, M.; El Ayeb, M. Hemicalcin, a new toxin from the Iranian scorpion Hemiscorpius lepturus which is active on ryanodine-sensitive Ca2+ channels. Biochem. J. 2007, 404, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Lundy, P.M.; Frew, R. Effect of omega-agatoxin-IVA on autonomic neurotransmission. Eur. J. Pharmacol. 1994, 261, 79–84. [Google Scholar] [CrossRef]
- Oppenheim, J.J.; Biragyn, A.; Kwak, L.W.; Yang, D. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann. Rheum. Dis. 2003, 62, ii17–ii21. [Google Scholar] [CrossRef] [PubMed]
- Hazlett, L.; Wu, M. Defensins in innate immunity. Cell Tissue Res. 2011, 343, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, R.C.R.; García, B.I.; D’Ambrosio, C.; Diego-García, E.; Scaloni, A.; Possani, L.D. Antimicrobial peptide induction in the haemolymph of the Mexican scorpion Centruroides limpidus limpidus in response to septic injury. Cell. Mol. Life Sci. 2004, 61, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Tan, C.; Chanhome, L.; Tan, N. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: Elucidating geographical venom variation and insights into sequence novelty. PeerJ 2017, 5, e3142. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, Y.; Wei, L.; Ye, H.; Liu, H.; Wang, L.; Liu, R.; Li, D.; Lai, R. Snake venom-like waprin from the frog of Ceratophrys calcarata contains antimicrobial function. Gene 2013, 514, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Wong, H.; Desai, M.; Moochhala, S.; Kuchel, P.; Kini, R. Identification of a novel family of proteins in snake venoms. Purification and structural characterization of nawaprin from Naja nigricollis snake venom. J. Biol. Chem. 2003, 278, 40097–40104. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H. Snake venom proteases inhibitors: Enhanced identification, expanding biological function, and promising future. In Snake Venoms, Toxinology; Springer: Dordrecht, The Netherland, 2017; pp. 161–186. [Google Scholar]
- Almaaytah, A.; Albalas, Q. Scorpion venom peptides with no disulfide bridges: A review. Peptides 2014, 51, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Carreto, S.; Jimenez-Vargas, J.M.; Rivas-Santiago, B.; Corzo, G.; Possani, L.D.; Becerril, B.; Ortiz, E. Peptides from the scorpion Vaejovis punctatus with broad antimicrobial activity. Peptides 2015, 73, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Carreto, S.; Quintero-Hernandez, V.; Jimenez-Vargas, J.M.; Corzo, G.; Possani, L.D.; Becerril, B.; Ortiz, E. Gene cloning and functional characterization of four novel antimicrobial-like peptides from scorpions of the family Vaejovidae. Peptides 2012, 34, 290–295. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, R.C.R.; Possani, L.D. Current views on scorpion toxins specific for K+-channels. Toxicon 2004, 43, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Vanuopadath, M.; Sajeev, N.; Murali, A.R.; Sudish, N.; Kangosseri, N.; Sebastian, I.R.; Jain, N.D.; Pal, A.; Raveendran, D.; Nair, B.G.; et al. Mass spectrometry-assisted venom profiling of Hypnale hypnale found in the Western Ghats of India incorporating de novo sequencing approaches. Int. J. Biol. Macromol. 2018, 118, 1736–1746. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Bechsgaard, J.; Scavenius, C.; Dyrlund, T.S.; Sanggaard, K.W.; Enghild, J.J.; Bilde, T. Characterisation of protein families in spider digestive fluids and their role in extra-oral digestion. BMC Genom. 2017, 18, 600. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Bjourson, A.J.; Orr, D.F.; Kwok, H.; Rao, P.; Ivanyi, C.; Shaw, C. Unmasking venom gland transcriptomes in reptile venoms. Anal. Biochem. 2002, 311, 152–156. [Google Scholar] [CrossRef]
- De Oliveira, U.C.; Candido, D.M.; Coronado Dorce, V.A.; Junqueira-De-Azevedo, I.D.L.M. The transcriptome recipe for the venom cocktail of Tityus bahiensis scorpion. Toxicon 2015, 95, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.; Rucavado, A.; Escalante, T.; Díaz, C. Hemorrhage induced by snake venom metalloproteinases: Biochemical and biophysical mechanisms involved in microvessel damage. Toxicon 2005, 45, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Hass, E.; Stanley, D. Phospholipases. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: New York, NY, USA, 2007; pp. 1–3. [Google Scholar]
- Hmed, B.; Serria, H.; Mounir, Z. Scorpion peptides: Potential use for new drug development. J. Toxicol. 2013, 2013, 958797. [Google Scholar] [CrossRef] [PubMed]
- Borchani, L.; Sassi, A.; Shahbazzadeh, D.; Strub, J.; Tounsi-Guetteti, H.; Boubaker, M.; Akbari, A.; Dorsselaer, A.; El Ayeb, M. Heminecrolysin, the first hemolytic dermonecrotic toxin purified from scorpion venom. Toxicon 2011, 58, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Vines, C.M.; Bill, C.A. Phospholipases. In eLS; Wiley Online Library: Hoboken, NJ, USA, 2015; pp. 1–9. [Google Scholar]
- Bordon, K.C.F.; Wiezel, G.A.; Amorim, F.G.; Arantes, E.C. Arthropod venom Hyaluronidases: Biochemical properties and potential applications in medicine and biotechnology. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 43. [Google Scholar] [CrossRef] [PubMed]
- King, T.P.; Wittkowski, K.M. Hyaluronidase and hyaluronan in insect venom allergy. Int. Arch. Allergy Immunol. 2011, 156, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Sutti, R.; Tamascia, M.L.; Hyslop, S.; Rocha-E-Silva, T.A.A. Purification and characterization of a hyaluronidase from venom of the spider Vitalius dubius (Araneae, Theraphosidae). J. Venom. Anim. Toxins Incl. Trop. Dis. 2014, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Juárez-González, V.R.; Possani, L.D. Whole transcriptome of the venom gland from Urodacus yaschenkoi scorpion. PLoS ONE 2015, 10, e0127883. [Google Scholar] [CrossRef] [PubMed]
- Amorim, F.G.; Costa, T.R.; Baiwir, D.; De Pauw, E.; Quinton, L.; Sampaio, S.V. Proteopeptidomic, functional and immunoreactivity characterization of Bothrops moojeni snake venom: Influence of snake gender on venom composition. Toxins 2018, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Kazuma, K.; Masuko, K.; Konno, K.; Inagaki, H. Combined venom gland transcriptomic and venom peptidomic analysis of the predatory ant Odontomachus monticola. Toxins 2017, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, K.; Marciniak, P.; Rosiñski, G.; Rychlik, L. Toxic activity and protein identification from the parotoid gland secretion of the common toad Bufo bufo. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 205, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Silva-Libério, M.; Bastos, I.; Júnior, O.; Fontes, W.; Santana, J.; Castro, M. The crude skin secretion of the pepper frog Leptodactylus labyrinthicus is rich in metallo and serine peptidases. PLoS ONE 2014, 9, e96893. [Google Scholar]
- León, D.; Fellay, I.; Mantel, P.; Walch, M. Killing bacteria with cytotoxic effector proteins of human killer immune cells: Granzymes, granulysin, and perforin. Methods Mol. Biol. 2017, 1535, 275–284. [Google Scholar] [PubMed]
- Vema, M.; Xavier, F.; Verma, Y.; Sobha, K. Evaluation of cytotoxic and anti-tumor activity of partially purified serine protease isolate from the Indian earthworm Pheretima posthuma. Asian Pac. J. Trop. Biomed. 2013, 3, 896–901. [Google Scholar]
- Menaldo, D.; Bernardes, C.; Pereira, J.; Silveira, D.; Mamede, C.; Stanziola, L.; de Oliveira, F.; Pereira-Crott, L.; Faccioli, L.; Sampaio, S. Effects of two serine proteases from Bothrops pirajai snake venom on the complement system and the inflammatory response. Int. Immunopharmacol. 2013, 15, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.; Sánchez, E. Natural protease inhibitors to hemorrhagins in snake venoms and their potential use in medicine. Toxicon 1999, 37, 703–728. [Google Scholar] [CrossRef]
- Ma, H.; Xiao-Peng, T.; Yang, S.; Lu, Q.; Lai, R. Protease inhibitor in scorpion (Mesobuthus eupeus) venom prolongs the biological activities of the crude venom. Chin. J. Nat. Med. 2016, 14, 607–614. [Google Scholar] [CrossRef]
- Proaño-Bolaños, C.; Li, R.; Zhou, M.; Wang, L.; Xi, X.; Tapia, E.; Coloma, L.; Chen, T.; Shaw, C. Novel Kazal-tyoe proteinase inhibitors from the skin secretion of the Splendid leaf frog, Cruziohyla calcarifer. EuPA Open Proteom. 2017, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Y.; Lu, Z.; Zhain, L.; Jiang, J.; Liu, J.; Yu, H. A novel serine protease inhibitor from the venom of Vespa bicolor Fabricius. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2009, 153, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, X.; Liu, H.; San, M.; Xu, Y.; Li, J.; Li, S.; Cao, Z.; Li, W.; Wu, Y.; et al. A new Kunitz-type plasmin inhibitor from scorpion venom. Toxicon 2015, 106, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Nagao, J.; Miyashita, M.; Nakagawa, Y.; Miyagawa, H. Chemical synthesis of La1 isolated from the venom of the scorpion Liocheles australasiae and determination of its disulfide bonding pattern. J. Pept. Sci. 2015, 21, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, M.; Otsuki, J.; Hanai, Y.; Nakagawa, Y.; Miyagawa, H. Characterization of peptide components in the venom of the scorpion Liocheles australasiae (Hemiscorpiidae). Toxicon 2007, 50, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Hograindleur, J.; Voisin, S.; Abi Nahed, R.; Abd El Aziz, T.; Escoffier, J.; Bessonnat, J.; Fovet, C.; De Waard, M.; Hennebicg, S.; et al. Spermaurin, an La1-like peptide from the venom of the scorpion Scorpio maurus plamatus, improves sperm motility and fertilization in different mammalian species. Mol. Hum. Reprod. 2017, 23, 116–131. [Google Scholar] [PubMed]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—Roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.; Vidal, N.; Norman, J.; Vonk, F.; Scheib, H.; Ramjan, S.; Kuruppu, S.; Fung, K.; Hedges, S.; Richardson, M.; et al. Early evolution of the venom system in lizards and snakes. Nature 2006, 439, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Reddy, T.; Gibbs, G.; Merriner, D.; Kerr, J.; O’Bryan, M. Cysteine-rich secretory proteins are not exclusively expressed in the male reproductive tract. Dev. Dyn. 2008, 237, 3313–3323. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.; Scortecci, K.; Kobashi, L.; Agnez-Lima, L.; Medeiros, S.; Silva-Junior, A.; Junqueira-de-Azevedo, I.; Fernandes-Pedrosa, M. Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey. BMC Genom. 2012, 13, 362. [Google Scholar] [CrossRef] [PubMed]
- Hwa, V.; Oh, Y.; Rosenfeld, R. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr. Rev. 1999, 20, 761–787. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [PubMed]
- Tolksdorf, S.; McCready, M.H. The turbidimetric assay of hyaluronidase. J. Lab. Clin. Med. 1949, 34, 74–89. [Google Scholar] [PubMed]
- Horta, C.C.R.; de Magalhães, B.F.; Oliveira-Mendes, B.B.R.; Carmo, A.O.D.; Duarte, C.G.; Felicori, L.F.; Machado-de-Ávila, R.A.; Chávez-Olórtegui, C.; Kalapothakis, E. Molecular, immunological, and biological characterization of Tityus serrulatus venom hyaluronidase: New insights into its role in envenomation. PLoS Negl. Trop. Dis. 2014, 8, e2693. [Google Scholar] [CrossRef] [PubMed]
- Habermann, E.; Hardt, K.L. A sensitive and specific plate test for the quantitation of phospholipases. Anal. Biochem. 1972, 50, 163–173. [Google Scholar] [CrossRef]
- Ramírez-Avila, J.; Quevedo, B.E.; López, E.; Renjifo, J.M. Purification and partial characterization of phospholipases A2 from Bothrops asper (barba amarilla) snake venom from Chiriguaná (Cesar, Colombia). J. Venom. Anim. Toxins Incl. Trop. Dis. 2004, 10, 242–259. [Google Scholar] [CrossRef]


| Retention Time (Min) | Molecular Mass (Da) |
|---|---|
| 0−20 | 444.26; 492.30; 673.37; 748.24; 775.41; 815.48; 888.44; 1018.52; 1105.66; 1158.64; 1234.61; 1245.64; 1374.74; 1434.69; 1460.76; 1484.79; 1573.83; 1633.82; 1694.86; 1731.90; 1832.95; 2290.88; 2350.31; 2452.07; 3464.44; 3565.49; 3616.63; 3648.56; 3701.77; 3773.73; 3928.89; 4052.58; 4361.81; 4544.96; 4744.10; 4815.13; 4879.28; 4882.28 |
| 20−40 | 2709.25; 3951.91; 4001.95; 4138.94; 4180.02; 4694.16 |
| 40−60 | 645.34; 683.30; 963.55; 1286.78; 1305.70; 1343.65; 1413.68; 1902.00; 2124.17; 3937.86; 4393.03; 5053.20; 5318.35 |
| 60−80 | 539.15; 722.39; 765.40; 836.46; 1004.54; 1193.63; 1264.68; 1408.59; 1643.90; 1750.81; 2036.03; 2103.22; 2229.20; 2411.28; 2467.34; 2580.42; 2629.31; 2757.39; 2855.56; 2837.54; 2984.55; 3338.73; 3350.48; 4068.67; 4348.99; 4444.81; 8029.68 |
| 80−100 | 707.35; 890.52; 978.51; 1518.92; 1647.01; 1744.06; 1859.09; 1972.17; 2561.42; 2950.62; 3274.43; 3421.02; 6230.62; 6418.87; 6568.88; 7088.10; 9023.88 |
| 100−120 | 1162.70; 1391.84; 1616.99; 1677.98; 1845.10; 1883.05; 2057.18; 2083.02; 2575.43; 3219.89; 3914.08; 6741.05; 6813.82; 7070.08 |
| 120−140 | 1380.75; 1495.78; 1582.81; 1683.86; 1721.82; 2248.22; 2361.30; 3001.49; 6457.78 |
| 140−160 | 804.39; 871.42; 1081.50; 1925.03; 2066.00; 3327.88; 3529.02; 6142.59; 6614.95; 6725.05 |
| 160−180 | 727.49; 876.43; 932.56; 1138.62; 1251.71; 1348.67; 1626.82; 1717.00; 1786.11; 1823.99; 2014.18; 2151.16; 2315.34; 2917.48; 3290.92; 6126.59; 6441.77; 6586.89 |
| 180−200 | 557.37; 989.52; 1046.54; 1119.63; 1567.83; 1603.92; 1807.94; 1888.12; 1956.00; 2146.18; 2485.44; 2674.46; 3031.66; 3310.88; 4256.28 |
| 200−220 | 599.40; 712.48; 1360.80; 1475.83; 1588.91; 1759.02; 2275.35; 2493.38; 2557.42; 2820.65; 3044.73; 3263.67; 3394.80; 3689.03; 4330.32; 4592.48; 4720.54; 4791.57; 4862.60; 4949.65; 12,869.20 |
| 220−240 | 657.36; 770.45; 883.51; 1027.60; 1096.62; 1185.67; 1298.75; 1438.81; 1525.84; 1838.02; 2371.36; 4428.40; 4652.41; 11,833.52; 12,386.79; 12,432.80 |
| 240−290 | N/D |
| Transcriptome ID | Score | Coverage | Protein Fragments | Xcorr of the Protein Fragments | Protein Type | Accession Number of the Reference Protein |
|---|---|---|---|---|---|---|
| SgeKTxScr02 | 1248.72 | 56.41 | HGCLADFDVGGGCEQHCR | 7.26 | Potassium channel toxin. Scorpine-like peptide. | API81325.1 |
| KIQDAIDR | 2.54 | |||||
| AWISEK | 1.91 | |||||
| SgeHDPND301 | 163.69 | 87.50 | FWGFLGK | 3.18 | HDP. NDBP-3 family. | ALG64975.1 |
| TIPSLLGGSK | 2.09 | |||||
| SgeHDPND204 | 153.72 | 36.67 | KAWNSNIGK | 3.61 | HDP. NDBP-2 family. | AGK88593.1 |
| AWNSNIGK | 2.31 | |||||
| SgeHDPND202 | 138.57 | 63.83 | AWNSDIGK | 3.59 | HDP. NDBP-2 family. | ALG64975.1 |
| GIWGTIK | 2.76 | |||||
| IGVTPSQAAS | 2.70 | |||||
| SgeEnzPA207 | 109.20 | 50.60 | YGLSnTGSYTLLNcDcEK | 6.04 | Enzyme. Phospholipase A2. | API81335.1 |
| WIYFTAYSPK | 2.85 | |||||
| cANPVGKWKADYK | 1.38 | |||||
| EGWIKK | 1.02 | |||||
| SgeCaTLio02 | 65.14 | 42.86 | DLPLSNEYETcVRPR | 3.74 | Calcium channel toxin. Liotoxin. | P0DJ08.1 |
| SgeEnzPA204 | 64.99 | 36.4 | TLLNcDcEEAFDHcLQTTADK | 4.38 | Enzyme. Phospholipase A2. | API81335.1 |
| TLLNcDcEEAFDHcLQTTADKLEGADKEDTK | 3.99 | |||||
| IIQNYYFNIFK | 3.18 | |||||
| cRmLnSTKEVAR | 1.17 | |||||
| SgeHDPND404 | 64.77 | 53.85 | DLWNGVK | 2.37 | HDP. NDBP-4 family. | I0DEB3.1 |
| SgeNaTBet02 | 49.29 | 47.54 | GSSYGYcYGFGcYcEGLNDDVK | 4.87 | Sodium channel toxin. Beta. | P01491.3 |
| FcQSIcK | 2.46 | |||||
| SgeOthLa104 | 14.30 | 65.82 | NVPGPVDAPFPDccPTSLcR | 4.24 | La1-like peptide | AOF40216.1 |
| SgeEnzPA206 | 13.61 | 40.08 | AFYFHLYGnGcYHVK | 3.79 | Enzyme. Phospholipase A2. | API81339.1 |
| cLDQVVDGTSWYDYHATLGLIK | 1.31 | |||||
| SENGRGLR | 1.09 | |||||
| SgeOthLa106 | 13.36 | 33.33 | TGQYLNEGEEWRDPNHcSIYQcR | 3.82 | La1-like peptide | API81334.1 |
| SgeNaTAlp02 | 12.80 | 64.79 | AmPPFGLPGGcFcPNIPK | 3.14 | Sodium channel toxin. Alpha. | Q5MJP5.1 |
| SgeCaTLio01 | 12.17 | 88.57 | NLPLSDEYGTcVRPR | 2.96 | Calcium channel toxin. Liotoxin. | P0DJ08.1 |
| cKPPLWcNPQQIcVYK | 2.86 | |||||
| SgeKTxScr01 | 10.51 | 71.08 | GVDALTNLIPAPVIGGIVNK | 4.70 | Potassium channel toxin. Scorpine-like peptide. | P0C8W5.1 |
| VQELcAFNK | 3.31 | |||||
| SgeOthCAP02 | 8.09 | 48.96 | STNDFIFYcDFSK | 4.15 | CAP superfamily | API81352.1 |
| VEHSGTAFWTIGmHMqFDQEMESTIKLEAYR | 1.33 | |||||
| VLYTcNYGPAGNmQGGTmYIKGEPcSQcPK | 1.21 | |||||
| SgeEnzHya01 | 4.11 | 31.30 | YFTDTTSVFDSK | 2.43 | Enzyme. Hyaluronidase. | API81375.1 |
| ILVNNK | 1.68 | |||||
| ILVNNKESFVGDK | 1.07 | |||||
| SgeEnzMtP02 | 3.85 | 27.14 | KFNYPHGTIK | 2.15 | Enzyme. Metalloproteinase. | AMO02513.1 |
| KEPKKPPVPK | 1.19 | |||||
| SgeCatClc02 | 2.27 | 24.24 | ADcLPHLK | 2.27 | Calcium channel toxin. Calcin. | API81327.1 |
| c15440_g1_i1 | 6.83 | 9.30 | ELTLEDVGTILR | 3.67 | N/D | N/D |
| c23802_g1_i1 | 7.08 | 10.48 | WYNDcIDYcDR | 3.59 | N/D | N/D |
| c27313_g1_i1 | 13.32 | 15.28 | FGDITLLGPEVTNR | 4.12 | N/D | N/D |
| IISSPFYK | 1.21 | N/D | N/D | |||
| c26154_g1_i1 | 7.74 | 10.53 | LAELETIVALAK | 3.85 | N/D | N/D |
| c26889_g1_i1 | 94.33 | 20.29 | EFFLGLLGLK | 3.63 | N/D | N/D |
| IDPKEFFLGLLGLK | 1.54 | N/D | N/D |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-Gutiérrez, M.T.; Santibáñez-López, C.E.; Jiménez-Vargas, J.M.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi. Toxins 2018, 10, 359. https://doi.org/10.3390/toxins10090359
Romero-Gutiérrez MT, Santibáñez-López CE, Jiménez-Vargas JM, Batista CVF, Ortiz E, Possani LD. Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi. Toxins. 2018; 10(9):359. https://doi.org/10.3390/toxins10090359
Chicago/Turabian StyleRomero-Gutiérrez, Maria Teresa, Carlos Eduardo Santibáñez-López, Juana María Jiménez-Vargas, Cesar Vicente Ferreira Batista, Ernesto Ortiz, and Lourival Domingos Possani. 2018. "Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi" Toxins 10, no. 9: 359. https://doi.org/10.3390/toxins10090359
APA StyleRomero-Gutiérrez, M. T., Santibáñez-López, C. E., Jiménez-Vargas, J. M., Batista, C. V. F., Ortiz, E., & Possani, L. D. (2018). Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi. Toxins, 10(9), 359. https://doi.org/10.3390/toxins10090359








