Abstract
Diet has a significant impact on fecal microbiota, which in turn plays an important role in human health. To evaluate the impact of dietary habits on fecal microbiota, we investigated the fecal microbial composition in vegetarians and omnivores using 16S rRNA gene sequencing, and estimated the correlation between fecal microbiota, body mass and diet. The dietary data showed that vegetarians consumed more plant-based foods rich in dietary fiber, omnivores consumed more animal-based foods rich in fat and overweight and obese people consumed more high-energy foods. Compared to omnivores, vegetarians had greater richness and diversity in their fecal microbiota. The Firmicutes/Bacteroidetes ratio was lower and the Prevotella/Bacteroides ratio was higher in vegetarians. The meat intake correlated positively with the proportion of Bacteroides and negatively with the proportion of Prevotella. The composition and diversity in fecal microbiota in the normal weight group, overweight group and obesity group were similar to that of vegetarians and omnivores, respectively. This paper revealed the distinctive characteristics of fecal microbiota in vegetarians and omnivores. The omnivorous diet contained more fat, which reduced the fecal microbial diversity, and was more likely to lead to being overweight or obese.
1. Introduction
The human intestine is inhabited by a great variety of microorganisms, which has profound effects on many aspects of human health such as the immune system [1], inflammatory disease [2] and obesity [3]. Increasing studies on the composition and diversity in fecal microbiota have been carried out in recent years [4,5,6,7]. Studies have shown that several factors such as host genotype, birth mode, age, use of antibiotics and dietary habits might alter the fecal microbial composition, and lead to chronic inflammation or metabolic dysfunction [8,9]. Additionally, the effects of the dietary factor on fecal microbial composition were dominant [10,11]. Long-term, relatively fixed dietary habits made the fecal microbial composition tend to be balanced and stable. It has been widely recognized that the vegetarian diet is a kind of healthy and therapeutic feeding type as it regulates the fecal microbial composition. In addition, it has been demonstrated that fecal microbiota regulate host energy metabolism and body mass [12]. Increasing evidence has supported the notion that vegetarians have a lower body mass index, lower serum cholesterol levels and lower prevalence rates of diabetes, cardiometabolic and other chronic diseases compared to omnivores [13,14]. Detailed comparative studies on fecal microbial composition among humans with different dietary habits are still scarce. To fill this knowledge gap, this study investigated the fecal microbial composition in vegetarians and omnivores and identified the distinctive characteristics of fecal microbiota that correlated with obesity.
2. Materials and Methods
2.1. Study Participants
In the present study, a total of 121 participants were recruited voluntarily via advertisement and telephone in the area of Harbin, China. All of the participants provided written informed consent before participating in this study. The participants were categorized as vegetarians (V, n = 46) and omnivores (R, n = 75). Vegetarians exclude meat and fish but may consume milk and eggs. Inclusion criteria: participants were required to be between the ages of 25 and 45, without chronic, infectious or intestinal diseases such as diabetes, irritable bowel syndrome, cancer and neurodegenerative disease; none had received any antibiotic treatment within at least six months prior to the study. Exclusion criteria: participants who are pregnant, breastfeeding, smoking or drinking. Vegetarians or omnivorous were those who had had this dietary lifestyle for at least one year before the study [15,16].
The trained interviewer collected information about the participants’ dietary and anthropometric data. The types of dietary habits were distinguished based on the consumption of food items in the last year. Quantitative and qualitative data on dietary intake were assessed using a semi-quantitative food frequency questionnaire (SQFFQ) as previously described in our works [17]. The energy and nutrient intake of each participant was calculated based on the Chinese food composition tables [18]. Participants’ height was measured without shoes within 0.1 cm using a research-grade digital stadiometer (HT-DM40, Faenza, Italy), and weight was measured without shoes in light clothing within 0.1 kg using an electronic scale (Yolanda-CS10A, Shenzhen, China). The nutritional status of all of the participants was checked based on their body mass index (BMI): underweight, BMI < 18.5; normal weight, 18.5 ≤ BMI < 24; overweight, 24 ≤ BMI < 28 or obese, BMI ≥ 28 [19]. BMI was calculated using the following formula: BMI = weight (kg)/height (m)2.
2.2. Fecal Sample Collection and DNA Extraction
The fecal samples of each participant were collected in sterile feces collection tubes with DNA stabilizer and stored at −80 °C. Microbial DNA was extracted from fecal samples using the E.Z.N.A. Stool DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s protocols. The 16S rDNA V3-V4 region of the Eukaryotic ribosomal RNA gene was amplified via PCR (95 °C for 2 min, followed by 27 cycles at 98 °C for 10 s, 62 °C for 30 s and 68 °C for 30 s and a final extension at 68 °C for 10 min) using bacterial primers 341F: CCTACGGGNGGCWGCAG and 806R: GGACTACHVGGGTATCTAAT, where the barcodes are eight-base sequences unique to each sample [20]. PCR reactions were performed in triplicate. A 50.0 μL mixture containing 5.0 μL of 10 × KOD Buffer, 5.0 μL of 2.5 mM dNTPs, 1.5 μL of each primer (5.0 μM), 1.0 μL of KOD Polymerase and 100.0 ng of template DNA was used [21].
2.3. Analysis of 16S rRNA Sequences
Amplicons were extracted from 2% agarose gels, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions and quantified using Quanti Fluor-ST (Promega, Madison, WI, USA), and purified amplicons were pooled in equimolar and paired-end sequenced (2 × 250) using an Illumina platform according to the standard protocols [16].
The raw data containing adapters or low-quality reads would affect the following reads’ assembly and analysis. Thus, to obtain high-quality clean reads, raw reads were further filtered according to the following rules: (1) removing reads containing more than 10% of unknown nucleotides (N); (2) removing reads containing less than 80% of bases with quality (Q-value) > 20. Paired-end clean reads were merged as raw tags using FLASH [22] (v 1.2.11) with a minimum overlap of 10 bp and mismatch error rates of 2%. Noisy sequences of raw tags were filtered using the QIIMF [23] (V1.9.1) pipeline under specific filtering conditions [24] to obtain high-quality clean tags. Clean tags were searched against the reference database [http://drive5.com/uchime/uchime_download.html (accessed on 12 August 2019)] to perform reference-based chimera checking using the UCHIME algorithm [http://www.drive5.com/usearch/manual/uchime_algo.html (accessed on 12 August 2019)]. All chimeric tags were removed and finally we obtained effective tags for further analysis. The effective tags were clustered into operational taxonomic units (OTUs) of ≥97% similarity using the UPARSE [25] pipeline.
2.4. Statistical Analysis
Statistical analysis was carried out using R [26] (version 3.5.0) software packages and in-house scripts. p < 0.05 was defined as a significant statistical difference. The age, height, weight and BMI data were represented as mean and standard deviation (mean ± SD).
The tag sequence with the highest abundance was selected as a reprehensive sequence within each cluster. Venn analysis between groups was performed in R to identify unique and common OTUs. The representative sequences were classified into organisms using a naive Bayesian model and an RDP classifier [27] (Version 2.2) based on the SILVA [28] database [https://www.arb-silva.de/ (accessed on 17 Aug 2019)]. The abundance statistics of each taxonomy and a phylogenetic tree were constructed in a Perl script and visualized using SVG. Chao1, Simpson and all other alpha diversity indexes were calculated in QIIME [29]. Statistical analyses of alpha diversity indexes between groups were calculated using Welch’s t-test and a Wilcoxon rank-sum test in R. Weighted and unweighted UniFrac distance matrixes were generated using QIIME. Multivariate statistical analyses, including principal component analysis (PCA), principal coordinates analysis (PCoA) and non-metric multidimensional scaling (NMDS) of (un)weighted UniFrac distances matrix, were conducted and plotted in R, as previously described [30].
3. Results
3.1. Characterization of Subjects
Twenty-one pieces of participants’ data were excluded from dietary analysis due to unqualified SQFFQ, and ultimately 100 participants (36 vegetarians and 64 omnivores) underwent dietary analysis and fecal microbiome analysis. According to the participants’ nutritional status, vegetarians were divided into a normal weight group (VN) and overweight group (VO); omnivores were divided into a normal weight group (RN), overweight group (RO) and obese group (RC).
Table 1 shows the characteristics of the 100 subjects. There were 48 males and 52 females. The average age, height, weight and BMI were 32.8 ± 4.8 years, 167.0 ± 7.4 cm, 65.1 ± 5.9 kg and 23.1 ± 3.1 kg/m2, respectively. The average BMI of omnivores was just numerically greater than that of the vegetarians. Similarly, the average age, height and weight were just numerically different between vegetarians and omnivores.

Table 1.
Characterization of subjects.
3.2. Dietary Profiles
The dietary habits and food intake of participants were surveyed using the SQFFQ. The survey results indicated that there was a significant difference in food intake between vegetarians and omnivores (Figure 1A). Compared to omnivores, vegetarians have significantly higher intakes of coarse cereals, vegetables and fruits, while they have significantly lower intakes of rice, meat and fish (p < 0.05).
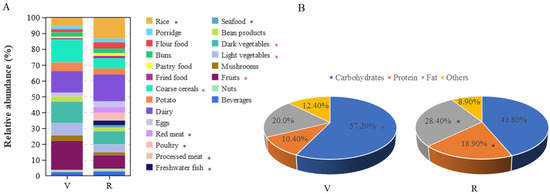
Figure 1.
Food intake and macronutrient energy supply ratio of vegetarians (V) and omnivores (R). (A) Relative content of food intake in vegetarians and omnivores; (B) energy supply ratio of macronutrients in vegetarians and omnivores; red asterisks represent being significantly higher in vegetarians; black asterisks represent being significantly higher in omnivores.
The difference in macronutrient energy supply ratio between vegetarians and omnivores is shown in Figure 1B. The energy supply ratio of fat (28.4% vs. 20.0%) and protein (18.9% vs. 10.4%) in omnivores was significantly higher than that of vegetarians (p < 0.05), while the energy supply ratio of carbohydrates (43.8% vs. 57.2%) was significantly lower than that of vegetarians (p < 0.05). Compared with the dietary guidelines for Chinese residents [31], the energy supply ratios of carbohydrates, fat and protein in vegetarians were within the recommended range, while the energy supply ratios of fat and protein in omnivores were within the recommended range, and the proportion was large, but the energy supply ratio of carbohydrates was far lower than the recommended range.
3.3. Analysis of Fecal Microbial Composition in Vegetarians and Omnivores
The gene sequencing results showed that a total of 7,824,057 reads were obtained from 100 fecal samples: 2,844,103 reads belonged to vegetarians with a mean value of 79,003 reads per sample, whereas 4,979,954 reads were from omnivores with a mean value of 77,812 reads per sample. From these reads, we identified an overall total of 769 OTUs, and of which 465 OTUs were common to all groups, while 179 and 125 OTUs were unique to vegetarians and omnivores, respectively. The identified OTUs were grouped into twelve phyla including Firmicute, Bacteroidetes, Proteobacteria, Actinobacteria, Verrucomicrobia, Fusobacteria, Cyanobacteria, Synergistetes, Saccharibacteria, Euryarchaeota, Lentisphaerae and Tenericutes (Figure 2A).
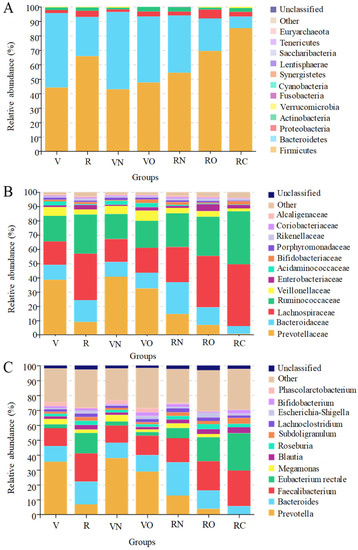
Figure 2.
Composition of fecal microbiota in vegetarians and omnivores. (A) Phylum level; (B) family level; (C) genus level. V represents vegetarians; R represents omnivores; VN and VO represent normal weight group and overweight group for vegetarians, respectively; RN, RO and RC represent normal weight group, overweight group and obesity group in omnivores, respectively.
The relative abundance of Firmicutes and Bacteroidetes was the highest in the fecal samples of vegetarians and omnivores followed by Proteobacteria and Actinobacteria with >1% relative abundance, and the relative abundance sum of the four phyla accounted for more than 99.47% of the total bacteria. Each taxon was tested for differential abundance between vegetarians and omnivores. From the perspective of dietary habits, the abundance of Actinobacteria, Firmicute and Proteobacteria in vegetarians was significantly lower than that in omnivores (p < 0.05), and the abundance of Bacteroides was significantly higher than that in omnivores (p < 0.05). From the perspective of body mass index, in vegetarians, the abundance of Firmicute, Proteobacteria and Actinobacteria of the normal weight group (VN) was significantly lower than that of the overweight group (VO) (p < 0.05), while the abundance of Bacteroides and Verrucomicrobia in the normal weight group (VN) was significantly higher than that of the overweight group (VO) (p < 0.05); in omnivores, the abundance of Firmicute, Proteobacteria, Actinobacteria and Verrucomicrobia of the normal weight group (RN) was significantly lower than that of the overweight group (RO) and the obesity group (RC) (p < 0.05), while the abundance of Bacteroides and Fusobacteria in the normal weight group (RN) was significantly higher than that of the overweight group (RO) and obesity group (RC) (p < 0.05). The abundance of Firmicute increased with increasing weight, while the abundance of Bacteroides decreased with increasing weight, leading to an increased Firmicutes/Bacteroides ratio with increased weight. The Firmicutes/Bacteroides ratio (0.86 vs. 2.43) in the vegetarians was smaller than that in the omnivores, and the results indicated a significant difference in the fecal microbial composition between vegetarians and omnivores.
A total of 46 families of bacteria were detected in the fecal samples of vegetarians and omnivores. Of these families, those with relative abundance greater than 1% in all of the groups included Ruminococcaceae, Lachnospiraceae, Bacteroidaceae, Veillonellaceae, Acidaminococcaceae, Porphyromonadaceae, Enterobacteriaceae, Rikenellaceae, Bifidobacteriaceae, Prevotellaceae, Coriobacteriaceae and Alcaligenaceae (Figure 2B). The sum of the relative abundance of Prevotellaceae, Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, Veillonellaceae and Enterobacteriaceae accounts for more than 90.65% of the total bacteria; these form the most dominant bacteria family in vegetarians and omnivores.
At the family level, there was more discrepancy in fecal microbial composition between vegetarians and omnivores. The abundance of Prevotellaceae, Veillonellaceae and Acidaminococcaceae in vegetarians was significantly higher compared with omnivores (p < 0.05), while the abundance of Bacteroidaceae, Ruminococcaceae, Lachnospiraceae, Enterobacteriaceae and Rikenellaceae was significantly lower compared with omnivores (p < 0.05).
From the perspective of body mass index, in vegetarians, the abundance of Prevotellaceae, Porphyromonadaceae and Rikenellaceae in the normal weight group (VN) was significantly higher compared with the overweight group (VO) (p < 0.05), and the abundance of Ruminococcaceae, Lachnospiraceae and Veillonellaceae in the normal weight group (VN) was significantly lower compared with the overweight group (VO) (p < 0.05); in omnivores, the abundance of Prevotellaceae, Bacteroidaceae, Veillonellaceae, Acidaminococcaceae, Porphyromonadaceae, Coriobacteriaceae and Veillonellaceae in the normal weight group (RN) was significantly higher compared with the overweight group (RO) and obesity group (RC) (p < 0.05), whilst the abundance of Ruminococcaceae, Lachnospiraceae, Enterobacteriaceae and Rikenellaceae in the normal weight group (RN) was significantly lower compared with the overweight group (RO) and obesity group (RC) (p < 0.05).
The genera with a high abundance (>1%) detected in all of the groups were shown in Figure 2C, including Prevotella, Faecalibacterium, Bacteroides, Eubacterium rectale, Megamonas, Blautia, Roseburia, Subdoligranulum, Lachnoclostridium, Escherichia-Shigella, Bifidobacterium and Phascolarctobacterium. Those with the highest abundance of 35.46% and 18.91% found in vegetarians and omnivores were Prevotella and Faecalibacterium, respectively. The abundance of Prevotella, Megamonas and Phascolarctobacterium in vegetarians was significantly higher than that in omnivores (p < 0.05), whereas the abundance of Faecalibacterium, Lachnoclostridium, Bacteroides, Eubacterium rectale, Blautia, Roseburia, Subdoligranulum and Escherichia-Shigella in vegetarians was significantly lower than that of omnivores (p < 0.05).
From the perspective of body mass index, in vegetarians, the abundance of Prevotella and Megamonas in the normal weight group (VN) was significantly higher than that of the overweight group (VO) (p < 0.05), and the abundance of Faecalibacterium and Lachnoclostridium in the normal weight group (VN) was significantly lower than that of the overweight group (VO) (p < 0.05); in omnivores, the abundance of Prevotella, Megamonas, Bacteroides and Phascolarctobacterium in the normal weight group (RN) was significantly higher compared with the overweight group (RO) and obesity group (RC) (p < 0.05), whilst the abundance of Faecalibacterium, Eubacterium rectale, Blautia and Roseburia in the normal weight group (RN) was significantly lower compared with the overweight group (RO) and obesity group (RC) (p < 0.05).
3.4. Analysis of Fecal Microbial Diversity in Vegetarians and Omnivores
To evaluate the overall differences in fecal microbial composition between vegetarians and omnivores, we calculated the alpha diversity indexes (Chao1, Ace, Shannon and Simpson). Compared to omnivores, the higher alpha diversity indexes were obtained for vegetarians (Table 2), and with an increase in BMI, the alpha diversity indexes decreased in vegetarians and omnivores, indicating that the alpha diversity in vegetarians was higher than that in omnivores and that the alpha diversity in the normal weight groups was higher compared with the overweight groups and obesity group.

Table 2.
Comparative analysis of alpha diversity index for each group.
To show the differences between microbial community structures in vegetarians and omnivores, we calculated the beta diversity parameter, including the unweighted and weighted UniFrac distance matrix (PCA, PCoA and NMDS). Due to the similar trend of all of the analysis results, only the results of the PCoA are shown in Figure 3. Figure 3A,B shows that the first coordinate (PCo1) explains 71.03% of the inter-sample variance (p < 0.05), while Figure 3C,D explains the 18.81% in vegetarians versus omnivores (p < 0.05). In subgroups based on BMI, we found that the samples in the same groups were gathered, the samples among groups were well differentiated and that there was a clear boundary between two vegetarian groups (VN and VO) and three omnivorous groups (RN, RO and RC) (p = 0.01; Figure 3B,D).
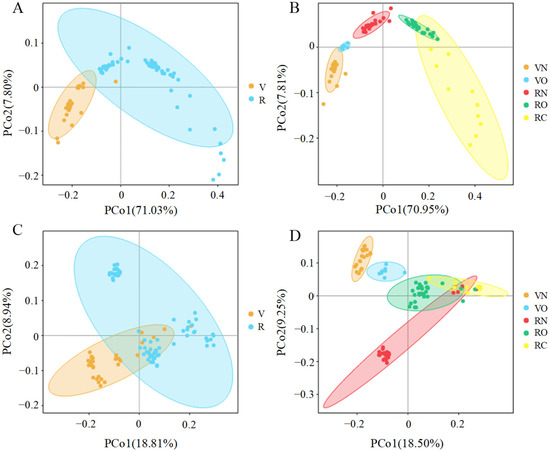
Figure 3.
Principal coordinates analysis (PCoA) plots of fecal microbiota in vegetarians (V) and omnivores (O). The plots show the first two principal coordinates axes using a weighted (A,B) and unweighted (C,D) UniFrac distance matrix. VN and VO represent the normal weight group and overweight group in vegetarians, respectively; RN, RO and RC represent the normal weight group, overweight group and obesity group in omnivores, respectively.
3.5. Correlation between Food and the Fecal Microbial Community
To investigate the correlation between food intake and fecal microbiota, we conducted a redundancy analysis (RDA) on vegetarians and omnivores. Variation in fecal microbiota correlated significantly with individual food intake variation (Figure 4). The result showed that omnivores consumed more rice and animal-based foods, and vegetarians consumed more coarse food and plant-based foods. Compared to the normal weight group, daily fat intake was significantly higher in the overweight group and obesity group, the intake of plant-based foods rich in dietary fiber correlated positively with the genera Prevotella, Megamonas and Phascolarctobacterium and the intake of animal-based foods rich in fat and protein correlated positively with the genera Faecalibacterium, Eubacterium rectale, Blautia and Escherichia-Shigella. The correlation between altered fecal microbiota and high BMI suggested that high-fat-diet-associated obesity was present among the omnivores.
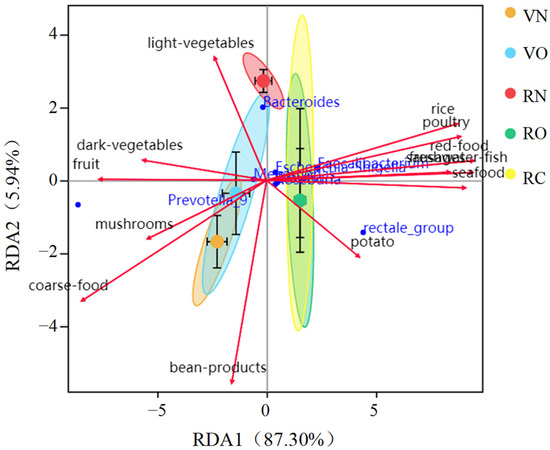
Figure 4.
Redundancy analysis (RDA) of the correlation between food intake and fecal microbiota. VN and VO represent the normal weight group and overweight group in vegetarians, respectively; RN, RO and RC represent the normal weight group, overweight group and obesity group in omnivores, respectively.
4. Discussion
The interaction between diet and fecal microbiota has been increasingly studied [32,33]. Thus, the beneficial use of diet to regulate the fecal microbial composition to improve human health has proven to be an effective nutritional treatment method [34]. Here, we investigated the distinctive influence of vegetarian diet and omnivorous diet on the fecal microbial composition.
The diversity in fecal microbiota reflected the stability of the microecology and the ability to resist the invasion of external pathogenic bacteria. Low diversity did not necessarily mean illness, but it meant one was more susceptible to factors such as diet, environment or disease. Higher diversity often led to healthier physical conditions [35]. A significant difference in the alpha diversity index between vegetarians and omnivores was found, with vegetarians displaying greater richness. This was related to the fact that vegetarians mainly consumed plant-based foods. A plant-based diet contains rich dietary fiber, which is the main nutrient source for fecal microbiota. The number of fecal microbiota increased due to the increase in nutrients, which in turn increased the abundance of fecal microbiota [35]. Additionally, richer fecal microbiota is advantageous to the host since greater taxonomic richness might also mean greater functional diversity. The current study found that the alpha diversity in the fecal microbiota in the normal weight group was higher compared with the overweight group and obesity group. The present results correspond to the proposals of Chen et al. [36] and Liu et al. [37]. The beta diversity in the fecal microbiota reflected the microbial overall composition and richness. The more similar the dietary structure of the study subjects, the more similar the composition of the fecal microbiota, and the closer the spatial distance of the samples. Thus, in the present research, the dietary habits of vegetarians and omnivores were different, leading to a significant difference in the fecal microbial composition. These results are congruent with the study of De Filippo et al. [38].
Vegetarians were reported to have lower frequencies of obesity, hypertension, diabetes and cardiovascular disease [39,40]. Vegetarians consumed more plant-based foods that were rich in dietary fiber and micronutrients, such as coarse grains, vegetables and fruits, while they ate fewer animal-based foods rich in fat such as meat, poultry and fish. Increasing evidence has shown that a vegetarian diet is beneficial to reduce oxidative stress. Additionally, a large amount of dietary fiber could stimulate fecal microbiota metabolism to produce more short-chain fatty acids (SCFAs), mainly acetate, propionate and butyrate. Diet provided a variety of nutrients and energy for the growth and reproduction of fecal microbiota. In turn, this caused differences in the fecal microbial composition. This study found that the fecal microbial composition of vegetarians was more favorable compared with omnivores with fewer Firmicutes and more Bacteroidetes, similar to the results of Wu et al. [41]. Up-to-date knowledge has suggested that a high abundance of Firmicutes leads to an imbalance of fecal microbial composition, which can cause metabolic dysfunction and induce obesity, hypertension, diabetes and other metabolic diseases [42]. The current research found a higher abundance of Firmicutes and a lower abundance of Bacteroidetes in omnivores compared with vegetarians, which might be attributed to the differences in body mass index. Evidence from animal and human studies has demonstrated that obesity is related to an increased Firmicutes/Bacteroidetes ratio [43,44]. A study reported by Ley et al. [45] showed that a lower Firmicutes/Bacteroidetes ratio was associated with the lean phenotype, which was generally considered to be beneficial for health. It was suggested that the content and proportion of Firmicutes and Bacteroidetes could be used as characteristics to distinguish between the microbial communities in lean and obese people [46]. Firmicutes and Bacteroidetes were the two more abundant bacteria in the intestine. Bacteroides has a high degree of functional redundancy, while Firmicutes was composed of core bacteria with multiple metabolic functions. The fecal microbiota in obesity could increase energy gain from food [47]. The metabolite of Bacteroides was mainly propionate, which was absorbed by the colon and transported to the liver as the substrate of gluconeogenesis to maintain energy balance, inhibited the synthesis of fat and cholesterol and had a lipid-lowering effect. Analyzing particularly the Firmicutes subpopulations, the results of the present study found an increase in the abundance of two acetate-producing bacteria—Faecalibacterium and Eubacterium rectale in omnivores and the obesity group in comparison with vegetarians and the normal weight group. Acetate was a major energy source for the body cells and stimulated the expression of adipocyte differentiation factors, contributing to adipocyte proliferation and fat deposition [48]. The abundance of Bacteroidetes mainly composed of the genera Prevotella and Bacteroides which were higher in vegetarians than in omnivores. Both Prevotella and Bacteroides were commonly presented in the human intestine. Our study observed that there was a higher Prevotella/Bacteroides ratio in vegetarians and the normal weight group than in omnivores and the obesity group. Likewise, several studies have suggested that there is a higher abundance of Prevotella in individuals who consume plant-based food and a predominance of Bacteroides in individuals who consume animal-based food [49,50]. This result might be due to colonic fermentation, which could inhibit some fecal microbiota. The reason for this might be that dietary fiber from plant-based food is fermented by fecal microbiota to produce more short-chain fatty acids, which in turn leads to a decrease in pH from 6.5 to 5.5. Bacteroides did not grow well under pH 5.5 conditions [51]. This might also be the reason why there was a lower abundance of Bacteroides in vegetarians and the normal weight group than in omnivores and the obesity group. Other studies have shown that legumes in a vegetarian diet can increase the proportion of Megamonas, Bifidobacterium and Phascolarctobacterium, but reduce the proportion of Bacteroides [52]. Phascolarctobacterium focus on utilizing succinate salts produced by Bacteroides, and the proportion of Bacteroides increased due to a high-fat diet and was positively correlated with body weight. Normal-weight individuals had a higher abundance of Phascolarctobacterium in their intestines, making it an indicator for predicting obesity [53]. Species related to obesity, such as Faecalibacterium, Bacteroides, Eubacterium rectale, Brucella, Roseburia and Escherichia-Shigella, have been increasingly confirmed in research. The ability of Faecalibacterium to utilize carbohydrates from dietary sources was quite limited, and it could not grow on starch or hemicellulose, while a high saturated fat diet could increase the proportion of Faecalibacterium. Brucella and Roseburia have an extraordinary ability to utilize carbon dioxide and hydrogen or formic acid to produce acetate, which is related to adipocyte differentiation and fat deposition. The diversity in fecal microbiota in obese individuals decreased, intestinal integrity was disrupted, intestinal permeability was enhanced, harmful substances such as endotoxins were absorbed more and the body’s ability to obtain energy from the diet was increased.
In this study, we found a significant difference in the fecal microbial composition between vegetarians and omnivores and a correlation between obesity and fecal microbiota. This study has limitations due to a relatively small sample size and a lack of in-depth biological and biochemical information, not allowing it to address the complex physiological link between diet, fecal microbiota and phenotypes.
5. Conclusions
The current research revealed the distinctive characteristics in fecal microbial diversity and composition in vegetarians and omnivores. A vegetarian diet with high fiber might increase fecal microbial diversity. An omnivorous diet containing more fat and calories might reduce fecal microbial diversity, and is likely to lead to being overweight or obese. This study provides a new theoretical basis for future research into a dietary intervention to regulate the balance of fecal microbial composition and the obesity phenotypes, and the development of precision nutrition.
Author Contributions
Conceptualization, C.S. and J.H.; methodology, C.S.; software, A.L.; validation, C.X., J.M. and H.W.; formal analysis, A.L.; investigation, C.S.; data curation, C.X., J.M. and H.W.; writing—original draft preparation, C.S.; writing—review and editing, Z.J.; visualization, C.S.; project administration, J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key Projects of the Natural Science Foundation of Heilongjiang Province, grant number ZD2021C007, and the National Key Research and Development Program of China, grant number 2016YFD0400605.
Institutional Review Board Statement
This study was approved by the Research Ethics Committee of Northeast Agricultural University (NEAU-1347, 12 June 2017).
Informed Consent Statement
Written informed consent was obtained from all participants before participating in this study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.
Acknowledgments
We thank all of the participants for their participation in this study, and we thank all of the authors for their contributions to the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Akagbosu, B.; Tayyebi, Z.; Shibu, G.; Paucar Iza, Y.A.; Deep, D.; Parisotto, Y.F.; Fisher, L.; Pasolli, H.A.; Thevin, V.; Elmentaite, R.; et al. Novel antigen-presenting cell imparts T-dependent tolerance to gut microbiota. Nature 2022, 610, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Federici, S.; Kredo-Russo, S.; Valdés-Mas, R.; Kviatcovsky, D.; Weinstock, E.; Matiuhin, Y.; Silberberg, Y.; Atarashi, K.; Furuichi, M.; Oka, X.; et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 2022, 185, 2879–2898. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Usyk, M.; Vázquez-Baeza, Y.; Chen, G.C.; Isasi, C.R.; Williams-Nguyen, J.S.; Hua, S.; McDonald, D.; Thyagarajan, B.; Daviglus, M.L.; et al. Microbial co-occurrence complicates associations of gut microbiome with US immigration, dietary intake and obesity. Genome Biol. 2021, 22, 336. [Google Scholar] [CrossRef] [PubMed]
- Davenport, E.R.; Sanders, J.G.; Song, S.J.; Amato, K.R.; Clark, A.G.; Knight, R. The human microbiome in evolution. BMC Biol. 2017, 15, 127. [Google Scholar] [CrossRef]
- Amamoto, R.; Shimamoto, K.; Suwa, T.; Park, S.; Matsumoto, H.; Shimizu, K.; Katto, M.; Makino, H.; Matsubara, S.; Aoyagi, Y. Relationships between dietary diversity and gut microbial diversity in the elderly. Benef. Microbes 2022, 13, 453–464. [Google Scholar] [CrossRef]
- Thingholm, L.B.; Ruehlemann, M.C.; Koch, M.; Fuqua, B.; Laucke, G.; Boehm, R.; Bang, C.; Franzosa, E.A.; Huebenthal, M.; Rahnavard, A. Obese Individuals with and without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe 2019, 26, 252–275. [Google Scholar] [CrossRef]
- Kesika, P.; Suganthy, N.; Sivamaruthi, B.S.; Chaiyasut, C. Role of gut-brain axis, gut microbial composition, and probiotic intervention in Alzheimer’s disease. Life Sci. 2020, 264, 118627. [Google Scholar] [CrossRef]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef]
- Šumilo, D.; Brocklehurst, P. What is the relationship between mode of birth, antibiotics, and childhood health? BMJ (Clin. Res. Ed.) 2022, 377, O1526. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- De Filippis, F.; Pasolli, E.; Tett, A.; Tarallo, S.; Naccarati, A.; De Angelis, M.; Neviani, E.; Cocolin, L.; Gobbetti, M.; Segata, N. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe 2019, 25, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.F.; Murphy, E.F.; Nilaweera, K.; Ross, P.R.; Shanahan, F.; O’Toole, P.W.; Cotter, P.D. The gut microbiota and its relationship to diet and obesity: New insights. Gut Microbes 2012, 3, 186–202. [Google Scholar] [CrossRef] [PubMed]
- Medawar, E.; Huhn, S.; Villringer, A.; Witte, A.V. The effects of plant-based diets on the body and the brain: A systematic review. Transl. Psychiatr. 2019, 9, 226. [Google Scholar] [CrossRef]
- Mohammed, A. Prevalence of vegan/vegetarian diet and eating behavior among Saudi adults and its correlation with body mass index: A cross-sectional study. Front. Nutr. 2022, 9, 966629. [Google Scholar]
- Xiao, W.; Zhang, Q.; Yu, L.; Tian, F.; Chen, W.; Zhai, Q. Effects of vegetarian diet-associated nutrients on gut microbiota and intestinal physiology. Food Sci. Hum. Wellness 2022, 11, 208–217. [Google Scholar] [CrossRef]
- Beisner, J.; Gonzalez-Granda, A.; Basrai, M.; Damms-Machado, A.; Bischoff, S.C. Fructose-Induced Intestinal Microbiota Shift Following Two Types of Short-Term High-Fructose Dietary Phases. Nutrients 2020, 12, 3444. [Google Scholar] [CrossRef]
- Sun, C.; Wang, Q.; Xu, C.; Wang, W.; Ma, J.; Gu, L.; Liu, Z.; Hou, J.; Jiang, Z. Reproducibility and Validity of a Semi-Quantitative Food Frequency Questionnaire for Assessing Dietary Intake of Vegetarians and Omnivores in Harbin, China. Nutrients 2022, 14, 3975. [Google Scholar] [CrossRef]
- Yang, Y. Chinese Food Composition Table, 6th ed.; Peking University Medical Press: Beijing, China, 2019. [Google Scholar]
- Xiao, L.; Zhou, J.; Galling, B.; Chen, R.S.; Wang, G. The association of body mass index (BMI) with treatment outcomes in patients with major depressive disorder. J. Affect. Disord. 2021, 281, 799–804. [Google Scholar] [CrossRef]
- Federica, D.C.; Francesca, A.; Alessandra, R.; Andrea, Q.; Sofia, R.; Danila, C.; Romina, C.; Stefano, G.C.; Valerio, N.; Francesco, D.P.; et al. Gut Microbiota Markers in Obese Adolescent and Adult Patients: Age-Dependent Differential Patterns. Front. Microbiol. 2018, 9, 1210. [Google Scholar]
- Choong-Kyun, N.; Sun, K.B.; Gana, H.; Youn, C.J.; Jae, L.K. Effects of the Administration of Probiotics on Fecal Microbiota Diversity and Composition in Healthy Individuals. J. Neurogastroenterol. Motil. 2018, 24, 452–459. [Google Scholar]
- Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Team, R.D.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.G.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef]
- Leong, C.; Haszard, J.J.; Heath, A.M.; Tannock, G.W.; Lawley, B.; Cameron, S.L.; Szymlek-Gay, E.A.; Gray, S.R.; Taylor, B.J.; Galland, B.C.; et al. Using compositional principal component analysis to describe children’s gut microbiota in relation to diet and body composition. Am. J. Clin. Nutr. 2020, 111, 70–78. [Google Scholar] [CrossRef]
- Nutrition Society, T.C. Dietary Guidelines for Chinese Residents (2022); People’s Medical Publishing House: Beijing, China, 2022. [Google Scholar]
- De, F.F.; Pellegrini, N.; Vannini, N.; Jeffery, I.B.; La, S.A.; Laghi, L.; Serrazanetti, D.I.; Di, C.R.; Ferrocino, I. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar]
- Simpson, R.C.; Shanahan, E.R.; Batten, M.; Reijers, I.L.M.; Read, M.; Silva, I.P.; Versluis, J.M.; Ribeiro, R.; Angelatos, A.S.; Tan, J.; et al. Diet-driven microbial ecology underpins associations between cancer immunotherapy outcomes and the gut microbiome. Nat. Med. 2022, 28, 2344–2352. [Google Scholar] [CrossRef]
- Arifuzzaman, M.; Won, T.H.; Li, T.T.; Yano, H.; Digumarthi, S.; Heras, A.F.; Zhang, W.; Parkhurst, C.N.; Kashyap, S.; Jin, W.B.; et al. Inulin fiber promotes microbiota-derived bile acids and type 2 inflammation. Nature 2022, 611, 578–584. [Google Scholar] [CrossRef]
- Caio, G.; Lungaro, L.; Segata, N.; Guarino, M.; Zoli, G.; Volta, U.; De Giorgio, R. Effect of Gluten-Free Diet on Gut Microbiota Composition in Patients with Celiac Disease and Non-Celiac Gluten/Wheat Sensitivity. Nutrients 2020, 12, 1832–1855. [Google Scholar] [CrossRef]
- Chen, X.; Sun, H.; Jiang, F.; Shen, Y.; Wei, P. Alteration of the gut microbiota associated with childhood obesity by 16S rRNA gene sequencing. PeerJ 2020, 8, e8317. [Google Scholar] [CrossRef]
- Liu, Y.; Tan, Y.F.; Zhang, J.Y.; Yu, M.; Li, M.W.; Sun, F.; Zhang, Y.; Qi, W. Difference of gut microbiota in people with overweight/obese and normal weight. Chin. J. Clin. Res. 2022, 35, 21–24. [Google Scholar]
- De, F.C.; Cavalieri, D.; Paola, M.D.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar]
- Orlich, M.J.; Singh, P.N.; Sabaté, J.; Jaceldo-Siegl, K.; Fan, J.; Knutsen, S.; Beeson, W.L.; Fraser, G.E. Vegetarian Dietary Patterns and Mortality in Adventist Health Study 2. JAMA Intern. Med. 2013, 173, 1230–1238. [Google Scholar] [CrossRef]
- Tharrey, M.; Mariotti, F.; Mashchak, A.; Barbillon, P.; Delattre, M.; Fraser, G.E. Patterns of plant and animal protein intake are strongly associated with cardiovascular mortality: The Adventist Health Study-2 cohort. Int. J. Epidemiol. 2018, 47, 1603–1612. [Google Scholar] [CrossRef]
- Wu, G.D.; Compher, C.; Chen, E.Z.; Smith, S.A.; Shah, R.D.; Bittinger, K.; Chehoud, C.; Albenberg, L.G.; Nessel, L.; Gilroy, E.; et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 2016, 65, 63–72. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Falalyeyeva, T.; Chornenka, N.; Cherkasova, L.; Tsyryuk, O.; Kobyliak, N. Gut Microbiota Interactions with Obesity. Ref. Modul. Food Sci. 2022, 147, 112678. [Google Scholar]
- Cao, S.Y.; Zhao, C.N.; Xu, X.Y.; Tang, G.Y.; Li, H.B. Dietary plants, gut microbiota, and obesity: Effects and mechanisms. Trends Food Sci. Technol. 2019, 92, 194–204. [Google Scholar] [CrossRef]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Grigor’Eva, I.N. Gallstone Disease, Obesity and the Firmicutes/Bacteroidetes Ratio as a Possible Biomarker of Gut Dysbiosis. J. Pers. Med. 2020, 11, 13. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Niu, J.; Cui, M.; Yang, Y.; Li, J.; Yao, Y.; Guo, C.; Lu, A.; Qi, X.; Zhou, D.; Zhang, C.; et al. Microbiota-derived acetate enhances host antiviral response via NLRP3. Nat. Commun. 2023, 14, 642. [Google Scholar] [CrossRef]
- Ilario, F.; Raffaella, D.C.; Maria, D.A.; Silvia, T. Fecal Microbiota in Healthy Subjects Following Omnivore, Vegetarian and Vegan Diets: Culturable Populations and rRNA DGGE Profiling. PLoS ONE 2015, 10, e128669. [Google Scholar]
- Carmen, L.; Eckert, E.M.; Eleonora, M.; Jorg, V.; Marzia, M.; Ilaria, P.; Andrea, D.C.; Veronica, C.; Federica, B.; Jakob, P. Assessing the Influence of Vegan, Vegetarian and Omnivore Oriented Westernized Dietary Styles on Human Gut Microbiota: A Cross Sectional Study. Front. Microbiol. 2018, 9, 317. [Google Scholar]
- Chelius, M.K.; Triplett, E.W. Immunolocalization of dinitrogenase reductase produced by Klebsiella pneumoniae in association with Zea mays L. Appl. Environ. Microbiol. 2019, 66, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Liang, T.; Geng, D.; Wang, L.; Liu, L.; Zhou, X.; Pu, H.; Huang, J.; Zhou, S.; Tong, L. Correction to: Dietary Proteins Alter Fermentation Characteristics of Human Gut Microbiota In Vitro. Plant Foods Hum. Nutr. 2021, 77, 157–158. [Google Scholar] [CrossRef]
- Ogata, Y.; Suda, W.; Ikeyama, N.; Hattori, M.; Ohkuma, M.; Sakamoto, M.; Gill, S.R. Complete Genome Sequence of Phascolarctobacterium faecium JCM 30894, a Succinate-Utilizing Bacterium Isolated from Human Feces. Microbiol. Resour. Ann. 2019, 8, e01487-18. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).