Association between Genetic Variation in the TAS2R38 Bitter Taste Receptor and Propylthiouracil Bitter Taste Thresholds among Adults Living in Japan Using the Modified 2AFC Procedure with the Quest Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Stimuli and Apparatus
2.3. Taste Detection Threshold Test
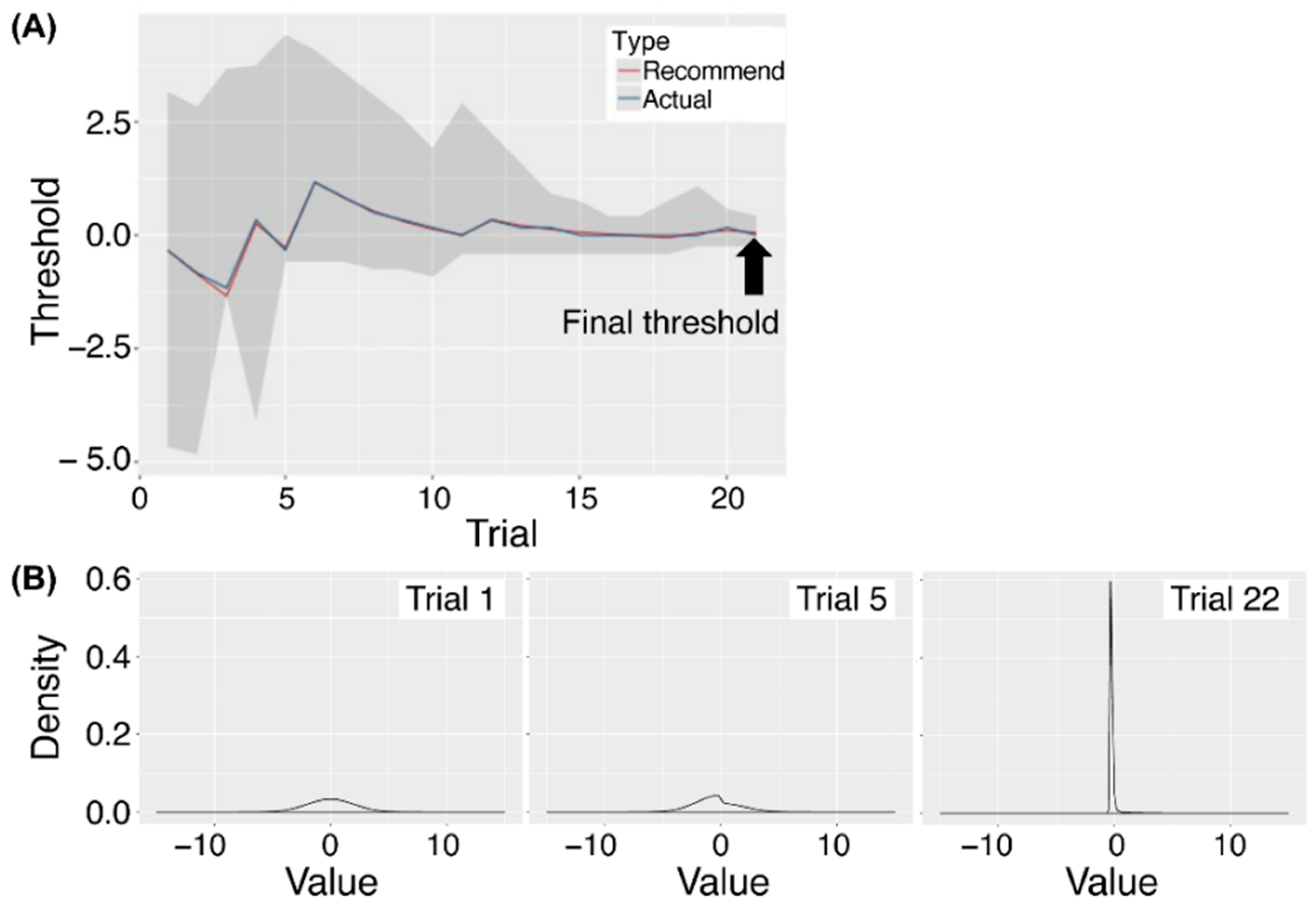
2.4. Threshold Estimation
2.5. TAS2R Genotyping
2.6. Statistical Analyses
3. Results
3.1. TAS2R Polymorphism
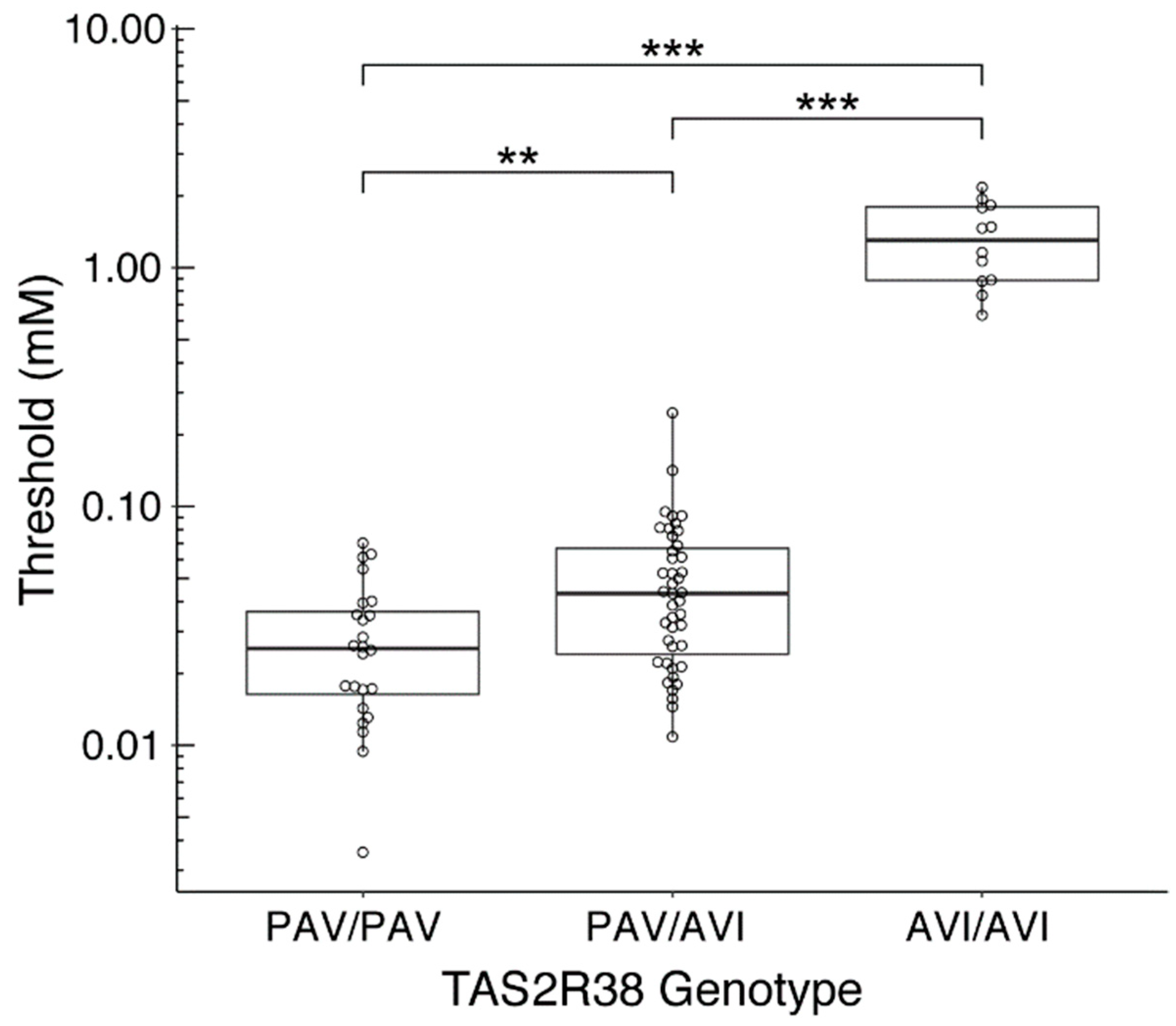
3.2. Relationship between the PROP Bitter Taste Threshold and the TAS2R38 Genotypes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilbertson, T.A.; Damak, S.; Margolskee, R.F. The molecular physiology of taste transduction. Curr. Opin. Neurobiol. 2000, 10, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, B. Taste reception. Physiol. Rev. 1996, 76, 719–766. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, B.; Ogiwara, Y.; Ninomiya, Y. The discovery of umami. Chem. Senses 2002, 27, 843–844. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.L.; McGeary, J.E.; Knopik, V.S.; Hayes, J.E. Bitterness of the non-nutritive sweetener acesulfame potassium varies with polymorphisms in TAS2R9 and TAS2R31. Chem. Senses 2013, 38, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Behrens, M.; Gunn, H.C.; Ramos, P.C.M.; Meyerhof, W.; Wooding, S.P. Genetic, functional, and phenotypic diversity in TAS2R38-mediated bitter taste perception. Chem. Senses 2013, 38, 475–484. [Google Scholar] [CrossRef]
- Campbell, M.C.; Ranciaro, A.; Froment, A.; Hirbo, J.; Omar, S.; Bodo, J.M.; Nyambo, T.; Lema, G.; Zinshteyn, D.; Drayna, D.; et al. Evolution of functionally diverse alleles associated with PTC bitter taste sensitivity in Africa. Mol. Biol. Evol. 2012, 29, 1141–1153. [Google Scholar] [CrossRef]
- Duffy, V.B.; Davidson, A.C.; Kidd, J.R.; Kidd, K.K.; Speed, W.C.; Pakstis, A.J.; Reed, D.R.; Snyder, D.J.; Bartoshuk, L.M. Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol. Clin. Exp. Res. 2004, 28, 1629–1637. [Google Scholar] [CrossRef]
- Kim, U.K.; Jorgenson, E.; Coon, H.; Leppert, M.; Risch, N.; Drayna, D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 2003, 299, 1221–1225. [Google Scholar] [CrossRef]
- Pronin, A.N.; Xu, H.; Tang, H.; Zhang, L.; Li, Q.; Li, X. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr. Biol. 2007, 17, 1403–1408. [Google Scholar] [CrossRef]
- Reed, D.R.; Zhu, G.; Breslin, P.A.S.; Duke, F.F.; Henders, A.K.; Campbell, M.J.; Montgomery, G.W.; Medland, S.E.; Martin, N.G.; Wright, M.J. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum. Mol. Genet. 2010, 19, 4278–4285. [Google Scholar] [CrossRef]
- Roudnitzky, N.; Bufe, B.; Thalmann, S.; Kuhn, C.; Gunn, H.C.; Xing, C.; Crider, B.P.; Behrens, M.; Meyerhof, W.; Wooding, S.P. Genomic, genetic and functional dissection of bitter taste responses to artificial sweeteners. Hum. Mol. Genet. 2011, 20, 3437–3449. [Google Scholar] [CrossRef] [PubMed]
- Soranzo, N.; Bufe, B.; Sabeti, P.C.; Wilson, J.F.; Weale, M.E.; Marguerie, R.; Meyerhof, W.; Goldstein, D.B. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr. Biol. 2005, 15, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Wooding, S.; Gunn, H.; Ramos, P.; Thalmann, S.; Xing, C.; Meyerhof, W. Genetics and bitter taste responses to goitrin, a plant toxin found in vegetables. Chem. Senses 2010, 35, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.G.; Eny, K.M.; Cockburn, M.; Chiu, W.; Nielsen, D.E.; Duizer, L.; El-Sohemy, A. Variation in the TAS1R2 gene, sweet taste perception and intake of sugars. J. Nutrigenet. Nutr. 2015, 8, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Fushan, A.A.; Simons, C.T.; Slack, J.P.; Manichaikul, A.; Drayna, D. Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr. Biol. 2009, 19, 1288–1293. [Google Scholar] [CrossRef] [PubMed]
- Meyerhof, W.; Mennella, J.A.; Finkbeiner, S.; Lipchock, S.V.; Hwang, L.-D.; Reed, D.R. Preferences for salty and sweet tastes are elevated and related to each other during childhood. PLoS ONE 2014, 9, e92201. [Google Scholar]
- Chen, Q.-Y.; Alarcon, S.; Tharp, A.; Ahmed, O.M.; Estrella, N.L.; Greene, T.A.; Rucker, J.; Breslin, P.A.S. Perceptual variation in umami taste and polymorphisms in TAS1R taste receptor genes. Am. J. Clin. Nutr. 2009, 90, 770S–779S. [Google Scholar] [CrossRef]
- Raliou, M.; Wiencis, A.; Pillias, A.M.; Planchais, A.; Eloit, C.; Boucher, Y.; Trotier, D.; Montmayeur, J.P.; Faurion, A. Nonsynonymous single nucleotide polymorphisms in human tas1r1, tas1r3, and mGluR1 and individual taste sensitivity to glutamate. Am. J. Clin. Nutr. 2009, 90, 789S–799S. [Google Scholar] [CrossRef]
- Shigemura, N.; Shirosaki, S.; Sanematsu, K.; Yoshida, R.; Ninomiya, Y. Genetic and molecular basis of individual differences in human umami taste perception. PLoS ONE 2009, 4, e6717. [Google Scholar] [CrossRef]
- Dias, A.G.; Rousseau, D.; Duizer, L.; Cockburn, M.; Chiu, W.; Nielsen, D.; El-Sohemy, A. Genetic variation in putative salt taste receptors and salt taste perception in humans. Chem. Senses 2013, 38, 137–145. [Google Scholar] [CrossRef]
- Green, B.G.; Dalton, P.; Cowart, B.; Shaffer, G.; Rankin, K.; Higgins, J. Evaluating the ‘labeled magnitude scale’ for measuring sensations of taste and smell. Chem. Senses 1996, 21, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Bartoshuk, L.M.; Duffy, V.; Green, B.G.; Hoffman, H.J.; Ko, C.W.; Lucchina, L.A.; Marks, L.E.; Snyder, D.J.; Weiffenbach, J.M. Valid across-group comparisons with labeled scales: The gLMS versus magnitude matching. Physiol. Behav. 2004, 82, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Ikeda, M.; Okuda, Y. Basis and practice of clinical taste examinations. Auris Nasus Larynx 1986, 13 (Suppl. S1), S1–S15. [Google Scholar] [CrossRef] [PubMed]
- E679-19; ASTM Standard Practice for Determination of Odor and Taste Threshold by a Forced Choice Method of Limits. ASTM International: West Conshohocken, PA, USA, 2019.
- Joseph, P.V.; Mennella, J.A.; Cowart, B.J.; Pepino, M.Y. Psychophysical tracking method to assess taste detection thresholds in children, adolescents, and adults: The taste detection threshold (TDT) test. J. Vis. Exp. 2021, 170, e62384. [Google Scholar]
- Watson, A.B.; Pelli, D.G. Quest: A Bayesian adaptive psychometric method. Percept. Psychophys. 1983, 33, 113–120. [Google Scholar] [CrossRef]
- King-Smith, P.E.; Grigsby, S.S.; Vingrys, A.J.; Benes, S.C.; Supowit, A. Efficient and unbiased modifications of the QUEST threshold method: Theory, simulations, experimental evaluation and practical implementation. Vis. Res. 1994, 34, 885–912. [Google Scholar] [CrossRef]
- Höchenberger, R.; Ohla, K. Repeatability of taste recognition threshold measurements with QUEST and Quick Yes-No. Nutrients 2020, 12, 24. [Google Scholar] [CrossRef]
- Hardikar, S.; Höchenberger, R.; Villringer, A.; Ohla, K. Higher sensitivity to sweet and salty taste in obese compared to lean individuals. Appetite 2017, 111, 158–165. [Google Scholar] [CrossRef]
- Höchenberger, R.; Ohla, K. Rapid estimation of gustatory sensitivity thresholds with SIAM and QUEST. Front. Psychol. 2017, 8, 981. [Google Scholar] [CrossRef]
- Green, D.M.; Swets, J.A. Signal Detection Theory and Psychophysics; Wiley: New York, NY, USA, 1966. [Google Scholar]
- Galindo-Cuspinera, V.; Waeber, T.; Antille, N.; Hartmann, C.; Stead, N.; Martin, N. Reliability of threshold and suprathreshold methods for taste phenotyping: Characterization with PROP and sodium chloride. Chem. Percept. 2009, 2, 214–228. [Google Scholar] [CrossRef]
- Höchenberger, R.; Ohla, K. Estimation of olfactory sensitivity using a Bayesian adaptive method. Nutrients 2019, 11, 1278. [Google Scholar] [CrossRef] [PubMed]
- Adler, E.; Hoon, M.A.; Mueller, K.L.; Chandrashekar, J.; Ryba, N.J.P.; Zuker, C.S. A novel family of mammalian taste receptors. Cell 2000, 100, 693–702. [Google Scholar] [CrossRef]
- Drayna, D. Human taste genetics. Annu. Rev. Genomics. Hum. Genet. 2005, 6, 217–235. [Google Scholar] [CrossRef] [PubMed]
- Kim, U.; Wooding, S.; Ricci, D.; Jorde, L.B.; Drayna, D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum. Mutat. 2005, 26, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Thomas, S.D.; Zhang, J.Z. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum. Mol. Genet. 2004, 13, 2671–2678. [Google Scholar] [CrossRef]
- Hayes, J.E.; Feeney, E.L.; Allen, A.L. Do polymorphisms in chemosensory genes matter for human ingestive behavior? Food Qual. Prefer. 2013, 30, 202–216. [Google Scholar] [CrossRef]
- Meyerhof, W.; Batram, C.; Kuhn, C.; Brockhoff, A.; Chudoba, E.; Bufe, B.; Appendino, G.; Behrens, M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses 2010, 35, 157–170. [Google Scholar] [CrossRef]
- Wooding, S.; Bufe, B.; Grassi, C.; Howard, M.T.; Stone, A.C.; Vazquez, M.; Dunn, D.M.; Meyerhof, W.; Weiss, R.B.; Bamshad, M.J. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature 2006, 440, 930–934. [Google Scholar] [CrossRef]
- Bufe, B.; Breslin, P.A.S.; Kuhn, C.; Reed, D.R.; Tharp, C.D.; Slack, J.P.; Kim, U.K.; Drayna, D.; Meyerhof, W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 2005, 15, 322–327. [Google Scholar] [CrossRef]
- Hayes, J.E.; Bartoshuk, L.M.; Kidd, J.R.; Duffy, V.B. Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem. Senses 2008, 33, 255–265. [Google Scholar] [CrossRef]
- Nolden, A.A.; McGeary, J.E.; Hayes, J.E. Predominant qualities evoked by quinine, sucrose, and capsaicin associate with PROP bitterness, but not TAS2R38 genotype. Chem. Senses 2020, 45, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Tepper, B.J. Nutritional implications of genetic taste variation: The role of PROP sensitivity and other taste phenotypes. Annu. Rev. Nutr. 2008, 28, 367–388. [Google Scholar] [CrossRef]
- Dinehart, M.E.; Hayes, J.E.; Bartoshuk, L.M.; Lanier, S.L.; Duffy, V.B. Bitter taste markers explain variability in vegetable sweetness, bitterness, and intake. Physiol. Behav. 2006, 87, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Duffy, V.B.; Bartoshuk, L.M. Food acceptance and genetic variation in taste. J. Am. Diet. Assoc. 2000, 100, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.E.; Duffy, V.B. Revisiting sugar-fat mixtures: Sweetness and creaminess vary with phenotypic markers of oral sensation. Chem. Senses 2007, 32, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, M.; Brainard, D.; Pelli, D.; Ingling, A.; Murray, R.; Broussard, C. What’s new in psychtoolbox-3? In ECVP abstract supplement. Perception 2007, 36, 14. [Google Scholar]
- Gordon, D.; Abajian, C.; Green, P. Consed: A graphical tool for sequence finishing. Genome Res. 1998, 8, 195–202. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 2nd ed.; Sage: Thousand Oaks, CA, USA, 2011. [Google Scholar]
- Boxer, E.E.; Garneau, N.L. Rare haplotypes of the gene TAS2R38 confer bitter taste sensitivity in humans. Springerplus 2015, 4, 505. [Google Scholar] [CrossRef]
- Risso, D.S.; Kozlitina, J.; Sainz, E.; Gutierrez, J.; Wooding, S.; Getachew, B.; Luiselli, D.; Berg, C.J.; Drayna, D. Genetic variation in the TAS2R38 bitter taste receptor and smoking behaviors. PLoS ONE 2016, 11, e0164157. [Google Scholar] [CrossRef]
- Risso, D.S.; Mezzavilla, M.; Pagani, L.; Robino, A.; Morini, G.; Tofanelli, S.; Carrai, M.; Campa, D.; Barale, R.; Caradonna, F.; et al. Global diversity in the TAS2R38 bitter taste receptor: Revisiting a classic evolutionary PROPosal. Sci. Rep. 2016, 6, 25506. [Google Scholar] [CrossRef]
- Yamaki, M.; Saito, H.; Isono, K.; Goto, T.; Shirakawa, H.; Shoji, N.; Satoh-Kuriwada, S.; Sasano, T.; Okada, R.; Kudoh, K.; et al. Genotyping analysis of bitter-taste receptor genes TAS2R38 and TAS2R46 in Japanese patients with gastrointestinal cancers. J. Nutr. Sci. Vitaminol. 2017, 63, 148–154. [Google Scholar] [CrossRef]
- Melis, M.; Atzori, E.; Cabras, S.; Zonza, A.; Calo, C.; Muroni, P.; Nieddu, M.; Padiglia, A.; Sogos, V.; Tepper, B.J.; et al. The gustin (CA6) gene polymorphism, rs2274333 (A/G), as a mechanistic link between PROP tasting and fungiform taste papilla density and maintenance. PLoS ONE 2013, 8, e74151. [Google Scholar] [CrossRef] [PubMed]
- Drewnowski, A.; Henderson, S.A.; Barratt-Fornell, A. Genetic taste markers and food preferences. Drug Metab. Dispos. 2001, 29, 535–538. [Google Scholar] [PubMed]
- Garcia-Bailo, B.; Toguri, C.; Eny, K.M.; El-Sohemy, A. Genetic variation in taste and its influence on food selection. OMICS—J. Integr. Biol. 2009, 13, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Gent, J.F.; Bartoshuk, L.M. Sweetness of sucrose, neohesperidin dihydrochalcone, and saccharin is related to genetic ability to taste the bitter substance 6-N-propylthiouracil. Chem. Senses 1983, 7, 265–272. [Google Scholar] [CrossRef]
- Keller, K.L.; Steinmann, L.; Nurse, R.J.; Tepper, B.J. Genetic taste sensitivity to 6-n-propylthiouracil influences food preference and reported intake in preschool children. Appetite 2002, 38, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Oter, B.; Ulukapi, I.; Ulukapi, H.; Topcuoglu, N.; Cildir, S. The relation between 6-n-propylthiouracil sensitivity and caries activity in schoolchildren. Caries Res. 2011, 45, 556–560. [Google Scholar] [CrossRef]
- Sacerdote, C.; Guarrera, S.; Smith, G.D.; Grioni, S.; Krogh, V.; Masala, G.; Mattiello, A.; Palli, D.; Panico, S.; Tumino, R.; et al. Lactase persistence and bitter taste response: Instrumental variables and Mendelian randomization in epidemiologic studies of dietary factors and cancer risk. Am. J. Epidemiol. 2007, 166, 576–581. [Google Scholar] [CrossRef]
- Turnbull, B.; Matisoo-Smith, E. Taste sensitivity to 6-n-propylthiouracil predicts acceptance of bitter-tasting spinach in 3-6-y-old children. Am. J. Clin. Nutr. 2002, 76, 1101–1105. [Google Scholar] [CrossRef]
- Wendell, S.; Wang, X.; Brown, M.; Cooper, M.E.; DeSensi, R.S.; Weyant, R.J.; Crout, R.; McNeil, D.W.; Marazita, M.L. Taste genes associated with dental caries. J. Dent. Res. 2010, 89, 1198–1202. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.E.; Wallace, M.R.; Knopik, V.S.; Herbstman, D.M.; Bartoshuk, L.M.; Duffy, V.B. Allelic variation in TAS2R bitter receptor genes associates with variation in sensations from and ingestive behaviors toward common bitter beverages in adults. Chem. Senses 2011, 36, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Ly, A.; Drewnowski, A. PROP (6-n-Propylthiouracil) tasting and sensory responses to caffeine, sucrose, neohesperidin dihydrochalcone and chocolate. Chem. Senses 2001, 26, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Horio, T. Relationship between PROP taste sensitivity and the preference for bitter food. Jpn. J. Nutr. Diet. 2009, 67, 15–19. [Google Scholar] [CrossRef]
- Duffy, V.B.; Hayes, J.E.; Davidson, A.C.; Kidd, J.R.; Kidd, K.K.; Bartoshuk, L.M. Vegetable intake in college-aged adults is explained by oral sensory phenotypes and TAS2R38 genotype. Chemosens. Percept. 2010, 3, 137–148. [Google Scholar] [CrossRef]
- Hoppu, U.; Laitinen, K.; Jaakkola, J.; Sandell, M. The hTAS2R38 genotype is associated with sugar and candy consumption in preschool boys. J. Hum. Nutr. Diet. 2015, 28, 45–51. [Google Scholar] [CrossRef]
- Sandell, M.; Hoppu, U.; Mikkila, V.; Mononen, N.; Kahonen, M.; Mannisto, S.; Ronnemaa, T.; Viikari, J.; Lehtimaki, T.; Raitakari, O.T. Genetic variation in the hTAS2R38 taste receptor and food consumption among Finnish adults. Genes. Nutr. 2014, 9, 433. [Google Scholar] [CrossRef]
- Sandell, M.A.; Breslin, P.A.S. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr. Biol. 2006, 16, R792–R794. [Google Scholar] [CrossRef]
- Ullrich, N.V.; Touger-Decker, R.; O’Sullivan-Maillet, J.; Tepper, B.J. PROP taster status and self-perceived food adventurousness influence food preferences. J. Am. Diet. Assoc. 2004, 104, 543–549. [Google Scholar] [CrossRef]
- Keller, M.; Liu, X.S.; Wohland, T.; Rohde, K.; Gast, M.T.; Stumvoll, M.; Kovacs, P.; Tonjes, A.; Bottcher, Y. TAS2R38 and its influence on smoking behavior and glucose homeostasis in the German sorbs. PLoS ONE 2013, 8, e80512. [Google Scholar] [CrossRef]
- Negri, R.; Smarrazzo, A.; Galatola, M.; Maio, A.; Iaccarino Idelson, P.; Sticco, M.; Biongiovanni, C.; Franzese, A.; Greco, L.; Risso, D.; et al. Age variation in bitter taste perception in relation to the Tas2r38 taste receptor phenotype. Int. J. Nutr. 2015, 1, 87–99. [Google Scholar] [CrossRef]
- Calò, C.; Padiglia, A.; Zonza, A.; Corrias, L.; Contu, P.; Tepper, B.J.; Barbarossa, I.T. Polymorphisrns in TAS2R38 and the taste bud trophic factor, gustin gene co-operate in modulating PROP taste phenotype. Physiol. Behav. 2011, 104, 1065–1071. [Google Scholar] [CrossRef] [PubMed]


| Nucleotide | Amino Acid | No. of Samples | Frequency | |||||
|---|---|---|---|---|---|---|---|---|
| Genotype | 145 | 785 | 886 | 49 | 262 | 296 | ||
| PAV/PAV | CC | CC | GG | Pro | Ala | Val | 37 | 0.314 |
| PAV/AVI | CG | CT | GA | P/A | A/V | V/I | 63 | 0.534 |
| AVI/AVI | GG | TT | AA | Ala | Val | Ile | 18 | 0.152 |
| Total | 118 | |||||||
| No. of Samples | Frequency | Sex/Age | Age | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Min | Max | M | SD | |||||||||||||||
| ~20 | ~30 | ~40 | ~50 | ~60 | 61~ | Total | ~20 | ~30 | ~40 | ~50 | ~60 | 61~ | Total | |||||||
| PAV/PAV | 24 | 0.304 | 1 | 6 | 3 | 3 | 1 | 0 | 14 | 0 | 2 | 3 | 1 | 4 | 0 | 10 | 20 | 60 | 37.33 | 11.94 |
| PAV/AVI | 43 | 0.544 | 0 | 5 | 5 | 9 | 6 | 1 | 26 | 0 | 4 | 3 | 6 | 3 | 1 | 17 | 21 | 65 | 41.74 | 12.00 |
| AVI/AVI | 12 | 0.152 | 0 | 2 | 1 | 1 | 2 | 0 | 6 | 0 | 0 | 3 | 2 | 1 | 0 | 6 | 22 | 60 | 41.42 | 12.15 |
| Total | 79 | 1.000 | 1 | 13 | 9 | 13 | 9 | 1 | 46 | 0 | 6 | 9 | 9 | 8 | 1 | 33 | 20 | 65 | 40.35 | 12.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aoki, K.; Mori, K.; Iijima, S.; Sakon, M.; Matsuura, N.; Kobayashi, T.; Takanashi, M.; Yoshimura, T.; Mori, N.; Katayama, T. Association between Genetic Variation in the TAS2R38 Bitter Taste Receptor and Propylthiouracil Bitter Taste Thresholds among Adults Living in Japan Using the Modified 2AFC Procedure with the Quest Method. Nutrients 2023, 15, 2415. https://doi.org/10.3390/nu15102415
Aoki K, Mori K, Iijima S, Sakon M, Matsuura N, Kobayashi T, Takanashi M, Yoshimura T, Mori N, Katayama T. Association between Genetic Variation in the TAS2R38 Bitter Taste Receptor and Propylthiouracil Bitter Taste Thresholds among Adults Living in Japan Using the Modified 2AFC Procedure with the Quest Method. Nutrients. 2023; 15(10):2415. https://doi.org/10.3390/nu15102415
Chicago/Turabian StyleAoki, Kyoko, Kanetaka Mori, Shohei Iijima, Masato Sakon, Nariaki Matsuura, Tsuneto Kobayashi, Masashi Takanashi, Takeshi Yoshimura, Norio Mori, and Taiichi Katayama. 2023. "Association between Genetic Variation in the TAS2R38 Bitter Taste Receptor and Propylthiouracil Bitter Taste Thresholds among Adults Living in Japan Using the Modified 2AFC Procedure with the Quest Method" Nutrients 15, no. 10: 2415. https://doi.org/10.3390/nu15102415
APA StyleAoki, K., Mori, K., Iijima, S., Sakon, M., Matsuura, N., Kobayashi, T., Takanashi, M., Yoshimura, T., Mori, N., & Katayama, T. (2023). Association between Genetic Variation in the TAS2R38 Bitter Taste Receptor and Propylthiouracil Bitter Taste Thresholds among Adults Living in Japan Using the Modified 2AFC Procedure with the Quest Method. Nutrients, 15(10), 2415. https://doi.org/10.3390/nu15102415







