1,25(OH)2D3 Promotes Macrophage Efferocytosis Partly by Upregulating ASAP2 Transcription via the VDR-Bound Enhancer Region and ASAP2 May Affect Antiviral Immunity
Abstract
1. Introduction
2. Material and Methods
2.1. Cells Culture and In Vitro Efferocytosis Assay
2.2. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.3. Western Blot Analysis
2.4. Stable Knockdown of ASAP2
2.5. Chromatin Immunoprecipitation (ChIP) Assay
2.6. Luciferase Assay
2.7. Pulldown Assay for Testing RAC1 Activity
2.8. Immunoprecipitation Assay
2.9. Analysis of Differentially Expressed Genes (DEGs) and Functional Enrichment
2.10. The Disease Model of Experimental Autoimmune Encephalomyelitis (EAE) and Peritonitis
2.11. Fluorescence-Activated Cell Sorting (FACS) Analysis
2.12. Histological Analysis and Immunofluorescence Staining
2.13. Statistical Analyses
3. Results
3.1. ASAP2 Was Significantly Upregulated in THP-1 Cells and Macrophages following 1,25(OH)2D3 Exposure
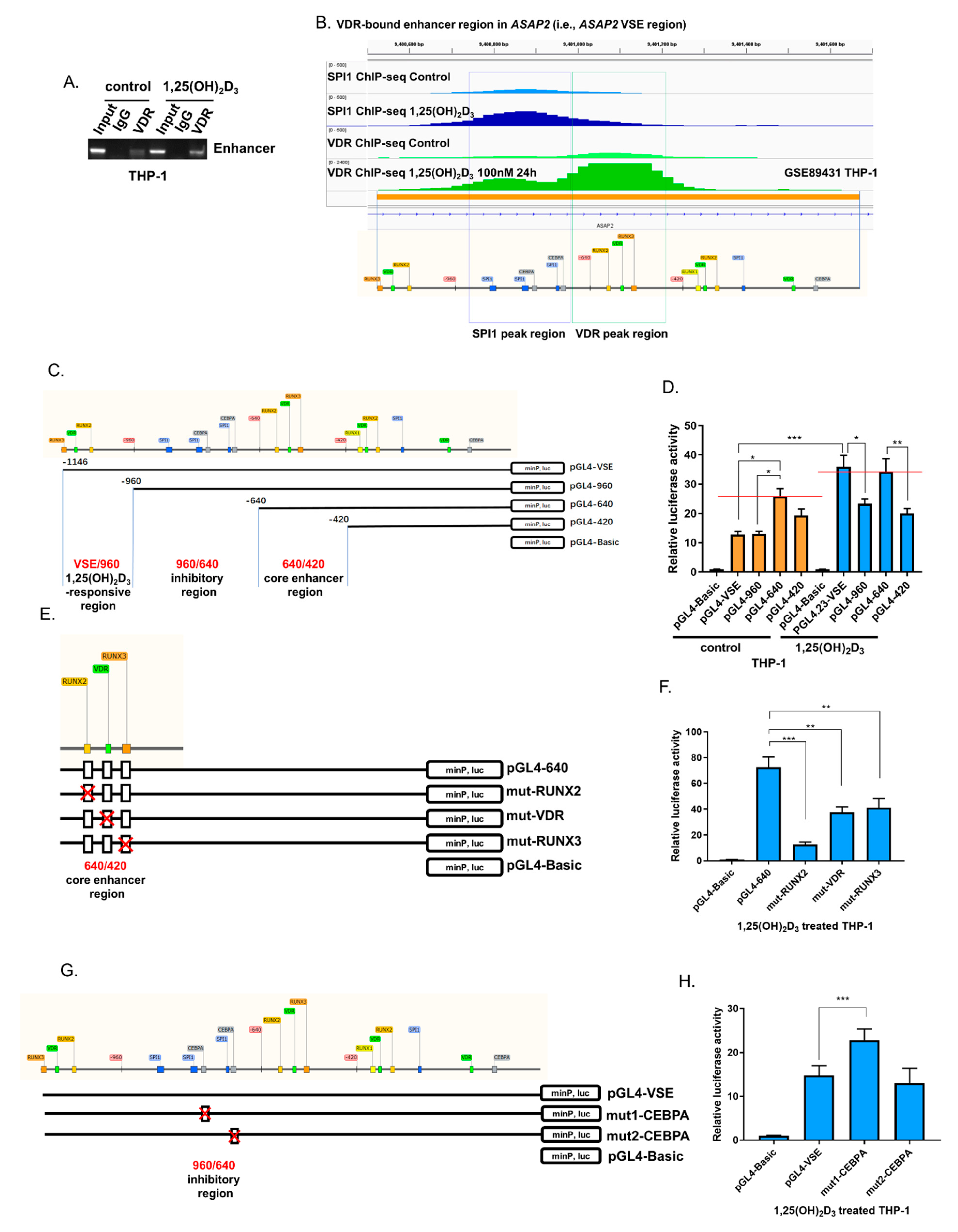
3.2. Three Regulatory Regions in the VDR-Bound Enhancer Region of ASAP2 Regulated the Transcription of ASAP2 in Response to 1,25(OH)2D3
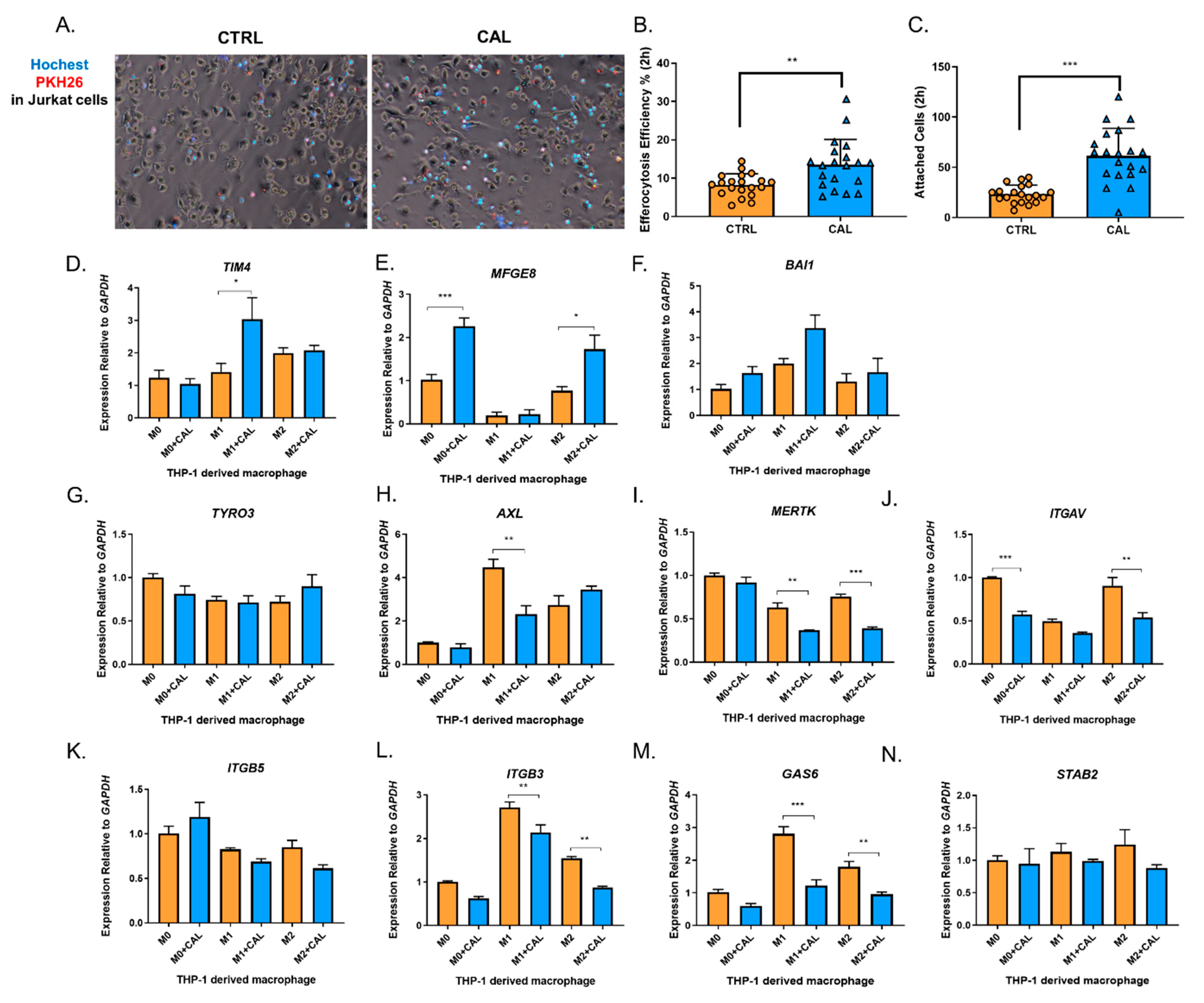
3.3. 1,25(OH)2D3 Promoted M2 Macrophage Efferocytosis and Transcription of Certain Genes Encoding the “Eat-me” Signals
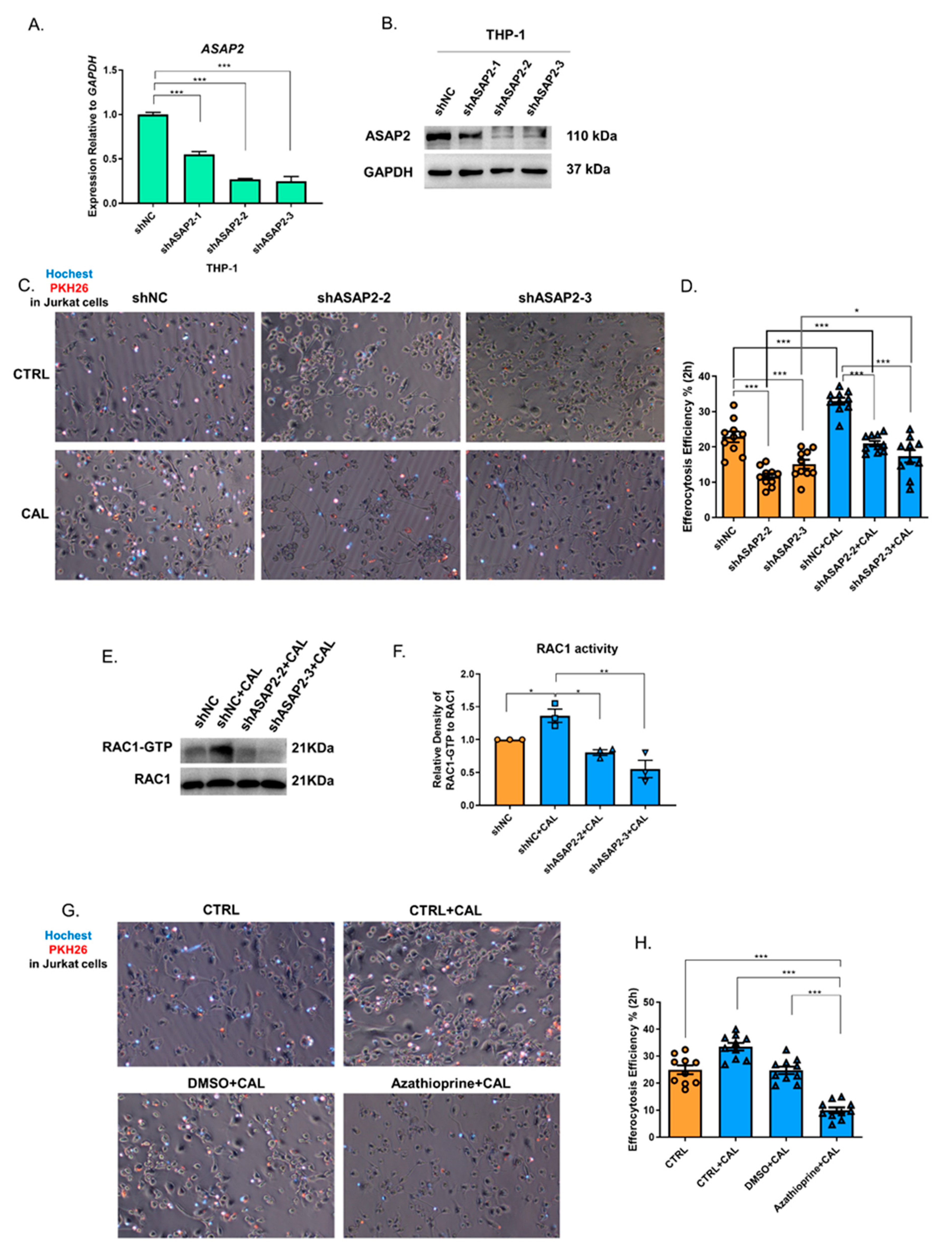
3.4. 1,25(OH)2D3 Promoted Macrophage Efferocytosis Partly via ASAP2
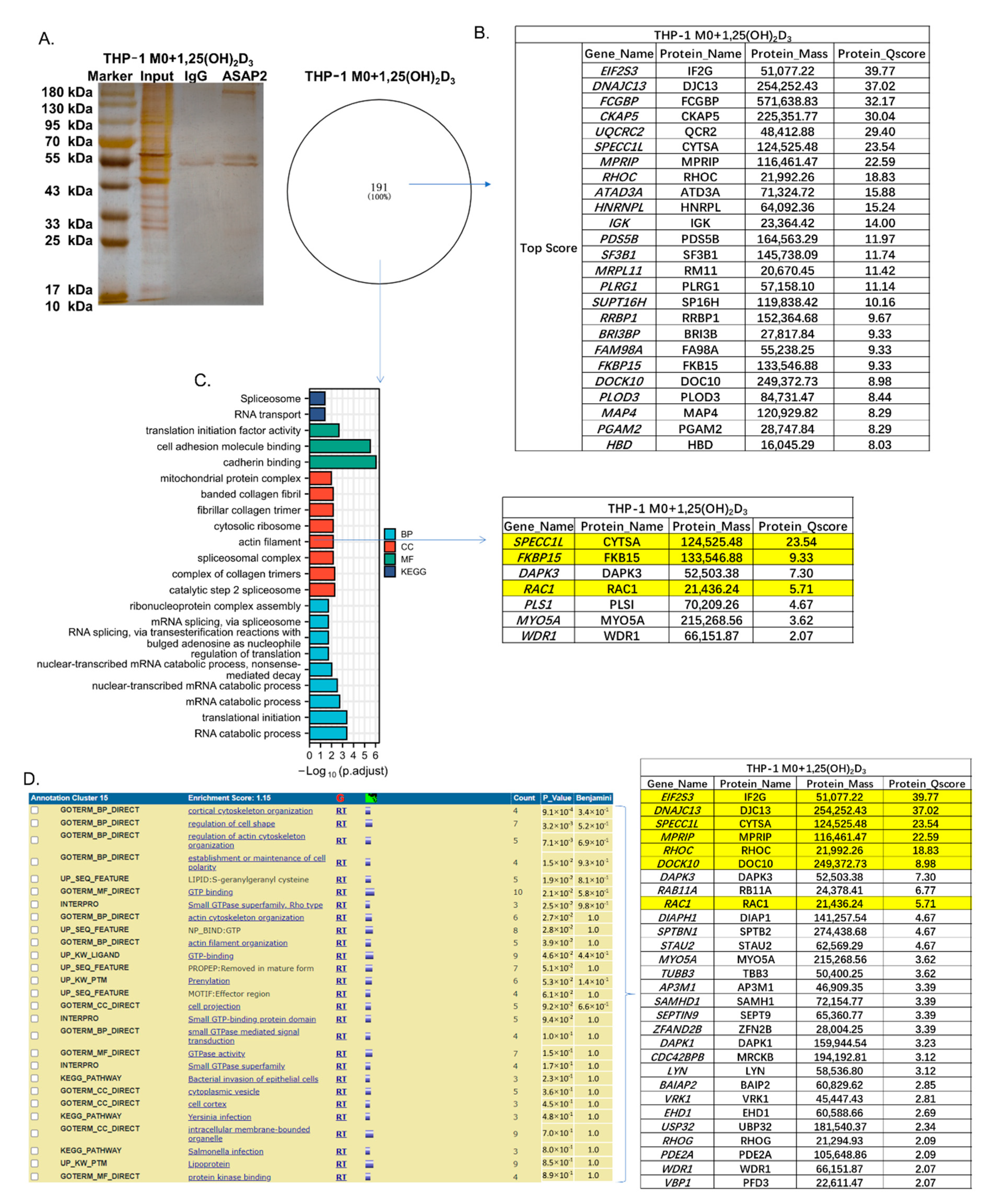
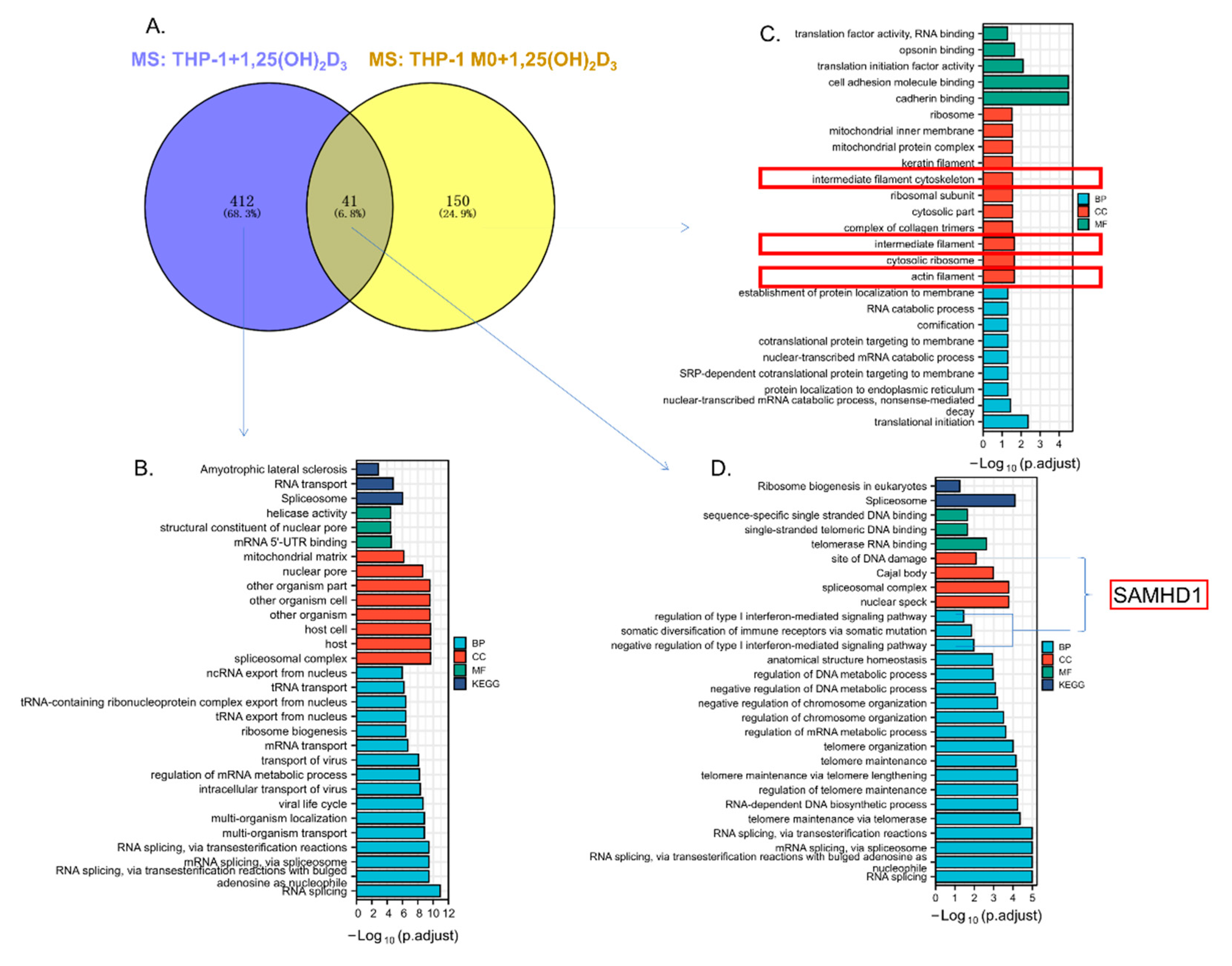
3.5. The Cytoskeletal-Associated Proteins and Interactors of ASAP2
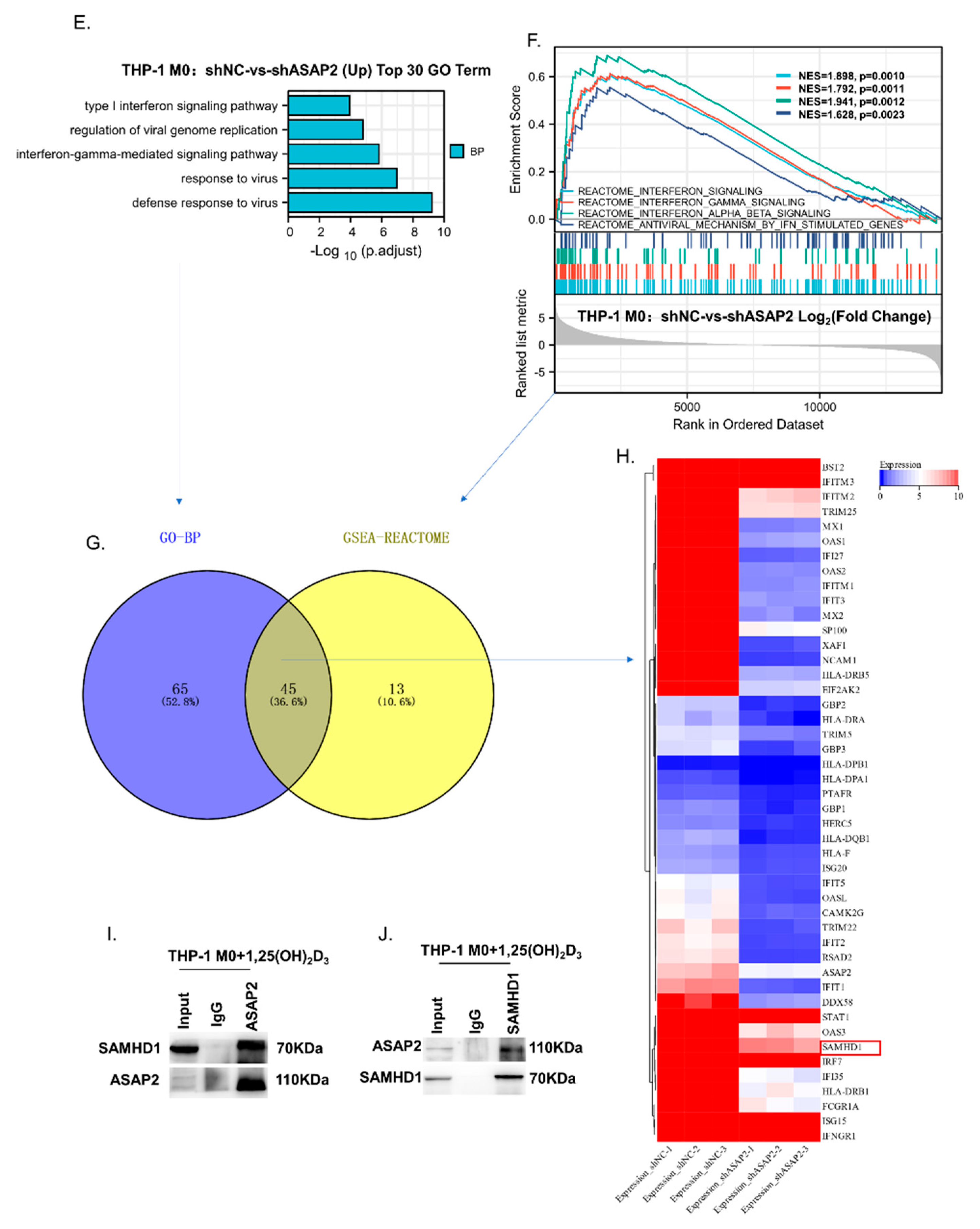
3.6. ASAP2 Promoted Interferon Signaling and Anti-Virus-Associated Pathways
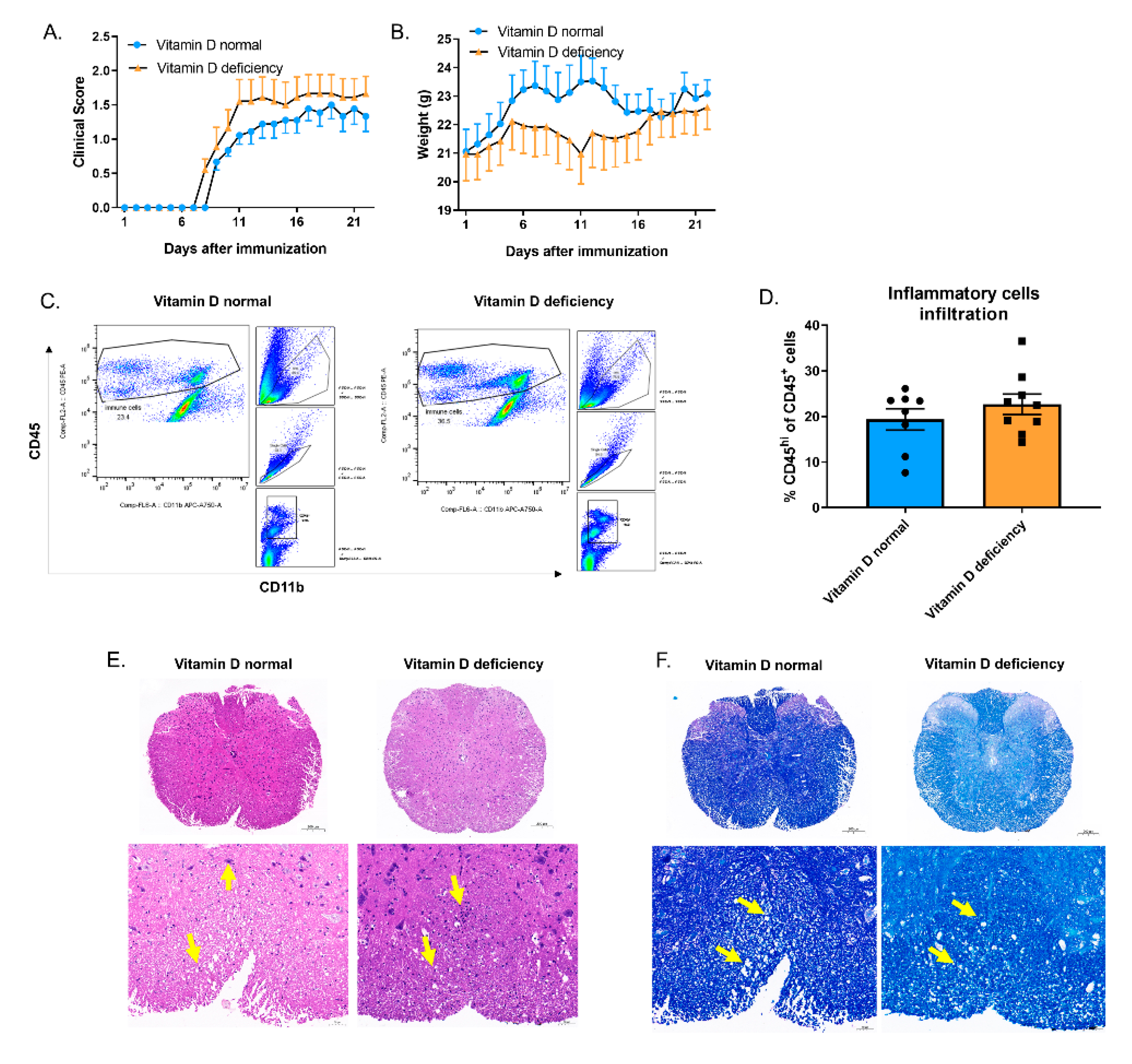
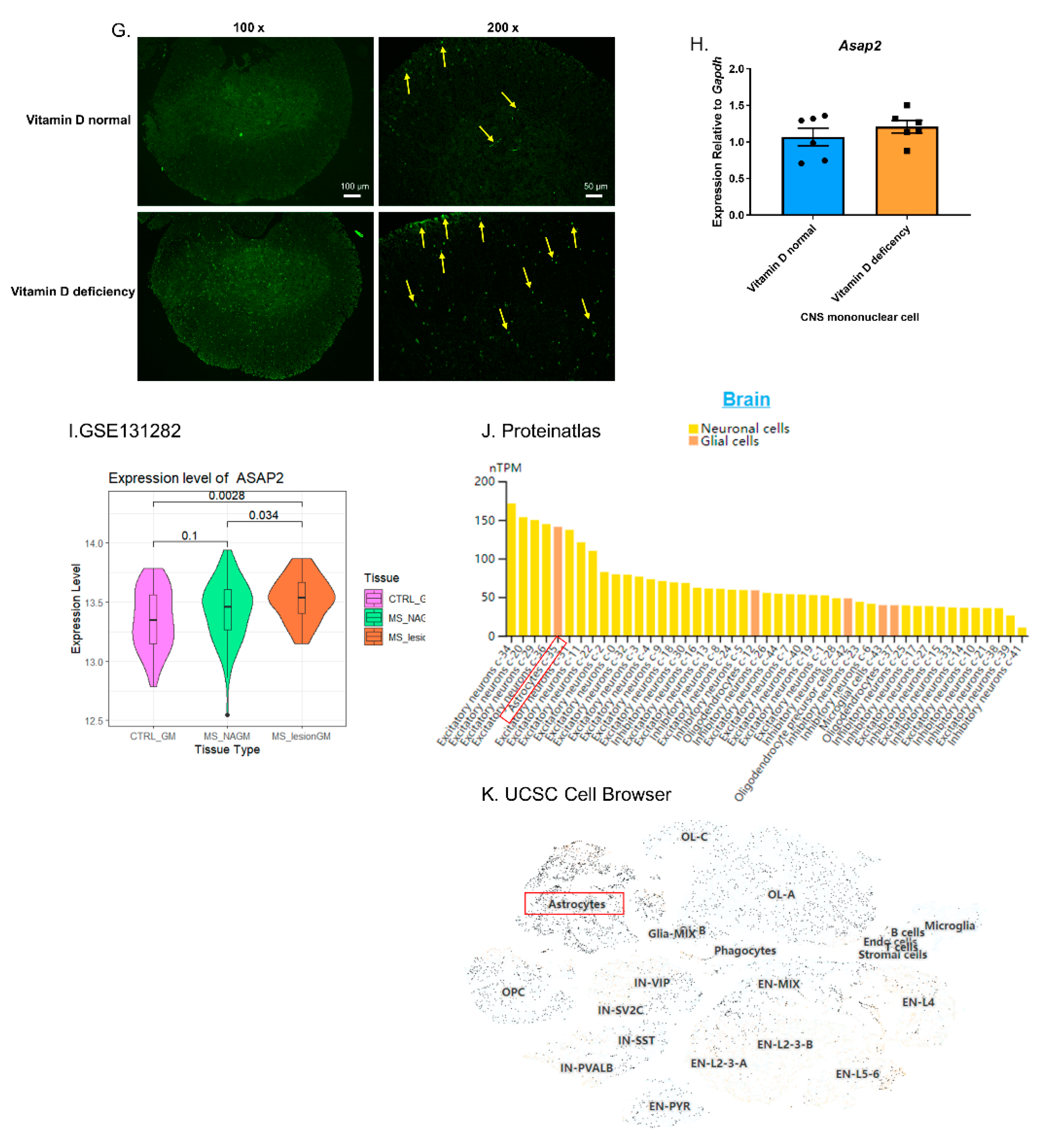
3.7. Vitamin D Reduced the Number of Apoptotic Cells in EAE and Promoted Macrophage Efferocytosis in Peritonitis without Changing the Total mRNA Level of Asap2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Leung, D.Y.; Richers, B.N.; Liu, Y.; Remigio, L.K.; Riches, D.W.; Goleva, E. Vitamin D inhibits monocyte/macrophage proinflammatory cytokine production by targeting MAPK phosphatase-1. J. Immunol. 2012, 188, 2127–2135. [Google Scholar] [CrossRef]
- Dauletbaev, N.; Herscovitch, K.; Das, M.; Chen, H.; Bernier, J.; Matouk, E.; Bérubé, J.; Rousseau, S.; Lands, L.C. Down-regulation of IL-8 by high-dose vitamin D is specific to hyperinflammatory macrophages and involves mechanisms beyond up-regulation of DUSP1. Br. J. Pharmacol. 2015, 172, 4757–4771. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Nestel, F.P.; Bourdeau, V.; Nagai, Y.; Wang, Q.; Liao, J.; Tavera-Mendoza, L.; Lin, R.; Hanrahan, J.W.; Mader, S. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 2004, 173, 2909–2912. [Google Scholar] [CrossRef]
- Lu, M.; Taylor, B.V.; Körner, H. Genomic Effects of the Vitamin D Receptor: Potentially the Link between Vitamin D, Immune Cells, and Multiple Sclerosis. Front. Immunol. 2018, 9, 477. [Google Scholar] [CrossRef]
- Pott, S.; Lieb, J.D. What Are Super-Enhancers? Nat. Genet. 2015, 47, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; McComish, B.J.; Burdon, K.P.; Taylor, B.V.; Körner, H. The Association Between Vitamin D and Multiple Sclerosis Risk: 1,25(OH)(2)D(3) Induces Super-Enhancers Bound by VDR. Front. Immunol. 2019, 10, 488. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, V.; Barbier, C.; Ismailova, A.; Wang, Y.; Dmowski, K.; Salehi-Tabar, R.; Memari, B.; Groulx-Boivin, E.; White, J.H. Vitamin D-regulated Gene Expression Profiles: Species-specificity and Cell-specific Effects on Metabolism and Immunity. Endocrinology 2021, 162, bqaa218. [Google Scholar] [CrossRef] [PubMed]
- Hanel, A.; Carlberg, C. Time-Resolved Gene Expression Analysis Monitors the Regulation of Inflammatory Mediators and Attenuation of Adaptive Immune Response by Vitamin D. Int. J. Mol. Sci. 2022, 23, 911. [Google Scholar] [CrossRef] [PubMed]
- Hanel, A.; Malmberg, H.R.; Carlberg, C. Genome-wide effects of chromatin on vitamin D signaling. J. Mol. Endocrinol. 2020, 64, R45–R56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cox, D.; Tseng, C.C.; Donaldson, J.G.; Greenberg, S. A Requirement for ARF6 in Fcγ Receptor-mediated Phagocytosis in Macrophages. J. Biol. Chem. 1998, 273, 19977–19981. [Google Scholar] [CrossRef]
- Tanna, C.E.; Goss, L.B.; Ludwig, C.G.; Chen, P.W. Arf GAPs as regulators of the actin cytoskeleton–An update. Int. J. Mol. Sci. 2019, 20, 442. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, T.; Tavernier, J.; Peelman, F. The Small GTPase Arf6: An Overview of Its Mechanisms of Action and of Its Role in Host-Pathogen Interactions and Innate Immunity. Int. J. Mol. Sci. 2019, 20, 2209. [Google Scholar] [CrossRef] [PubMed]
- Uchida, H.; Kondo, A.; Yoshimura, Y.; Mazaki, Y.; Sabe, H. PAG3/Papalpha/KIAA0400, a GTPase-activating protein for ADP-ribosylation factor (ARF), regulates ARF6 in Fcgamma receptor-mediated phagocytosis of macrophages. J. Exp. Med. 2001, 193, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Kondo, A.; Hashimoto, S.; Yano, H.; Nagayama, K.; Mazaki, Y.; Sabe, H. A New Paxillin-binding Protein, PAG3/Papα/KIAA0400, Bearing an ADP-Ribosylation Factor GTPase-activating Protein Activity, Is Involved in Paxillin Recruitment to Focal Adhesions and Cell Migration. Mol. Biol. Cell 2000, 11, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.L.; Fredericks, G.J.; Huang, Z.; Fay, J.D.; Hoffmann, F.W.; Hoffmann, P.R. Selenoprotein K regulation of palmitoylation and calpain cleavage of ASAP2 is required for efficient FcγR-mediated phagocytosis. J. Leukoc. Biol. 2017, 101, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S. Apoptosis and Clearance of Apoptotic Cells. Annu. Rev. Immunol. 2018, 36, 489–517. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Seuter, S.; Ryynänen, J.; Carlberg, C. The ASAP2 gene is a primary target of 1,25-dihydroxyvitamin D3 in human monocytes and macrophages. J. Steroid Biochem. Mol. Biol. 2014, 144, 12–18. [Google Scholar] [CrossRef]
- Warwick, T.; Schulz, M.H.; Günther, S.; Gilsbach, R.; Neme, A.; Carlberg, C.; Brandes, R.P.; Seuter, S. A hierarchical regulatory network analysis of the vitamin D induced transcriptome reveals novel regulators and complete VDR dependency in monocytes. Sci. Rep. 2021, 11, 6518. [Google Scholar] [CrossRef]
- Neme, A.; Seuter, S.; Carlberg, C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta. Gene Regul. Mech. 2017, 1860, 952–961. [Google Scholar] [CrossRef]
- Marcellini, S.; Bruna, C.; Henríquez, J.P.; Albistur, M.; Reyes, A.E.; Barriga, E.H.; Henríquez, B.; Montecino, M. Evolution of the interaction between Runx2 and VDR, two transcription factors involved in osteoblastogenesis. BMC Evol. Biol. 2010, 10, 78. [Google Scholar] [CrossRef][Green Version]
- Nagata, K.; Hojo, H.; Chang, S.; Okada, H.; Yano, F.; Chijimatsu, R.; Omata, Y.; Mori, D.; Makii, Y.; Kawata, M.; et al. Runx2 and Runx3 Differentially Regulate Articular chondrocytes during surgically induced osteoarthritis development. Nat. Commun. 2022, 13, 6187. [Google Scholar] [CrossRef]
- Gheibi Hayat, S.M.; Bianconi, V.; Pirro, M.; Sahebkar, A. Efferocytosis: Molecular mechanisms and pathophysiological perspectives. Immunol. Cell Biol. 2019, 97, 124–133. [Google Scholar] [CrossRef]
- Zhong, X.; Lee, H.N.; Kim, S.H.; Park, S.A.; Kim, W.; Cha, Y.N.; Surh, Y.J. Myc-nick promotes efferocytosis through M2 macrophage polarization during resolution of inflammation. Faseb. J. 2018, 32, 5312–5325. [Google Scholar] [CrossRef]
- Sun, L.; Zhou, H.; Zhu, Z.; Yan, Q.; Wang, L.; Liang, Q.; Richard, D.Y. Ex vivo and in vitro effect of serum amyloid a in the induction of macrophage M2 markers and efferocytosis of apoptotic neutrophils. J. Immunol. 2015, 194, 4891–4900. [Google Scholar] [CrossRef]
- Yang, M.; Liu, J.; Piao, C.; Shao, J.; Du, J. ICAM-1 suppresses tumor metastasis by inhibiting macrophage M2 polarization through blockade of efferocytosis. Cell Death Dis. 2015, 6, e1780. [Google Scholar] [CrossRef]
- Singh, V.; Davidson, A.C.; Hume, P.J.; Koronakis, V. Arf6 Can Trigger Wave Regulatory Complex-Dependent Actin Assembly Independent of Arno. Int. J. Mol. Sci. 2020, 21, 2457. [Google Scholar] [CrossRef]
- Marchesin, V.; Montagnac, G.; Chavrier, P. ARF6 promotes the formation of Rac1 and WAVE-dependent ventral F-actin rosettes in breast cancer cells in response to epidermal growth factor. PLoS ONE 2015, 10, e0121747. [Google Scholar] [CrossRef]
- Tarricone, C.; Xiao, B.; Justin, N.; Walker, P.; Rittinger, K.; Gamblin, S.; Smerdon, S. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 2001, 411, 215–219. [Google Scholar] [CrossRef]
- Honda, A.; Nogami, M.; Yokozeki, T.; Yamazaki, M.; Nakamura, H.; Watanabe, H.; Kawamoto, K.; Nakayama, K.; Morris, A.J.; Frohman, M.A. Phosphatidylinositol 4-phosphate 5-kinase α is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 1999, 99, 521–532. [Google Scholar] [CrossRef]
- Proto, J.D.; Doran, A.C.; Gusarova, G.; Yurdagul, A., Jr.; Sozen, E.; Subramanian, M.; Islam, M.N.; Rymond, C.C.; Du, J.; Hook, J.; et al. Regulatory T Cells Promote Macrophage Efferocytosis during Inflammation Resolution. Immunity 2018, 49, 666–677.e6. [Google Scholar] [CrossRef]
- Tao, H.; Yancey, P.G.; Babaev, V.R.; Blakemore, J.L.; Zhang, Y.; Ding, L.; Fazio, S.; Linton, M.F. Macrophage SR-BI mediates efferocytosis via Src/PI3K/Rac1 signaling and reduces atherosclerotic lesion necrosis. J. Lipid. Res. 2015, 56, 1449–1460. [Google Scholar] [CrossRef]
- Ravichandran, S.K.; Lorenz, U. Engulfment of apoptotic cells: Signals for a good meal. Nat. Rev. Immunol. 2007, 7, 964–974. [Google Scholar] [CrossRef]
- Schirmer, L.; Velmeshev, D.; Holmqvist, S.; Kaufmann, M.; Werneburg, S.; Jung, D.; Vistnes, S.; Stockley, J.H.; Young, A.; Steindel, M.; et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 2019, 573, 75–82. [Google Scholar] [CrossRef]
- Ungerbäck, J.; Hosokawa, H.; Wang, X.; Strid, T.; Williams, B.A.; Sigvardsson, M.; Rothenberg, E.V. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1). Genome Res. 2018, 28, 1508–1519. [Google Scholar] [CrossRef]
- Ha, S.D.; Cho, W.; DeKoter, R.P.; Kim, S.O. The transcription factor PU.1 mediates enhancer-promoter looping that is required for IL-1β eRNA and mRNA transcription in mouse melanoma and macrophage cell lines. J. Biol. Chem. 2019, 294, 17487–17500. [Google Scholar] [CrossRef]
- Nurminen, V.; Neme, A.; Seuter, S.; Carlberg, C. Modulation of vitamin D signaling by the pioneer factor CEBPA. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 96–106. [Google Scholar] [CrossRef]
- Hrecka, K.; Hao, C.; Gierszewska, M.; Swanson, S.K.; Kesik-Brodacka, M.; Srivastava, S.; Florens, L.; Washburn, M.P.; Skowronski, J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 2011, 474, 658–661. [Google Scholar] [CrossRef]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Ségéral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef]
- Ismailova, A.; White, J.H. Vitamin D, infections and immunity. Rev. Endocr. Metab. Disord. 2022, 23, 265–277. [Google Scholar] [CrossRef]
- White, J.H. Emerging Roles of Vitamin D-Induced Antimicrobial Peptides in Antiviral Innate Immunity. Nutrients 2022, 14, 284. [Google Scholar] [CrossRef]
- Bist, P.; Kim, S.S.; Pulloor, N.K.; McCaffrey, K.; Nair, S.K.; Liu, Y.; Lin, R.; Krishnan, M.N. ArfGAP Domain-Containing Protein 2 (ADAP2) Integrates Upstream and Downstream Modules of RIG-I Signaling and Facilitates Type I Interferon Production. Mol. Cell Biol. 2017, 37, e00537-16. [Google Scholar] [CrossRef]
- Shu, Q.; Lennemann, N.J.; Sarkar, S.N.; Sadovsky, Y.; Coyne, C.B. ADAP2 Is an Interferon Stimulated Gene That Restricts RNA Virus Entry. PLoS Pathog. 2015, 11, e1005150. [Google Scholar] [CrossRef]
- International Multiple Sclerosis Genetics Consortium (IMSGC). Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef]
- Cantorna, T.M.; Hayes, C.E.; DeLuca, H.F. 1,25-Dihydroxyvitamin D3 reversibly blocks the progression of relapsing encephalomyelitis, a model of multiple sclerosis. Proc. Natl. Acad. Sci. USA 1996, 93, 7861–7864. [Google Scholar] [CrossRef]
- DeLuca, F.H.; Plum, L.A. Vitamin D deficiency diminishes the severity and delays onset of experimental autoimmune encephalomyelitis. Arch. Biochem. Biophys. 2011, 513, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Fernandes de Abreu, D.A.; Ibrahim, E.C.; Boucraut, J.; Khrestchatisky, M.; Féron, F. Severity of experimental autoimmune encephalomyelitis is unexpectedly reduced in mice born to vitamin D-deficient mothers. J. Steroid. Biochem. Mol. Biol. 2010, 121, 250–253. [Google Scholar] [CrossRef]
- Adzemovic, M.Z.; Zeitelhofer, M.; Hochmeister, S.; Gustafsson, S.A.; Jagodic, M. Efficacy of vitamin D in treating multiple sclerosis-like neuroinflammation depends on developmental stage. Exp. Neurol. 2013, 249, 39–48. [Google Scholar] [CrossRef]
- Wang, Y.; Marling, S.; Zhu, J.; Severson, K.; DeLuca, H. Development of experimental autoimmune encephalomyelitis (EAE) in mice requires vitamin D and the vitamin D receptor. Proc. Natl. Acad. Sci. USA 2012, 109, 8501–8504. [Google Scholar] [CrossRef] [PubMed]
- Caturano, A.; Brunelli, V.; Galiero, R.; Spiezia, S.; Ferrara, R.; Sasso, F.C. Comment on: Effect of Vitamin D supplementation in patients with liver cirrhosis having spontaneous bacterial peritonitis: A randomized controlled study. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 2220–2221. [Google Scholar]
- Kerschbaum, J.; Vychytil, A.; Lhotta, K.; Prischl, F.C.; Wiesholzer, M.; Machhold-Fabrizii, V.; Kopriva-Altfahrt, G.; Schwarz, C.; Balcke, P.; Oberbauer, R.; et al. Treatment with oral active vitamin D is associated with decreased risk of peritonitis and improved survival in patients on peritoneal dialysis. PLoS ONE 2013, 8, e67836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-h.; Xu, X.; Pi, H.-c.; Yang, Z.-k.; Johnson, D.W.; Dong, J. The effects of oral vitamin D supplementation on the prevention of peritoneal dialysis-related peritonitis: Study protocol for a randomized controlled clinical trial. Trials 2019, 20, 657. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; Duan, J.; Wang, J.; Li, H.; Wu, Z.; Wang, S.; Wu, X.; Lu, M. 1,25(OH)2D3 Promotes Macrophage Efferocytosis Partly by Upregulating ASAP2 Transcription via the VDR-Bound Enhancer Region and ASAP2 May Affect Antiviral Immunity. Nutrients 2022, 14, 4935. https://doi.org/10.3390/nu14224935
Shi H, Duan J, Wang J, Li H, Wu Z, Wang S, Wu X, Lu M. 1,25(OH)2D3 Promotes Macrophage Efferocytosis Partly by Upregulating ASAP2 Transcription via the VDR-Bound Enhancer Region and ASAP2 May Affect Antiviral Immunity. Nutrients. 2022; 14(22):4935. https://doi.org/10.3390/nu14224935
Chicago/Turabian StyleShi, Hui, Jiangling Duan, Jiayu Wang, Haohao Li, Zhiheng Wu, Shuaideng Wang, Xueyan Wu, and Ming Lu. 2022. "1,25(OH)2D3 Promotes Macrophage Efferocytosis Partly by Upregulating ASAP2 Transcription via the VDR-Bound Enhancer Region and ASAP2 May Affect Antiviral Immunity" Nutrients 14, no. 22: 4935. https://doi.org/10.3390/nu14224935
APA StyleShi, H., Duan, J., Wang, J., Li, H., Wu, Z., Wang, S., Wu, X., & Lu, M. (2022). 1,25(OH)2D3 Promotes Macrophage Efferocytosis Partly by Upregulating ASAP2 Transcription via the VDR-Bound Enhancer Region and ASAP2 May Affect Antiviral Immunity. Nutrients, 14(22), 4935. https://doi.org/10.3390/nu14224935





