Curcumae Radix Decreases Neurodegenerative Markers through Glycolysis Decrease and TCA Cycle Activation
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Curcumae Radix Extract (CRE)
2.2. Animals and Treatment
2.3. Cell Culture
2.4. Western Blotting
2.5. Total RNA Extraction and Real-Time Quantitative PCR
2.6. Measurements of Cellular Glycolysis
2.7. Statistical Analysis
3. Results
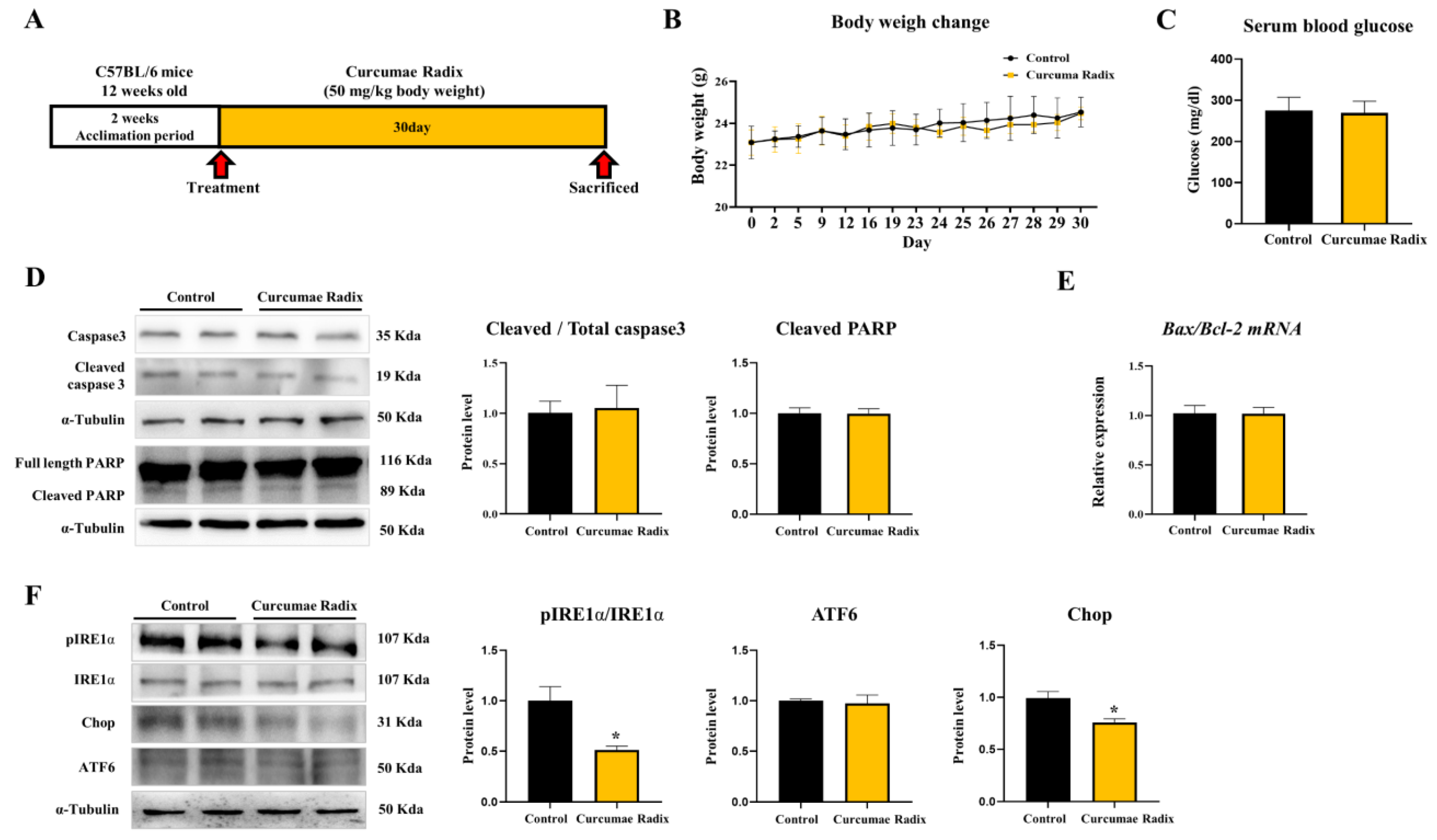
3.1. Curcumae Radix Extract Reduced Markers of the ER Stress in Mouse Cerebrum
3.2. Curcumae Radix Extract Reduced Neurodegenerative Markers
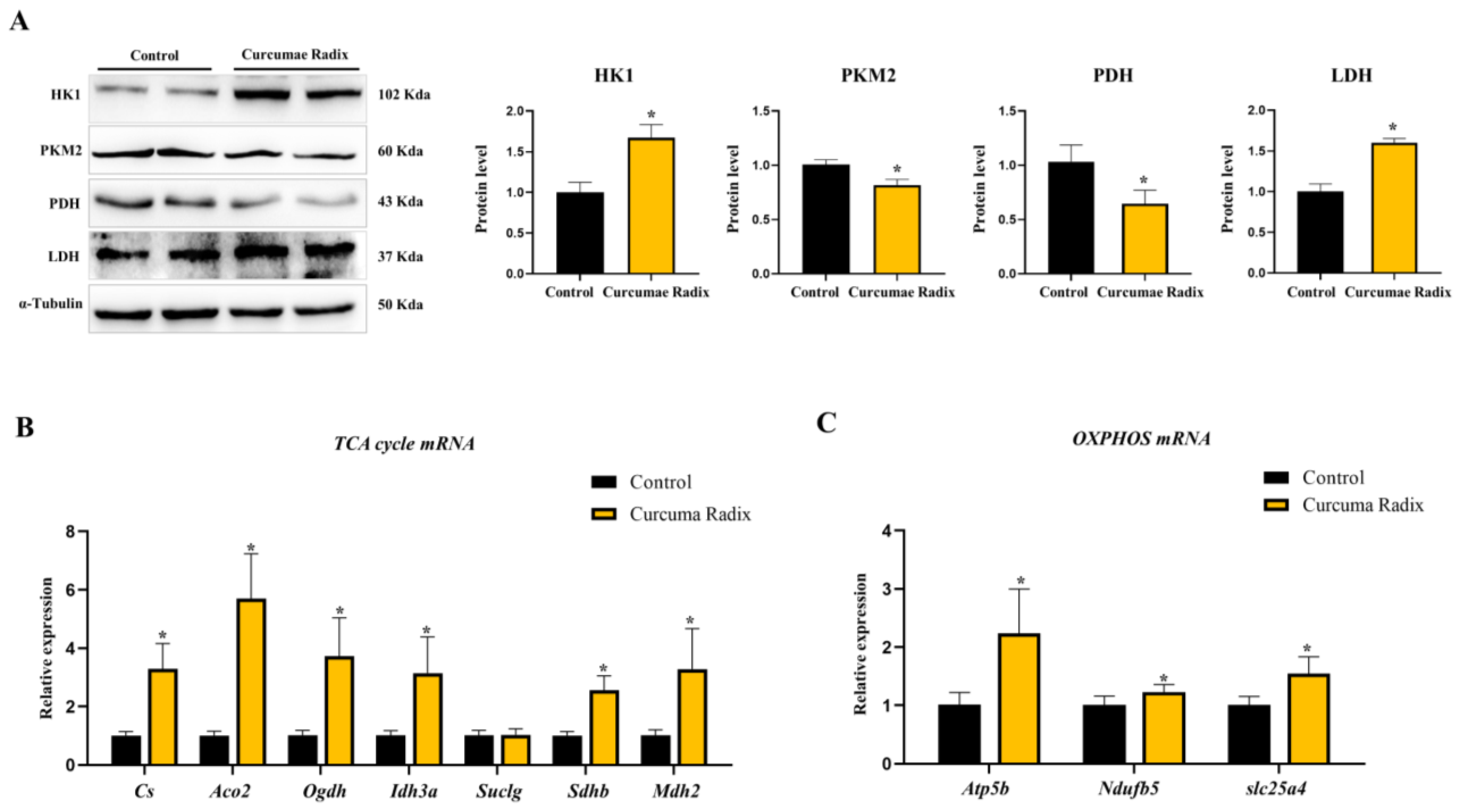
3.3. Curcumae Radix Extract Decreased Glycolysis and Compensatively Increased the TCA Cycle in Mouse Cerebrum
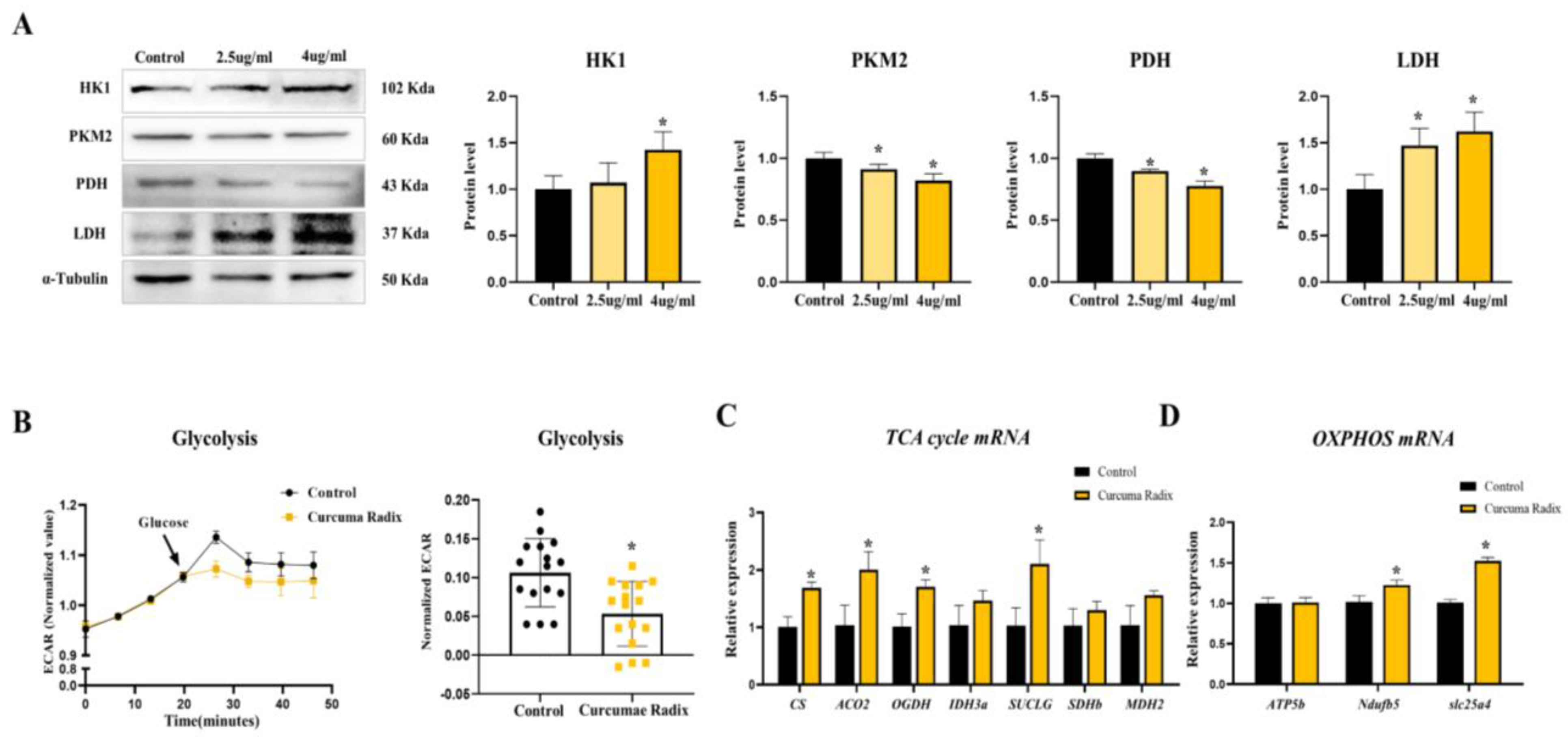
3.4. Curcumae Radix Extract Decreased Glycolysis Markers and Compensatively Increased the TCA Cycle in DBT Cells
3.5. Neurodegenerative Markers Decreased in Glycolysis Inhibition and TCA Activation State of DBT Cells
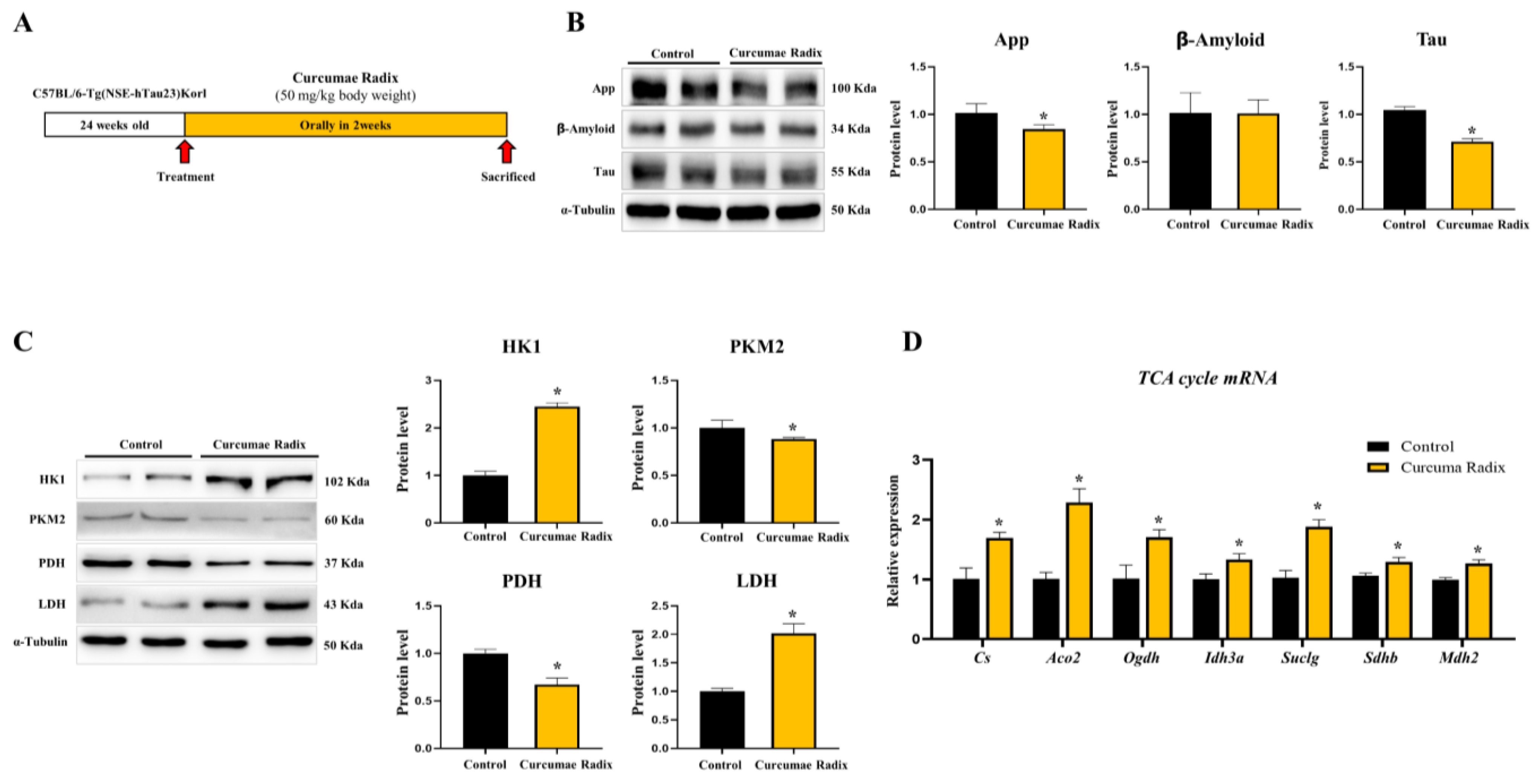
3.6. Curcumae Radix Extracts Have Neuroprotective Effects in Tau-Overexpressing Mouse Cerebrum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Twohig, D.; Nielsen, H.M. alpha-synuclein in the pathophysiology of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.; Iliffe, S. Alzheimer’s disease. BMJ 2009, 338, b158. [Google Scholar] [CrossRef] [PubMed]
- Mohandas, E.; Rajmohan, V.; Raghunath, B. Neurobiology of Alzheimer’s disease. Indian J. Psychiatry 2009, 51, 55–61. [Google Scholar] [CrossRef]
- Gomez-Benito, M.; Granado, N.; Garcia-Sanz, P.; Michel, A.; Dumoulin, M.; Moratalla, R. Modeling Parkinson’s Disease with the Alpha-Synuclein Protein. Front. Pharm. 2020, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Hou, J.; Ping, J.; Cai, D. Advances in developing novel therapeutic strategies for Alzheimer’s disease. Mol. Neurodegener. 2018, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Liang, J.; Zhou, B. Glucose Metabolic Dysfunction in Neurodegenerative Diseases-New Mechanistic Insights and the Potential of Hypoxia as a Prospective Therapy Targeting Metabolic Reprogramming. Int. J. Mol. Sci. 2021, 22, 5887. [Google Scholar] [CrossRef]
- Cleland, N.R.W.; Al-Juboori, S.I.; Dobrinskikh, E.; Bruce, K.D. Altered substrate metabolism in neurodegenerative disease: New insights from metabolic imaging. J. Neuroinflamm. 2021, 18, 248. [Google Scholar] [CrossRef]
- Mergenthaler, P.; Lindauer, U.; Dienel, G.A.; Meisel, A. Sugar for the brain: The role of glucose in physiological and pathological brain function. Trends Neurosci. 2013, 36, 587–597. [Google Scholar] [CrossRef]
- Rao, J.; Oz, G.; Seaquist, E.R. Regulation of cerebral glucose metabolism. Minerva Endocrinol. 2006, 31, 149–158. [Google Scholar]
- Kaya, P.; Lee, S.R.; Lee, Y.H.; Kwon, S.W.; Yang, H.; Lee, H.W.; Hong, E.J. Curcumae Radix Extract Decreases Mammary Tumor-Derived Lung Metastasis via Suppression of C-C Chemokine Receptor Type 7 Expression. Nutrients 2019, 11, 410. [Google Scholar] [CrossRef]
- Zhou, Y.; Xie, M.; Song, Y.; Wang, W.; Zhao, H.; Tian, Y.; Wang, Y.; Bai, S.; Zhao, Y.; Chen, X.; et al. Two Traditional Chinese Medicines Curcumae Radix and Curcumae Rhizoma: An Ethnopharmacology, Phytochemistry, and Pharmacology Review. Evid. Based Complement. Altern. Med. 2016, 2016, 4973128. [Google Scholar] [CrossRef] [PubMed]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A Review of Its Effects on Human Health. Foods 2017, 6, 92. [Google Scholar] [CrossRef]
- Jurenka, J.S. Anti-inflammatory properties of curcumin, a major constituent of Curcuma longa: A review of preclinical and clinical research. Altern. Med. Rev. 2009, 14, 141–153. [Google Scholar] [PubMed]
- Zhou, J.; Miao, H.; Li, X.; Hu, Y.; Sun, H.; Hou, Y. Curcumin inhibits placental inflammation to ameliorate LPS-induced adverse pregnancy outcomes in mice via upregulation of phosphorylated Akt. Inflamm. Res. 2017, 66, 177–185. [Google Scholar] [CrossRef]
- Moghadamtousi, S.Z.; Kadir, H.A.; Hassandarvish, P.; Tajik, H.; Abubakar, S.; Zandi, K. A review on antibacterial, antiviral, and antifungal activity of curcumin. Biomed. Res. Int. 2014, 2014, 186864. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Zhang, J.; Xia, Y.; Kanchana, K.; Guo, G.; Chen, W.; Huang, Y.; Wang, Z.; Yang, S.; Liang, G. ROS generation mediates the anti-cancer effects of WZ35 via activating JNK and ER stress apoptotic pathways in gastric cancer. Oncotarget 2015, 6, 5860–5876. [Google Scholar] [CrossRef] [PubMed]
- Rajitha, B.; Belalcazar, A.; Nagaraju, G.P.; Shaib, W.L.; Snyder, J.P.; Shoji, M.; Pattnaik, S.; Alam, A.; El-Rayes, B.F. Inhibition of NF-kappaB translocation by curcumin analogs induces G0/G1 arrest and downregulates thymidylate synthase in colorectal cancer. Cancer Lett. 2016, 373, 227–233. [Google Scholar] [CrossRef]
- Ren, B.; Luo, S.; Tian, X.; Jiang, Z.; Zou, G.; Xu, F.; Yin, T.; Huang, Y.; Liu, J. Curcumin inhibits liver cancer by inhibiting DAMP molecule HSP70 and TLR4 signaling. Oncol. Rep. 2018, 40, 895–901. [Google Scholar] [CrossRef]
- Huang, T.; Zhao, J.; Guo, D.; Pang, H.; Zhao, Y.; Song, J. Curcumin mitigates axonal injury and neuronal cell apoptosis through the PERK/Nrf2 signaling pathway following diffuse axonal injury. Neuroreport 2018, 29, 661–677. [Google Scholar] [CrossRef]
- Zhu, H.; Fan, Y.; Sun, H.; Chen, L.; Man, X. Curcumin inhibits endoplasmic reticulum stress induced by cerebral ischemia-reperfusion injury in rats. Exp. Med. 2017, 14, 4047–4052. [Google Scholar] [CrossRef]
- Thani, N.A.A.; Sallis, B.; Nuttall, R.; Schubert, F.R.; Ahsan, M.; Davies, D.; Purewal, S.; Cooper, A.; Rooprai, H.K. Induction of apoptosis and reduction of MMP gene expression in the U373 cell line by polyphenolics in Aronia melanocarpa and by curcumin. Oncol. Rep. 2012, 28, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Song, Q.; Wang, P.; Li, Y.; Yang, M.; Yu, S.Y. Neuroprotective Effects of Curcumin on IL-1beta-Induced Neuronal Apoptosis and Depression-Like Behaviors Caused by Chronic Stress in Rats. Front. Cell. Neurosci. 2018, 12, 516. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Park, J.M.; Kim, E.K.; Lee, J.O.; Lee, S.K.; Jung, J.H.; You, G.Y.; Park, S.H.; Suh, P.G.; Kim, H.S. Curcumin stimulates glucose uptake through AMPK-p38 MAPK pathways in L6 myotube cells. J. Cell. Physiol 2010, 223, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.C.; Lin, C.S.; Hsu, C.C.; Chen, L.J.; Cheng, K.C.; Cheng, J.T. Activation of muscarinic M-1 cholinoceptors by curcumin to increase glucose uptake into skeletal muscle isolated from Wistar rats. Neurosci. Lett. 2009, 465, 238–241. [Google Scholar] [CrossRef]
- Lian, N.; Jin, H.; Zhang, F.; Wu, L.; Shao, J.; Lu, Y.; Zheng, S. Curcumin inhibits aerobic glycolysis in hepatic stellate cells associated with activation of adenosine monophosphate-activated protein kinase. IUBMB Life 2016, 68, 589–596. [Google Scholar] [CrossRef]
- Das, L.; Vinayak, M. Long term effect of curcumin in regulation of glycolytic pathway and angiogenesis via modulation of stress activated genes in prevention of cancer. PLoS ONE 2014, 9, e99583. [Google Scholar] [CrossRef]
- Soni, V.K.; Shukla, D.; Kumar, A.; Vishvakarma, N.K. Curcumin circumvent lactate-induced chemoresistance in hepatic cancer cells through modulation of hydroxycarboxylic acid receptor-1. Int. J. Biochem. Cell Biol. 2020, 123, 105752. [Google Scholar] [CrossRef]
- Lian, N.; Jiang, Y.; Zhang, F.; Jin, H.; Lu, C.; Wu, X.; Lu, Y.; Zheng, S. Curcumin regulates cell fate and metabolism by inhibiting hedgehog signaling in hepatic stellate cells. Lab. Investig. 2015, 95, 790–803. [Google Scholar] [CrossRef]
- Perugini, J.; Di Mercurio, E.; Tossetta, G.; Severi, I.; Monaco, F.; Reguzzoni, M.; Tomasetti, M.; Dani, C.; Cinti, S.; Giordano, A. Biological Effects of Ciliary Neurotrophic Factor on hMADS Adipocytes. Front. Endocrinol. 2019, 10, 768. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Z.; Wang, Y.; Tweardy, D.J.; Mitch, W.E. Stat3 activation induces insulin resistance via a muscle-specific E3 ubiquitin ligase Fbxo40. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E625–E635. [Google Scholar] [CrossRef]
- Bianconi, V.; Pirro, M.; Moallem, S.M.H.; Majeed, M.; Bronzo, P.; D’Abbondanza, M.; Jamialahmadi, T.; Sahebkar, A. The Multifaceted Actions of Curcumin in Obesity. Adv. Exp. Med. Biol. 2021, 1328, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Tossetta, G.; Fantone, S.; Giannubilo, S.R.; Marzioni, D. The Multifaced Actions of Curcumin in Pregnancy Outcome. Antioxidants 2021, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.; Mahalakshmi, A.M.; Ray, B.; Tuladhar, S.; Hediyal, T.A.; Manthiannem, E.; Padamati, J.; Chandra, R.; Chidambaram, S.B.; Sakharkar, M.K. Benefits of curcumin in brain disorders. Biofactors 2019, 45, 666–689. [Google Scholar] [CrossRef]
- Magistretti, P.J.; Allaman, I. Lactate in the brain: From metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 2018, 19, 235–249. [Google Scholar] [CrossRef]
- Araque, A.; Navarrete, M. Glial cells in neuronal network function. Philos. Trans. R. Soc. Lond B Biol. Sci. 2010, 365, 2375–2381. [Google Scholar] [CrossRef]
- Wang, J.B.; Qi, L.L.; Zheng, S.D.; Wu, T.X. Curcumin induces apoptosis through the mitochondria-mediated apoptotic pathway in HT-29 cells. J. Zhejiang Univ. Sci. B 2009, 10, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, H.S.; Jung, E.J.; Lee, J.Y.; Tsang, B.K.; Lim, J.M.; Song, Y.S. Curcumin induces ER stress-mediated apoptosis through selective generation of reactive oxygen species in cervical cancer cells. Mol. Carcinog. 2016, 55, 918–928. [Google Scholar] [CrossRef]
- Ramkumar, M.; Rajasankar, S.; Gobi, V.V.; Dhanalakshmi, C.; Manivasagam, T.; Justin Thenmozhi, A.; Essa, M.M.; Kalandar, A.; Chidambaram, R. Neuroprotective effect of Demethoxycurcumin, a natural derivative of Curcumin on rotenone induced neurotoxicity in SH-SY 5Y Neuroblastoma cells. BMC Complement. Altern. Med. 2017, 17, 217. [Google Scholar] [CrossRef]
- Randino, R.; Grimaldi, M.; Persico, M.; De Santis, A.; Cini, E.; Cabri, W.; Riva, A.; D’Errico, G.; Fattorusso, C.; D’Ursi, A.M.; et al. Investigating the Neuroprotective Effects of Turmeric Extract: Structural Interactions of beta-Amyloid Peptide with Single Curcuminoids. Sci. Rep. 2016, 6, 38846. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, R.; He, D.; Zhou, G.; Wu, H.; Xu, C.; He, B.; Wu, L.; Wang, Y.; Chang, Y.; et al. Bisdemethoxycurcumin inhibits oxidative stress and antagonizes Alzheimer’s disease by up-regulating SIRT1. Brain Behav. 2020, 10, e01655. [Google Scholar] [CrossRef]
- He, D.; Chen, S.; Xiao, Z.; Wu, H.; Zhou, G.; Xu, C.; Chang, Y.; Li, Y.; Wang, G.; Xie, M. Bisdemethoxycurcumin exerts a cell-protective effect via JAK2/STAT3 signaling in a rotenone-induced Parkinson’s disease model in vitro. Folia Histochem. Cytobiol. 2020, 58, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Narlawar, R.; Baumann, K.; Schubenel, R.; Schmidt, B. Curcumin derivatives inhibit or modulate beta-amyloid precursor protein metabolism. Neurodegener. Dis. 2007, 4, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Darvesh, A.S.; Carroll, R.T.; Bishayee, A.; Novotny, N.A.; Geldenhuys, W.J.; Van der Schyf, C.J. Curcumin and neurodegenerative diseases: A perspective. Expert Opin. Investig. Drugs 2012, 21, 1123–1140. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, H.; Ghasemi, F.; Barreto, G.E.; Rafiee, R.; Sathyapalan, T.; Sahebkar, A. Effects of curcumin on mitochondria in neurodegenerative diseases. Biofactors 2020, 46, 5–20. [Google Scholar] [CrossRef]
- Wang, X.; Eno, C.O.; Altman, B.J.; Zhu, Y.; Zhao, G.; Olberding, K.E.; Rathmell, J.C.; Li, C. ER stress modulates cellular metabolism. Biochem. J. 2011, 435, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Groenendyk, J.; Sreenivasaiah, P.K.; Kim, D.H.; Agellon, L.B.; Michalak, M. Biology of endoplasmic reticulum stress in the heart. Circ. Res. 2010, 107, 1185–1197. [Google Scholar] [CrossRef]
- Cardenas, M.L.; Cornish-Bowden, A.; Ureta, T. Evolution and regulatory role of the hexokinases. Biochim. Biophys. Acta 1998, 1401, 242–264. [Google Scholar] [CrossRef]
- Dong, G.; Mao, Q.; Xia, W.; Xu, Y.; Wang, J.; Xu, L.; Jiang, F. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol. Lett. 2016, 11, 1980–1986. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Du, Z.; Li, G.; Chen, M.; Chen, X.; Liang, G.; Chen, T. Curcumin suppresses gastric tumor cell growth via ROS-mediated DNA polymerase gamma depletion disrupting cellular bioenergetics. J. Exp. Clin. Cancer Res. 2017, 36, 47. [Google Scholar] [CrossRef]
- Salah, A.S.; Ahmed-Farid, O.A.; Nassan, M.A.; El-Tarabany, M.S. Dietary Curcumin Improves Energy Metabolism, Brain Monoamines, Carcass Traits, Muscle Oxidative Stability and Fatty Acid Profile in Heat-Stressed Broiler Chickens. Antioxidants 2021, 10, 1265. [Google Scholar] [CrossRef]
- Huang, W.C.; Chiu, W.C.; Chuang, H.L.; Tang, D.W.; Lee, Z.M.; Wei, L.; Chen, F.A.; Huang, C.C. Effect of curcumin supplementation on physiological fatigue and physical performance in mice. Nutrients 2015, 7, 905–921. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, E.; Musich, P.R.; Lin, F. Mitochondrial dysfunction in neurodegenerative diseases and the potential countermeasure. CNS Neurosci. 2019, 25, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Oz, G.; Seaquist, E.R.; Kumar, A.; Criego, A.B.; Benedict, L.E.; Rao, J.P.; Henry, P.G.; Van De Moortele, P.F.; Gruetter, R. Human brain glycogen content and metabolism: Implications on its role in brain energy metabolism. Am. J. Physiol Endocrinol. Metab. 2007, 292, E946–E951. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Du, W.; Wu, M. Regulation of the pentose phosphate pathway in cancer. Protein Cell 2014, 5, 592–602. [Google Scholar] [CrossRef]
- Staricha, K.; Meyers, N.; Garvin, J.; Liu, Q.; Rarick, K.; Harder, D.; Cohen, S. Effect of high glucose condition on glucose metabolism in primary astrocytes. Brain Res. 2020, 1732, 146702. [Google Scholar] [CrossRef]
- Cho, E.S.; Cha, Y.H.; Kim, H.S.; Kim, N.H.; Yook, J.I. The Pentose Phosphate Pathway as a Potential Target for Cancer Therapy. Biomol. Ther. 2018, 26, 29–38. [Google Scholar] [CrossRef]
- Kujundzic, R.N.; Stepanic, V.; Milkovic, L.; Gasparovic, A.C.; Tomljanovic, M.; Troselj, K.G. Curcumin and its Potential for Systemic Targeting of Inflamm-Aging and Metabolic Reprogramming in Cancer. Int. J. Mol. Sci. 2019, 20, 1180. [Google Scholar] [CrossRef]
- Yao, J.; Chen, S.; Mao, Z.; Cadenas, E.; Brinton, R.D. 2-Deoxy-D-glucose treatment induces ketogenesis, sustains mitochondrial function, and reduces pathology in female mouse model of Alzheimer’s disease. PLoS ONE 2011, 6, e21788. [Google Scholar] [CrossRef]
- O’Hara, D.; Davis, G.M.; Adlesic, N.A.; Hayes, J.M.; Davey, G.P. Dichloroacetate Stabilizes Mitochondrial Fusion Dynamics in Models of Neurodegeneration. Front. Mol. Neurosci. 2019, 12, 219. [Google Scholar] [CrossRef]
- Jensen, N.J.; Wodschow, H.Z.; Nilsson, M.; Rungby, J. Effects of Ketone Bodies on Brain Metabolism and Function in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 8767. [Google Scholar] [CrossRef]
- Mitchell, R.W.; On, N.H.; Del Bigio, M.R.; Miller, D.W.; Hatch, G.M. Fatty acid transport protein expression in human brain and potential role in fatty acid transport across human brain microvessel endothelial cells. J. Neurochem. 2011, 117, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Koczwara, J.B.; Gallelli, C.A.; Vergara, D.; Micioni Di Bonaventura, M.V.; Gaetani, S.; Giudetti, A.M. Fats for thoughts: An update on brain fatty acid metabolism. Int. J. Biochem. Cell Biol. 2017, 84, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Tracy, T.E.; Madero-Perez, J.; Swaney, D.L.; Chang, T.S.; Moritz, M.; Konrad, C.; Ward, M.E.; Stevenson, E.; Huttenhain, R.; Kauwe, G.; et al. Tau interactome maps synaptic and mitochondrial processes associated with neurodegeneration. Cell 2022, 185, 712–728.e14. [Google Scholar] [CrossRef]
- Jadhav, S.; Avila, J.; Scholl, M.; Kovacs, G.G.; Kovari, E.; Skrabana, R.; Evans, L.D.; Kontsekova, E.; Malawska, B.; de Silva, R.; et al. A walk through tau therapeutic strategies. Acta Neuropathol. Commun. 2019, 7, 22. [Google Scholar] [CrossRef]
- Padmaraju, V.; Indi, S.S.; Rao, K.S. New evidences on Tau-DNA interactions and relevance to neurodegeneration. Neurochem. Int. 2010, 57, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.M.; Park, S.J.; Kang, H.I.; Kim, B.G.; Shim, S.B.; Jee, S.W.; Lee, S.H.; Sin, J.S.; Bae, C.J.; Jang, M.K.; et al. Characterization of changes in global gene expression in the brain of neuron-specific enolase/human Tau23 transgenic mice in response to overexpression of Tau protein. Int. J. Mol. Med. 2010, 25, 667–675. [Google Scholar] [CrossRef][Green Version]
- Li, L.; Liu, S.; Zhou, Y.; Zhao, M.; Wang, Y.; Wang, C.; Lou, P.; Huang, R.; Ma, L.; Lu, Y.; et al. Indispensable role of mitochondria in maintaining the therapeutic potential of curcumin in acute kidney injury. J. Cell. Mol. Med. 2021, 25, 9863–9877. [Google Scholar] [CrossRef]
- Hamidie, R.D.R.; Yamada, T.; Ishizawa, R.; Saito, Y.; Masuda, K. Curcumin treatment enhances the effect of exercise on mitochondrial biogenesis in skeletal muscle by increasing cAMP levels. Metabolism 2015, 64, 1334–1347. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.B.; Lim, H.J.; Chae, K.R.; Kim, C.K.; Hwang, D.Y.; Jee, S.W.; Lee, S.H.; Sin, J.S.; Leem, Y.H.; Lee, S.H.; et al. Tau overexpression in transgenic mice induces glycogen synthase kinase 3beta and beta-catenin phosphorylation. Neuroscience 2007, 146, 730–740. [Google Scholar] [CrossRef]


| Primary Antibodies | Type | Lot. | Inc. |
|---|---|---|---|
| PRAP | Rabbit monoclonal | 9532 | Cell signaling technology |
| Phospho-IRE1α | Rabbit polyclonal | Ab37073 | Abcam PLC |
| IRE1α | Rabbit polyclonal | Ab48187 | Abcam PLC |
| Chop | Mouse monoclonal | MA1-250 | Invitrogen |
| ATF6 | Rabbit polyclonal | Ab65838 | Abcam PLC |
| Beta-amyloid- | Mouse monoclonal | sc-28365 | Santa Cruz biotechology |
| Tau | Rabbit monoclonal | A1103 | Company ABclonal, Inc. |
| HK1ǀ | Rabbit monoclonal | 2024 | Cell signaling technology |
| PKM2 | Rabbit monoclonal | 4053 | Cell signaling technology |
| PDH | Rabbit monoclonal | 3205 | Cell signaling technology |
| LDHA | Rabbit monoclonal | 3582 | Cell signaling technology |
| Alpha-Tubulin | Mouse monoclonal | 66031-1-Ig | Proteintech Group Inc |
| AMPKα | Rabbit monoclonal | 5831 | Cell signaling technology |
| Phospho-AMPKα | Rabbit monoclonal | 2535 | Cell signaling technology |
| Secondary antibodies | Type | Lot. | Inc. |
| Anti-Mouse IgG | Goat | 121507 | Jackonimmuno |
| Anti-Rabbit IgG | Mouse | 123213 | Jackonimmuno |
| IRE1α | Rabbit polyclonal | Ab48187 | Abcam PLC |
| Gene | Upper Primer (5′-3′) | Lower Primer (5′-3′) | Species |
|---|---|---|---|
| Bax | TGA AGA CAG GGG CCT TTT TG | AAT TCG CCG GAG ACA CTC | Mouse |
| Bcl-2 | ATG CCT TTG TGG AAC TAT ATG GC | GGT ATG CAC CCA GAG TGA TGC | Mouse |
| Cs | CCT GAG TGC CAG AAA ATG CTG | CCA CAT GAG AAG GCA GAG CT | Mouse |
| Aco2 | ACA AGT GGG ACG GCA AAG AC | AGC ATT GCG TAC AGA GTT GGC | Mouse |
| Ogdh | AAT GCT GAG CTG GCC TGG TG | TCA GGT GTG TTT TCT TGT TGC C | Mouse |
| Idh3a | TGC TTC GCC ACA TGG GAC TT | CGT TGC CTC CCA GAT CTT TT | Mouse |
| Suclg2 | CTG TGC CAT CAT TGC CAA CG | ATG GGG AGT CCG CTG CTC TT | Mouse |
| Sdhb | CTC TGT CTA CCG CTG CCA C | GGC ACA CTC AGC ACG GAC T | Mouse |
| Mdh2 | ATG CTG GAG CCC GCT TTG TC | CAG GGA TAG CCT CGG CAA TC | Mouse |
| Atp5b | CCC TGA AGG AGA CCA TCA AA | AAG ACC CCT CAC GAT GAA TG | Mouse |
| Ndufb5 | CTT CCT CAC TCG TGG CTT TC | CGC ACT TCC AGC TCC TTT AC | Mouse |
| Slc25a4 | ATG GTC TGG GCG ACT GTA TC | TCA AAG GGG TAG GAC ACC AG | Mouse |
| RPLP0 | GCA GCA GAT CCG CAT GTC GCT CCG | GAG CTG GCA CAG TGA CCT CAC ACG G | Mouse |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, S.-L.; Yang, H.; Lee, S.R.; Heo, J.H.; Lee, H.-W.; Hong, E.-J. Curcumae Radix Decreases Neurodegenerative Markers through Glycolysis Decrease and TCA Cycle Activation. Nutrients 2022, 14, 1587. https://doi.org/10.3390/nu14081587
Jo S-L, Yang H, Lee SR, Heo JH, Lee H-W, Hong E-J. Curcumae Radix Decreases Neurodegenerative Markers through Glycolysis Decrease and TCA Cycle Activation. Nutrients. 2022; 14(8):1587. https://doi.org/10.3390/nu14081587
Chicago/Turabian StyleJo, Seong-Lae, Hyun Yang, Sang R. Lee, Jun H. Heo, Hye-Won Lee, and Eui-Ju Hong. 2022. "Curcumae Radix Decreases Neurodegenerative Markers through Glycolysis Decrease and TCA Cycle Activation" Nutrients 14, no. 8: 1587. https://doi.org/10.3390/nu14081587
APA StyleJo, S.-L., Yang, H., Lee, S. R., Heo, J. H., Lee, H.-W., & Hong, E.-J. (2022). Curcumae Radix Decreases Neurodegenerative Markers through Glycolysis Decrease and TCA Cycle Activation. Nutrients, 14(8), 1587. https://doi.org/10.3390/nu14081587






