Influence of Dietary Fiber and Polyphenols During Pre-Gestation, Gestation, or Lactation on Intestinal Gene Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Diets and Experimental Design
2.3. Sample Collection and Preparation
2.4. RNA Extraction
2.5. Microarray Procedure
2.6. Microarray Data Analysis
2.7. Validation of Gene Expression by Real Time PCR
2.8. Ferritin and Adipsin Quantification
2.9. Real Time PCR and ELISA Statistics
3. Results
3.1. Effect on Overall Intestinal Rat Gene Expression
3.2. Gene Expression Changes Due to the HFP Diet During Pre-Gestation
3.3. Gene Expression Changes Due to the HFP Diet During Gestation
3.4. Gene Expression Changes Due to the HFP Diet During Lactation
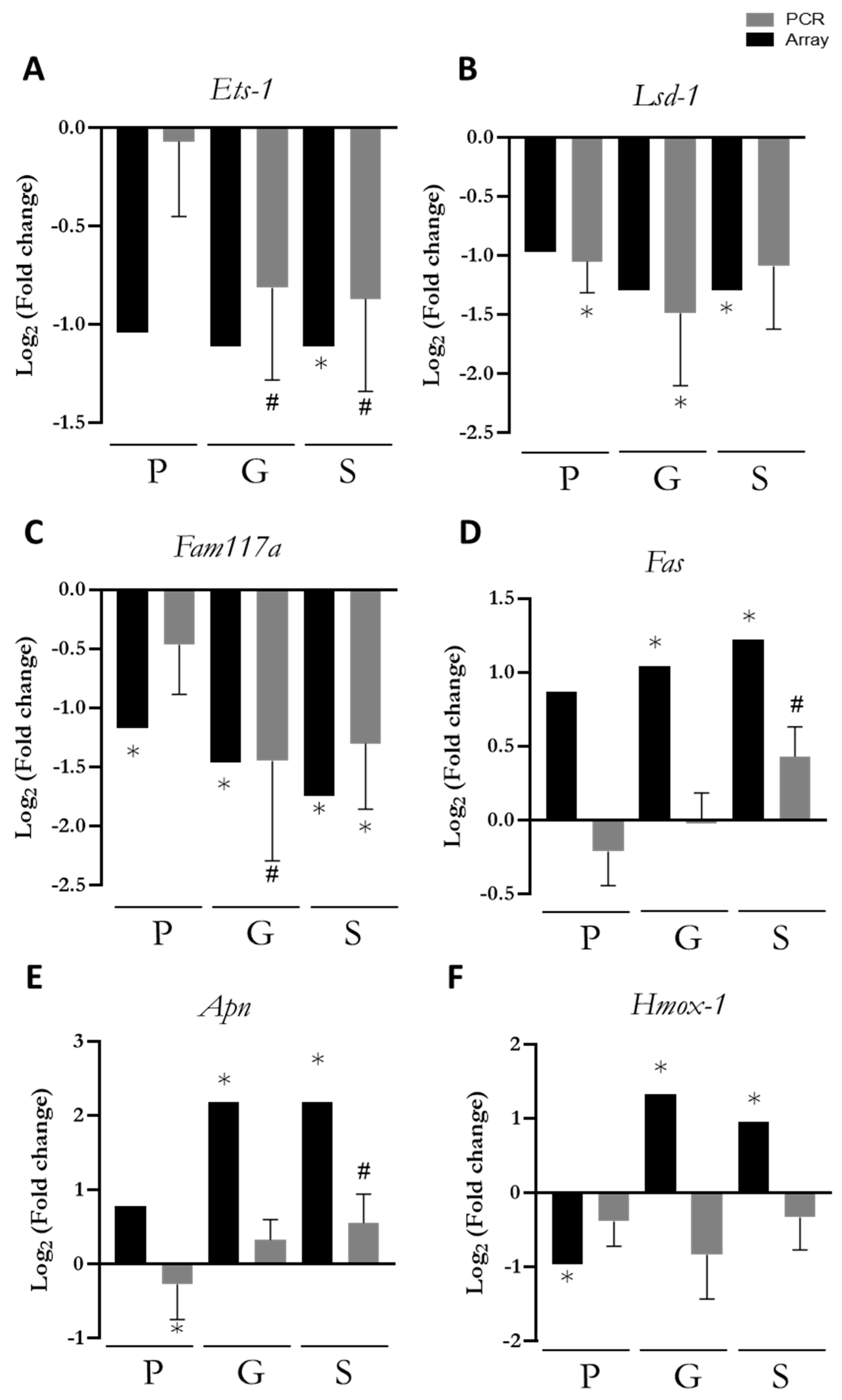
3.5. PCR Confirmation of Key Genes
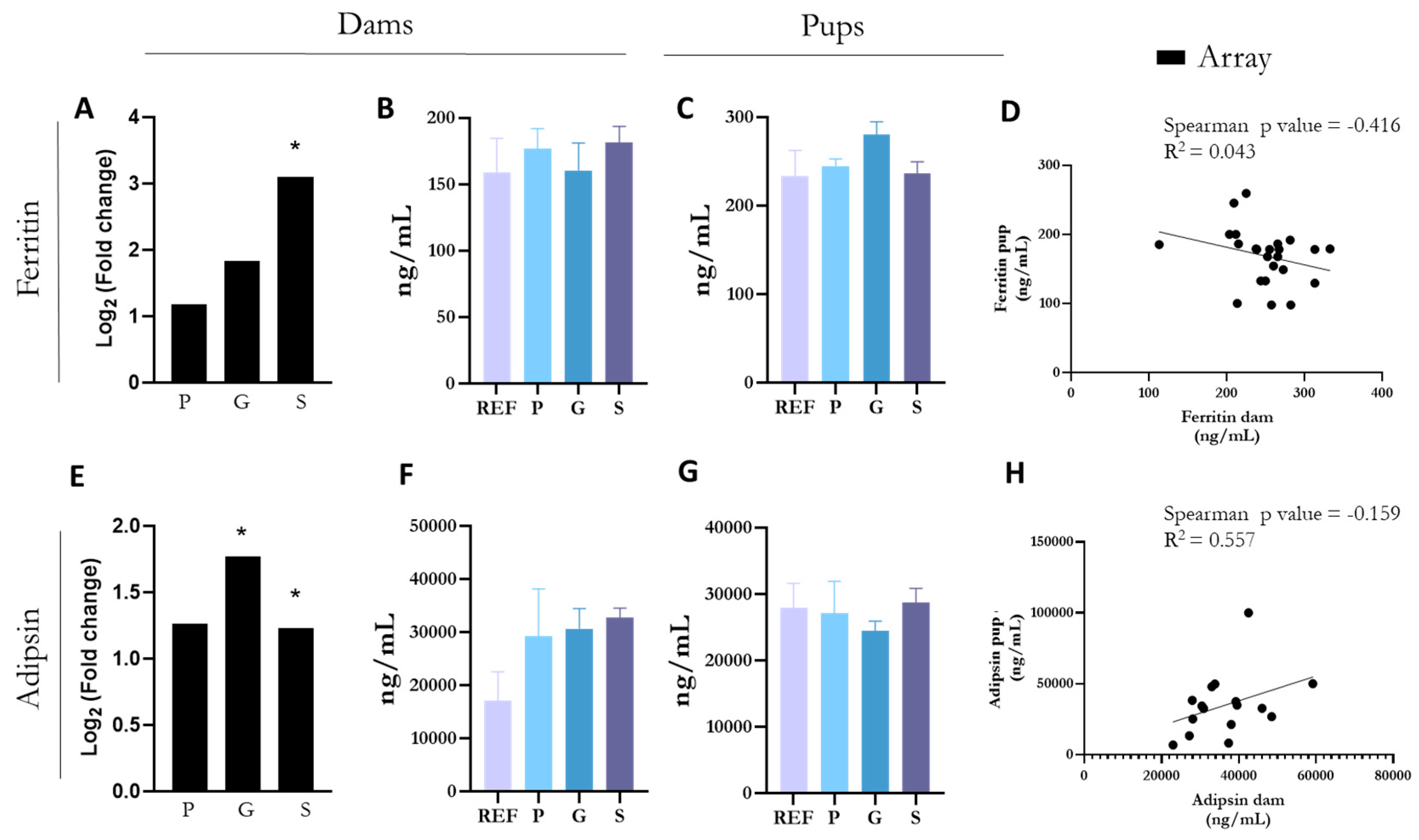
3.6. ELISA Confirmation of Upregulated Changes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vermeulen, R.; Schymanski, E.L.; Barabási, A.L.; Miller, G.W. The exposome and health: Where chemistry meets biology. Science 2020, 367, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Maitre, L.; Bustamante, M.; Hernández-Ferrer, C.; Thiel, D.; Lau, C.H.E.; Siskos, A.P.; Vives-Usano, M.; Ruiz-Arenas, C.; Pelegrí-Sisó, D.; Robinson, O.; et al. Multi-omics signatures of the human early life exposome. Nat. Commun. 2022, 13, 7024. [Google Scholar] [CrossRef]
- Panzella, L. Polyphenols and Their Impact on Human Health. Int. J. Mol. Sci. 2023, 24, 16683. [Google Scholar] [CrossRef] [PubMed]
- Cronin, P.; Joyce, S.A.; O’toole, P.W.; O’connor, E.M. Dietary Fibre Modulates the Gut Microbiota. Nutrients 2021, 13, 1655. [Google Scholar] [CrossRef]
- Li, M.; Ma, S. A review of healthy role of dietary fiber in modulating chronic diseases. Food Res. Int. 2024, 191, 114682. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Zhang, Y.; Chen, Q.; Fu, X.; Huang, Q.; Zhang, B.; Dong, H.; Li, C. Exploring the synergistic benefits of insoluble dietary fiber and bound phenolics: Unveiling the role of bound phenolics in enhancing bioactivities of insoluble dietary fiber. Trends Food Sci. Technol. 2024, 149, 104554. [Google Scholar] [CrossRef]
- He, J.; Zhang, P.; Shen, L.; Niu, L.; Tan, Y.; Chen, L.; Zhao, Y.; Bai, L.; Hao, X.; Li, X.; et al. Short-Chain Fatty Acids and Their Association with Signalling Pathways in Inflammation, Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2020, 21, 6356. [Google Scholar] [CrossRef]
- Pheiffer, C.; Riedel, S.; Dias, S.; Adam, S. Gestational Diabetes and the Gut Microbiota: Fibre and Polyphenol Supplementation as a Therapeutic Strategy. Microorganisms 2024, 12, 633. [Google Scholar] [CrossRef]
- Nacka-Aleksić, M.; Pirković, A.; Vilotić, A.; Bojić-Trbojević, Ž.; Jovanović Krivokuća, M.; Giampieri, F.; Battino, M.; Dekanski, D. The Role of Dietary Polyphenols in Pregnancy and Pregnancy-Related Disorders. Nutrients 2022, 14, 5246. [Google Scholar] [CrossRef] [PubMed]
- Tain, Y.L.; Hsu, C.N. Maternal Polyphenols and Offspring Cardiovascular-Kidney-Metabolic Health. Nutrients 2024, 16, 3168. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, Mechanisms and Clinical Perspective. Semin. Reprod. Med. 2009, 27, 351. [Google Scholar] [CrossRef] [PubMed]
- Larue, A.E.M.; Atlasi, Y. The epigenetic landscape in intestinal stem cells and its deregulation in colorectal cancer. Stem Cells 2024, 42, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Carlos-Reyes, Á.; López-González, J.S.; Meneses-Flores, M.; Gallardo-Rincón, D.; Ruíz-García, E.; Marchat, L.A.; la Vega, H.A.-D.; de la Cruz, O.N.H.; López-Camarillo, C. Dietary Compounds as Epigenetic Modulating Agents in Cancer. Front. Genet. 2019, 10, 79. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Milagro, F.I.; Riezu-Boj, J.I.; Martinez, J.A. Epigenetic signatures underlying inflammation: An interplay of nutrition, physical activity, metabolic diseases, and environmental factors for personalized nutrition. Inflamm. Res. 2020, 70, 29. [Google Scholar] [CrossRef]
- Selvakumar, S.C.; Preethi, K.A.; Thomas, P.; Ameya, K.P.; Sekar, D. Non-Coding RNAs and Diet. Epigenetics Hum. Health 2024, 12, 31–48. [Google Scholar]
- García-Mantrana, I.; Alcántara, C.; Selma-Royo, M.; Boix-Amorós, A.; Dzidic, M.; Gimeno-Alcañiz, J.; Úbeda-Sansano, I.; Sorribes-Monrabal, I.; Escuriet, R.; Gil-Raga, F.; et al. MAMI: A birth cohort focused on maternal-infant microbiota during early life. BMC Pediatr. 2019, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Fuertes, L.; Azagra-Boronat, I.; Massot-Cladera, M.; Knipping, K.; Garssen, J.; Franch, À.; Castell, M.; Pérez-Cano, F.J.; Rodríguez-Lagunas, M.J. Effect of Rotavirus Infection and 2′-Fucosyllactose Administration on Rat Intestinal Gene Expression. Nutrients 2023, 15, 1996. [Google Scholar] [CrossRef] [PubMed]
- Bieller, A.; Pasche, B.; Frank, S.; Gläser, B.; Kunz, J.; Witt, K.; Zoll, B. Isolation and Characterization of the Human Forkhead Gene FOXQ1. DNA Cell Biol. 2004, 20, 555–561. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, J.; Wang, K.; Fan, M.; Hu, Y. FAM117A Is a New Prognostic Marker of Lung Adenocarcinoma and Predicts Sensitivity to PD0332991. Evid Based Complement Altern. Med. 2022, 2022, 3945446. [Google Scholar] [CrossRef] [PubMed]
- Rio-Aige, K.; Azagra-Boronat, I.; Castell, M.; Selma-Royo, M.; Collado, M.C.; Rodríguez-Lagunas, M.J.; Pérez-Cano, F.J. The Breast Milk Immunoglobulinome. Nutrients 2021, 13, 1810. [Google Scholar] [CrossRef]
- Bai, M.; Liu, X.; Wang, L. Identification of potential tumor antigens and immune subtypes for lung adenocarcinoma. Med. Oncol. 2023, 40, 100. [Google Scholar] [CrossRef]
- Kim, D.; Kim KIl Baek, S.H. Roles of lysine-specific demethylase 1 (LSD1) in homeostasis and diseases. J. Biomed. Sci. 2021, 28, 41. [Google Scholar] [CrossRef]
- Wojtala, M.; Rybaczek, D.; Wielgus, E.; Sobalska-Kwapis, M.; Strapagiel, D.; Balcerczyk, A. The Role of Lysine-Specific Demethylase 1 (LSD1) in Shaping the Endothelial Inflammatory Response. Cell Physiol. Biochem. 2021, 55, 569–589. [Google Scholar]
- Zwiggelaar, R.T.; Lindholm, H.T.; Fosslie, M.; Pedersen, M.T.; Ohta, Y.; Díez-Sánchez, A.; Martín-Alonso, M.; Ostrop, J.; Matano, M.; Parmar, N.; et al. LSD1 represses a neonatal/reparative gene program in adult intestinal epithelium. Sci. Adv. 2020, 6, eabc0367. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H.; Hsieh, H.L. Roles of Heme Oxygenase-1 in Neuroinflammation and Brain Disorders. Antioxidants 2022, 11, 923. [Google Scholar] [CrossRef]
- Padda, I.; Sethi, Y.; Das, M.; Fabian, D.; Ralhan, T.; Aziz, D.; Sexton, J.; Johal, G. Heme Oxygenase-1, Cardiac Senescence, and Myocardial Infarction: A Critical Review of the Triptych. Cardiovasc. Drugs Ther. 2024. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, W. The Nuclear Translocation of Heme Oxygenase-1 in Human Diseases. Front. Cell Dev. Biol. 2022, 10, 890186. [Google Scholar] [CrossRef] [PubMed]
- Czibik, G.; Derumeaux, G.; Sawaki, D.; Valen, G.; Motterlini, R. Heme oxygenase-1: An emerging therapeutic target to curb cardiac pathology. Basic. Res. Cardiol. 2014, 109, 450. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Song, J.; Jeong, H.W.; Grönloh, M.L.B.; Koh, B.I.; Bovay, E.; Kim, K.-P.; Klotz, L.; Thistlethwaite, P.A.; van Buul, J.D.; et al. Apelin modulates inflammation and leukocyte recruitment in experimental autoimmune encephalomyelitis. Nat. Commun. 2024, 15, 6282. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wang, D.; Liu, W.; Wang, Y.; Chen, J.; Cai, X. Apelin receptor dimer: Classification, future prospects, and pathophysiological perspectives. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2024, 1870, 167257. [Google Scholar] [CrossRef] [PubMed]
- Winkle, P.; Goldsmith, S.; Koren, M.J.; Lepage, S.; Hellawell, J.; Trivedi, A.; Tsirtsonis, K.; Abbasi, S.A.; Kaufman, A.; Troughton, R.; et al. A First-in-Human Study of AMG 986, a Novel Apelin Receptor Agonist, in Healthy Subjects and Heart Failure Patients. Cardiovasc. Drugs Ther. 2023, 37, 743–755. [Google Scholar] [CrossRef]
- Song, J.; Tang, J.; Zhang, Z.; Liu, Y.; Zhong, J. Targeting the elabela/apelin-apelin receptor axis as a novel therapeutic approach for hypertension. Chin. Med. J. 2021, 135, 1019. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, L.; Li, P.; Zheng, Y.; Yang, Y.; Ji, S. Apelin/APJ system in inflammation. Int. Immunopharmacol. 2022, 109, 108822. [Google Scholar] [CrossRef]
- Milman, N.T. A Review of Nutrients and Compounds, Which Promote or Inhibit Intestinal Iron Absorption: Making a Platform for Dietary Measures That Can Reduce Iron Uptake in Patients with Genetic Haemochromatosis. J. Nutr. Metab. 2020, 2020, 7373498. [Google Scholar] [CrossRef]
- Lynch, S.R. Interaction of iron with other nutrients. Nutr. Rev. 1997, 55, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chang, Q.; Du, Q.; Liu, X.; Dang, S.; Tian, X. Maternal iron nutrition during pregnancy and fetal intrauterine growth. Nutr. J. 2024, 23, 140. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, J.; Fang, M.; Yu, B. Pregnancy immune tolerance at the maternal-fetal interface. Int. Rev. Immunol. 2020, 39, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Bhat, N.K.; Fisher, R.J.; Fujiwara, S.; Ascione, R.; Papas, T.S. Temporal and tissue-specific expression of mouse ets genes. Proc. Natl. Acad. Sci. USA 1987, 84, 3161–3165. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, J. The role of the transcription factor Ets1 in carcinoma. Semin. Cancer Biol. 2015, 35, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chan, Y.T.; Tan, H.Y.; Zhang, C.; Guo, W.; Xu, Y.; Sharma, R.; Chen, Z.-S.; Zheng, Y.-C.; Wang, N.; et al. Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates sorafenib resistance in human hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 3. [Google Scholar] [CrossRef]
- Rajendran, P.; Abdelsalam, S.A.; Renu, K.; Veeraraghavan, V.; Ben Ammar, R.; Ahmed, E.A. Polyphenols as Potent Epigenetics Agents for Cancer. Int. J. Mol. Sci. 2022, 23, 11712. [Google Scholar] [CrossRef]
- Carroll, R.G.; Zasłona, Z.; Galván-Peña, S.; Koppe, E.L.; Sévin, D.C.; Angiari, S.; Triantafilou, M.; Triantafilou, K.; Modis, L.K.; O’neill, L.A. An unexpected link between fatty acid synthase and cholesterol synthesis in proinflammatory macrophage activation. J. Biol. Chem. 2018, 293, 5509–5521. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Sun, K.Y.; Zhu, Y.; Zhang, X.; Zhou, Y.H.; Zou, X. Metformin alleviates inflammation through suppressing FASN-dependent palmitoylation of Akt. Cell Death Dis. 2021, 12, 934. [Google Scholar] [CrossRef] [PubMed]
- Young, K.E.; Flaherty, S.; Woodman, K.M.; Sharma-Walia, N.; Reynolds, J.M. Fatty acid synthase regulates the pathogenicity of Th17 cells. J. Leukoc. Biol. 2017, 102, 1229–1235. [Google Scholar] [CrossRef]
- Lim, S.A.; Wei, J.; Nguyen, T.L.M.; Shi, H.; Su, W.; Palacios, G.; Dhungana, Y.; Chapman, N.M.; Long, L.; Saravia, J.; et al. Lipid signalling enforces functional specialization of Treg cells in tumours. Nature 2021, 591, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Wang, N.; Tong, Z.; Wang, D.; Wang, P.; Yang, Q.; Yan, X.; Song, W.; Jin, Z.; Zhang, M. Role of complement factor D in cardiovascular and metabolic diseases. Front. Immunol. 2024, 15, 1453030. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, K.; Bari, M.F.; Adaikalakoteswari, A.; Guller, S.; Weickert, M.O.; Randeva, H.S.; Grammatopoulos, D.K.; Bastie, C.C.; Vatish, M. Elevated fetal adipsin/acylation-stimulating protein (ASP) in obese pregnancy: Novel placental secretion via Hofbauer cells. J. Clin. Endocrinol. Metab. 2013, 98, 4113–4122. [Google Scholar] [CrossRef] [PubMed]


| Components | REF Diet (g/kg) | HFP Diet (g/kg) |
|---|---|---|
| Casein | 200 | 200 |
| L-Cysteine | 3 | 3 |
| Cornstarch Flour | 379.186 | 289.186 |
| Inulin | 0 | 80 |
| Pectin | 0 | 10 |
| Maltodextrin | 132 | 132 |
| Sucrose | 100 | 100 |
| Soybean Oil | 70 | 70 |
| Cellulose | 50 | 50 |
| Mineral Mix (TD94049) | 48 | 48 |
| Ferric Citrate | 0.3 | 0.3 |
| Vitamin Mix (TD94047) | 15 | 15 |
| Choline Bitartrate | 2.5 | 2.5 |
| Tertiary Butylhydroquinone | 0.014 | 0.014 |
| Polyphenols | 0 | 5 |
| Catechin | 0 | 1 |
| Epicatechin | 0 | 1 |
| Hesperidin | 0 | 1.5 |
| Naringenin | 0 | 0.75 |
| Quercetin | 0 | 0.75 |
| (A) Downregulated Genes | |||
|---|---|---|---|
| Gene | Name | Log2FC | padj |
| ENSRNOG00000022136 | Loc685680 | −1.38 | 1 × 10−3 |
| ENSRNOG00000024543 | Foxq1 | −1.39 | 3 × 10−3 |
| ENSRNOG00000062314 | Mfsd4b1 | −1.05 | 3 × 10−3 |
| ENSRNOG00000004417 | Fam117a | −1.17 | 5 × 10−3 |
| ENSRNOG00000003681 | Lct | −1.20 | 2 × 10−2 |
| ENSRNOG00000019813 | Ppp4 | −1.36 | 4 × 10−2 |
| (B) Upregulated genes | |||
| Gene | Name | Log2FC | padj |
| ENSRNOG00000046165 | Mptx1 | 5.69 | 6 × 10−3 |
| (A) Downregulated Genes | |||
|---|---|---|---|
| Gene | Name | Log2FC | padj |
| ENSRNOG00000006137 | Arid1a | −1.02 | 8 × 10−6 |
| ENSRNOG00000027193 | Ppp1r26 | −1.07 | 2 × 10−4 |
| ENSRNOG00000042983 | Nudt21 | −1.04 | 2 × 10−4 |
| ENSRNOG00000027739 | Cndp1 | −1.16 | 2 × 10−4 |
| ENSRNOG00000017332 | Dapk2 | −1.01 | 1 × 10−3 |
| ENSRNOG00000011677 | Slc39a10 | −1.29 | 2 × 10−3 |
| ENSRNOG00000042758 | Tmem243 | −1.03 | 6 × 10−3 |
| ENSRNOG00000004417 | Fam117a | −1.46 | 7 × 10−3 |
| ENSRNOG00000021053 | Lsr | −1.03 | 1 × 10−2 |
| ENSRNOG00000006663 | Uch2 | −1.08 | 1 × 10−2 |
| ENSRNOG00000008584 | Rnaseh1 | −1.17 | 3 × 10−2 |
| ENSRNOG00000006600 | Unk | −1.06 | 4 × 10−2 |
| (B) Upregulated genes | |||
| Gene | Name | Log2FC | padj |
| ENSRNOG00000033564 | Cfd | 1.77 | 1 × 10−3 |
| ENSRNOG00000023493 | Creb314 | 1.02 | 6 × 10−3 |
| ENSRNOG00000006033 | Spon-2 | 1.03 | 6 × 10−3 |
| ENSRNOG00000045636 | Fasn | 1.04 | 8 × 10−3 |
| ENSRNOG00000014117 | Hmox1 | 1.32 | 8 × 10−3 |
| ENSRNOG00000012181 | Lpl | 1.41 | 2 × 10−2 |
| ENSRNOG00000068020 | Uqcc5 | 1.01 | 3 × 10−2 |
| ENSRNOG00000003984 | Apln | 2.18 | 2 × 10−3 |
| (A) Downregulated Genes | |||
|---|---|---|---|
| Gene | Name | Log2FC | padj |
| ENSRNOG00000006137 | Arid1a | −1.05 | 1 × 10−16 |
| ENSRNOG00000027193 | Ppp1r26 | −1.28 | 1 × 10−11 |
| ENSRNOG00000021081 | Vps72 | −1.27 | 6 × 10−11 |
| ENSRNOG00000019671 | Rsbn1 | −1.09 | 1 × 10−10 |
| ENSRNOG00000042983 | Nudt21 | −1.26 | 2 × 10−9 |
| ENSRNOG00000004417 | Fam117a | −1.74 | 1 × 10−8 |
| ENSRNOG00000042758 | Tmem243 | −1.21 | 2 × 10−8 |
| ENSRNOG00000011677 | Slc39a10 | −1.29 | 4 × 10−6 |
| ENSRNOG00000002032 | Ifngr2 | −1.02 | 1 × 10−5 |
| ENSRNOG00000053599 | Pogz | −1.00 | 1 × 10−5 |
| (B) Upregulated genes | |||
| Gene | Name | Log2FC | padj |
| ENSRNOG00000008118 | Sync | 1.31 | 2 × 109 |
| ENSRNOG00000045636 | Fasn | 1.22 | 5 × 109 |
| ENSRNOG00000017178 | Hydin | 5.91 | 1 × 10−4 |
| ENSRNOG00000063281 | 60S ribosomal protein L29 | 5.78 | 4 × 10−4 |
| ENSRNOG00000003984 | Apln | 2.18 | 2 × 10−3 |
| ENSRNOG00000022724 | Cnih3 | 5.26 | 3 × 10−3 |
| ENSRNOG00000043103 | Frrs1l | 2.49 | 3 × 10−3 |
| ENSRNOG00000003741 | Nptx1 | 1.97 | 5 × 10−3 |
| ENSRNOG00000033564 | Cfd | 1.22 | 5 × 10−3 |
| ENSRNOG00000003785 | Usp43 | 1.18 | 6 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceballos-Sánchez, D.; Sáez-Fuertes, L.; Casanova-Crespo, S.; Rodríguez-Lagunas, M.J.; Castell, M.; Pérez-Cano, F.J.; Massot-Cladera, M. Influence of Dietary Fiber and Polyphenols During Pre-Gestation, Gestation, or Lactation on Intestinal Gene Expression. Nutrients 2025, 17, 341. https://doi.org/10.3390/nu17020341
Ceballos-Sánchez D, Sáez-Fuertes L, Casanova-Crespo S, Rodríguez-Lagunas MJ, Castell M, Pérez-Cano FJ, Massot-Cladera M. Influence of Dietary Fiber and Polyphenols During Pre-Gestation, Gestation, or Lactation on Intestinal Gene Expression. Nutrients. 2025; 17(2):341. https://doi.org/10.3390/nu17020341
Chicago/Turabian StyleCeballos-Sánchez, Daniela, Laura Sáez-Fuertes, Sergi Casanova-Crespo, Maria J. Rodríguez-Lagunas, Margarida Castell, Francisco J. Pérez-Cano, and Malen Massot-Cladera. 2025. "Influence of Dietary Fiber and Polyphenols During Pre-Gestation, Gestation, or Lactation on Intestinal Gene Expression" Nutrients 17, no. 2: 341. https://doi.org/10.3390/nu17020341
APA StyleCeballos-Sánchez, D., Sáez-Fuertes, L., Casanova-Crespo, S., Rodríguez-Lagunas, M. J., Castell, M., Pérez-Cano, F. J., & Massot-Cladera, M. (2025). Influence of Dietary Fiber and Polyphenols During Pre-Gestation, Gestation, or Lactation on Intestinal Gene Expression. Nutrients, 17(2), 341. https://doi.org/10.3390/nu17020341










