Investigating Effects of Plasma Apolipoprotein E on Ischemic Heart Disease Using Mendelian Randomization Study
Abstract
1. Background
2. Materials & Methods
2.1. Study Design
2.2. Data Sources
2.2.1. Genetic Predictors of Plasma ApoE Isoforms
2.2.2. Genetic Associations with IHD, LDL Cholesterol, HDL Cholesterol, Triglycerides and ApoB
2.3. Statistical Aanalyses
2.3.1. Univariable MR Analyses
Main Analyses
Sensitivity Analyses
2.3.2. Multivariable MR Analyses
3. Results
3.1. Instrument Strength and Power Calculations
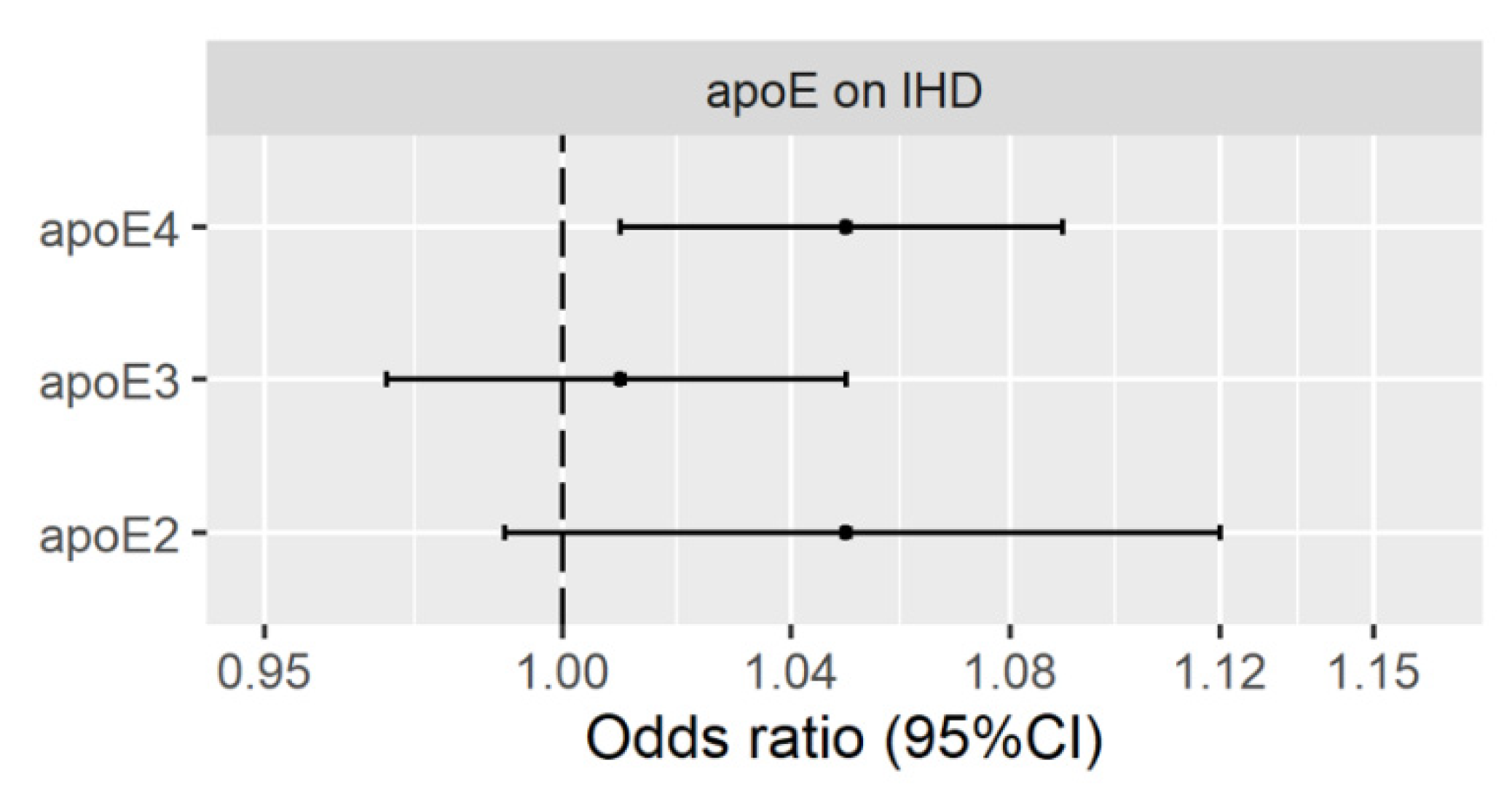
3.2. Genetically Predicted Plasma ApoE (ApoE2, ApoE3 and ApoE4) on IHD
Validation Using Genetic Instruments from the INTERVAL Study
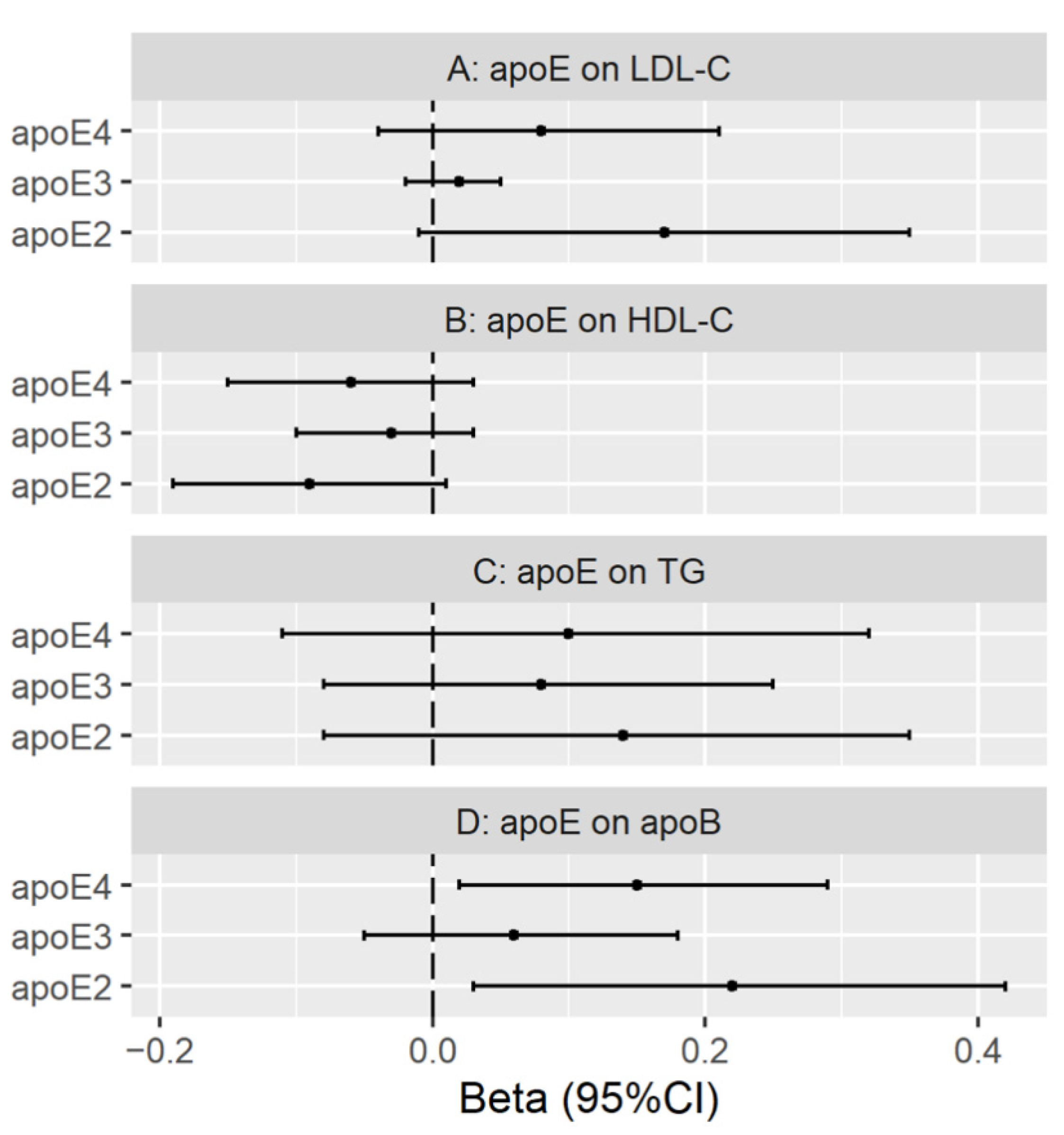
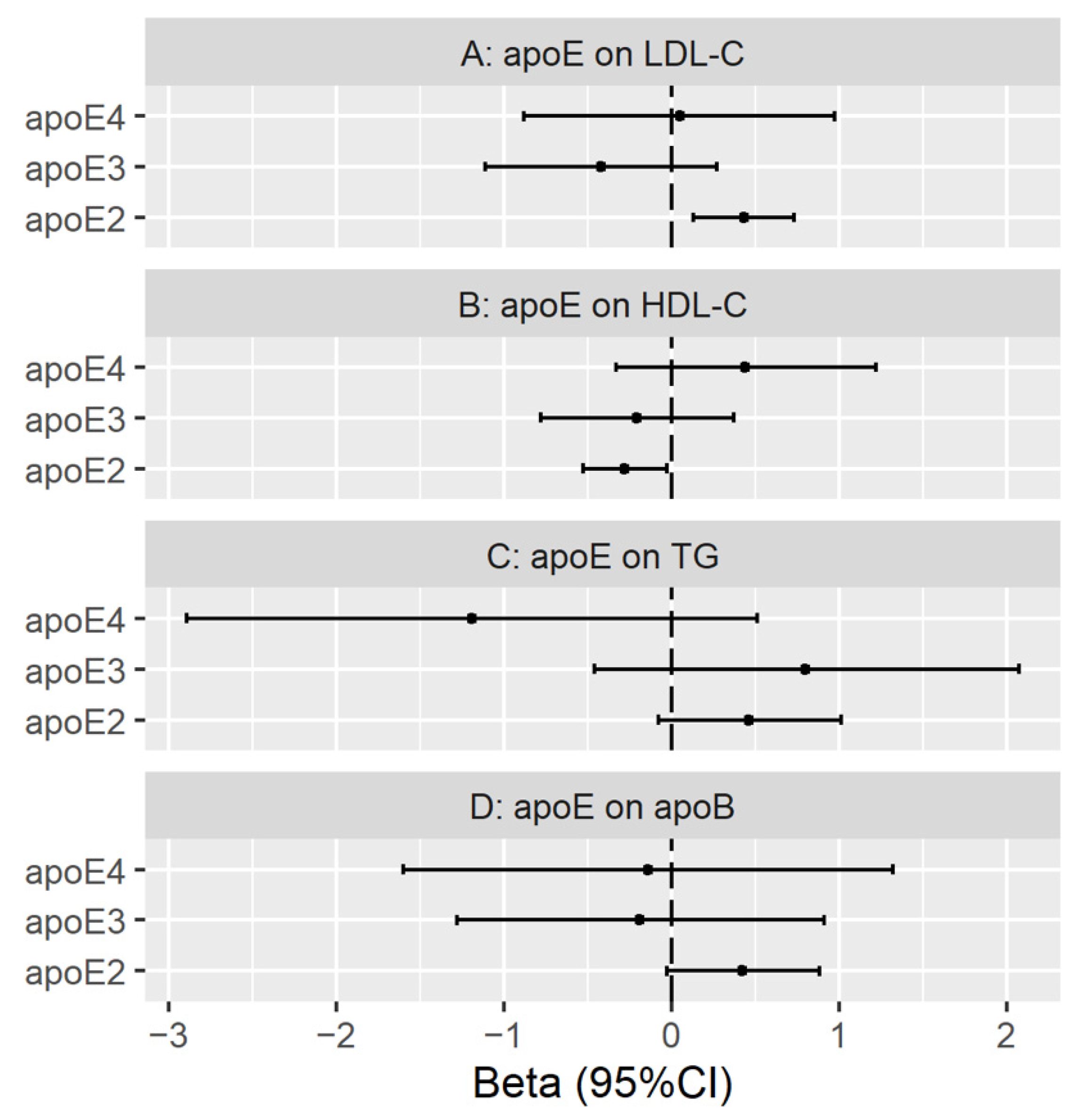
3.3. Genetically Predicted Plasma ApoE (ApoE2, ApoE3 and ApoE4) on LDL Cholesterol, HDL Cholesterol, Triglycerides and ApoB
Validation with Genetic Instruments from the INTERVAL Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Consent for Publication
Acknowledgments
Conflicts of Interest
References
- Mahley, R.W. Apolipoprotein E: From cardiovascular disease to neurodegenerative disorders. J. Mol. Med. 2016, 94, 739–746. [Google Scholar] [CrossRef]
- Boerwinkle, E.; Utermann, G. Simultaneous effects of the apolipoprotein E polymorphism on apolipoprotein E, apolipoprotein B, and cholesterol metabolism. Am. J. Hum. Genet. 1988, 42, 104–112. [Google Scholar]
- Smit, M.; de Knijff, P.; Rosseneu, M.; Bury, J.; Klasen, E.; Frants, R.; Havekes, L. Apolipoprotein E polymorphism in the Netherlands and its effect on plasma lipid and apolipoprotein levels. Qual. Life Res. 1988, 80, 287–292. [Google Scholar] [CrossRef]
- Richardson, T.G.; Sanderson, E.; Palmer, T.M.; Ala-Korpela, M.; Ference, B.A.; Smith, G.D.; Holmes, M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020, 17, e1003062. [Google Scholar] [CrossRef]
- Ference, B.A.; Kastelein, J.J.P.; Ray, K.K.; Ginsberg, H.N.; Chapman, M.J.; Packard, C.J.; Laufs, U.; Oliver-Williams, C.; Wood, A.M.; Butterworth, A.S.; et al. Association of Triglyceride-Lowering LPL Variants and LDL-C–Lowering LDLR Variants With Risk of Coronary Heart Disease. JAMA 2019, 321, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Ference, B.A.; Kastelein, J.J.P.; Ginsberg, H.N.; Chapman, M.J.; Nicholls, S.; Ray, K.K.; Packard, C.J.; Laufs, U.; Brook, R.D.; Oliver-Williams, C.; et al. Association of Genetic Variants Related to CETP Inhibitors and Statins With Lipoprotein Levels and Cardiovascular Risk. JAMA 2017, 318, 947–956. [Google Scholar] [CrossRef]
- Zuber, V.; Gill, D.; Ala-Korpela, M.; Langenberg, C.; Butterworth, A.; Bottolo, L.; Burgess, S. High-throughput multivariable Mendelian randomization analysis prioritizes apolipoprotein B as key lipid risk factor for coronary artery disease. Int. J. Epidemiol. 2020. [Google Scholar] [CrossRef]
- Kronfeldner, M. Commentary: How norms make causes. Int. J. Epidemiol. 2014, 43, 1707–1713. [Google Scholar] [CrossRef]
- Hubacek, J.A.; Vrablik, M. Effect of apolipoprotein E polymorphism on statin-induced decreases in plasma lipids and cardiovascular events. Drug Metab. Drug Interact. 2011, 26, 13–20. [Google Scholar] [CrossRef]
- Cohn, J.S.; Tremblay, M.; Batal, R.; Jacques, H.; Veilleux, L.; Rodriguez, C.; Barrett, P.H.; Dubreuil, D.; Roy, M.; Bernier, L.; et al. Effect of atorvastatin on plasma apoE metabolism in patients with combined hyperlipidemia. J. Lipid Res. 2002, 43, 1464–1471. [Google Scholar] [CrossRef]
- Ooi, E.; Ng, T.W.; Watts, G.; Chan, D.C.; Barrett, P.H.R. Effect of fenofibrate and atorvastatin on VLDL apoE metabolism in men with the metabolic syndrome. J. Lipid Res. 2012, 53, 2443–2449. [Google Scholar] [CrossRef] [PubMed]
- Hatters, D.M.; Peters-Libeu, C.A.; Weisgraber, K.H. Apolipoprotein E structure: Insights into function. Trends Biochem. Sci. 2006, 31, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.T.; Kuzawa, C.W.; Hayes, M.G. Worldwide allele frequencies of the human apolipoprotein E gene: Climate, local adaptations, and evolutionary history. Am. J. Phys. Anthr. 2010, 143, 100–111. [Google Scholar] [CrossRef]
- Bennet, A.M.; Di Angelantonio, E.; Ye, Z.; Wensley, F.; Dahlin, A.; Ahlbom, A.; Keavney, B.; Collins, R.; Wiman, B.; De Faire, U.; et al. Association of Apolipoprotein E Genotypes with Lipid Levels and Coronary Risk. JAMA 2007, 298, 1300–1311. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, H.-Q.; Peng, W.-J.; Zhang, B.-B.; Liu, M. Meta-analysis for the Association of Apolipoprotein E ε2/ε3/ε4 Polymorphism with Coronary Heart Disease. Chin. Med. J. 2015, 128, 1391–1398. [Google Scholar] [CrossRef]
- Xu, H.; Li, H.; Liu, J.; Zhu, D.; Wang, Z.; Chen, A.; Zhao, Q. Meta-Analysis of Apolipoprotein E Gene Polymorphism and Susceptibility of Myocardial Infarction. PLoS ONE 2014, 9, e104608. [Google Scholar] [CrossRef][Green Version]
- Xu, M.; Zhao, J.; Zhang, Y.; Ma, X.; Dai, Q.; Zhi, H.; Wang, B.; Wang, L. Apolipoprotein E Gene Variants and Risk of Coronary Heart Disease: A Meta-Analysis. BioMed Res. Int. 2016, 2016, 1–12. [Google Scholar] [CrossRef]
- Zhao, Q.R.; Lei, Y.Y.; Li, J.; Jiang, N.; Shi, J.P. Association between apolipoprotein E polymorphisms and premature coronary artery disease: A meta-analysis. Clin. Chem. Lab. Med. 2017, 55, 284–298. [Google Scholar] [CrossRef]
- Griffin, B.A.; Walker, C.G.; Jebb, S.A.; Moore, C.; Frost, G.S.; Goff, L.; Sanders, T.A.B.; Lewis, F.; Griffin, M.; Gitau, R.; et al. APOE4 Genotype Exerts Greater Benefit in Lowering Plasma Cholesterol and Apolipoprotein B than Wild Type (E3/E3), after Replacement of Dietary Saturated Fats with Low Glycaemic Index Carbohydrates. Nutrients 2018, 10, 1524. [Google Scholar] [CrossRef]
- Khan, T.; Shah, T.; Prieto-Merino, D.; Zhang, W.; Price, J.; Fowkes, G.R.; Cooper, J.; Talmud, P.J.; Humphries, S.E.; Sundstrom, J.; et al. Apolipoprotein E genotype, cardiovascular biomarkers and risk of stroke: Systematic review and meta-analysis of 14 015 stroke cases and pooled analysis of primary biomarker data from up to 60 883 individuals. Int. J. Epidemiol. 2013, 42, 475–492. [Google Scholar] [CrossRef]
- Soares, H.D. Plasma Biomarkers Associated With the Apolipoprotein E Genotype and Alzheimer Disease. Arch. Neurol. 2012, 69, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Lumsden, A.L.; Mulugeta, A.; Zhou, A.; Hyppönen, E. Apolipoprotein E (APOE) genotype-associated disease risks: A phenome-wide, registry-based, case-control study utilising the UK Biobank. EBioMedicine 2020, 59, 102954. [Google Scholar] [CrossRef]
- Wilson, P.W.; Schaefer, E.J.; Larson, M.; Ordovas, J.M. Apolipoprotein E Alleles and Risk of Coronary Disease. Arter. Thromb. Vasc. Biol. 1996, 16, 1250–1255. [Google Scholar] [CrossRef]
- Phillips, M.C. Apolipoprotein E isoforms and lipoprotein metabolism. IUBMB Life 2014, 66, 616–623. [Google Scholar] [CrossRef]
- Feussner, G.; Wagner, A.; Ziegler, R. Relation of cardiovascular risk factors to atherosclerosis in type III hyperlipoproteinemia. Qual. Life Res. 1993, 92, 122–126. [Google Scholar] [CrossRef]
- Rasmussen, K.L.; Tybjærg-Hansen, A.; Nordestgaard, B.G.; Frikke-Schmidt, R. Plasma levels of apolipoprotein E and risk of ischemic heart disease in the general population. Atherosclerosis 2016, 246, 63–70. [Google Scholar] [CrossRef]
- Reilly, M.; Rader, D.J. Apolipoprotein E and Coronary Disease: A Puzzling Paradox. PLoS Med. 2006, 3, e258. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Harbord, R.; Sterne, J.; Timpson, N.; Smith, G.D. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef]
- Rasmussen, K.L.; Tybjærg-Hansen, A.; Nordestgaard, B.G.; Frikke-Schmidt, R. Plasma levels of apolipoprotein E, APOE genotype, and all-cause and cause-specific mortality in 105 949 individuals from a white general population cohort. Eur. Hear. J. 2019, 40, 2813–2824. [Google Scholar] [CrossRef]
- Suhre, K.; Arnold, M.; Bhagwat, A.M.; Cotton, R.J.; Engelke, R.; Raffler, J.; Sarwath, H.; Thareja, G.; Wahl, A.; Delisle, R.K.; et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017, 8, 14357. [Google Scholar] [CrossRef]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic atlas of the human plasma proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef]
- Nelson, C.P.; Goel, A.; Butterworth, A.S.; Kanoni, S.; Webb, T.; Marouli, E.; Zeng, L.; Ntalla, I.; Lai, F.Y.; Hopewell, J.C.; et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017, 49, 1385–1391. [Google Scholar] [CrossRef]
- Neale lab UK Biobank GWAS round 2 results. Available online: http://www.nealelab.is/uk-biobank/ (accessed on 15 October 2019).
- Kettunen, J.; Demirkan, A.; Würtz, P.; Draisma, H.H.M.; Haller, T.; Rawal, R.; Vaarhorst, A.; Kangas, A.J.; Lyytikaeinen, L.-P.; Pirinen, M.; et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 2016, 7, 11122. [Google Scholar] [CrossRef]
- Kraemer, S.; Vaught, J.D.; Bock, C.; Gold, L.; Katilius, E.; Keeney, T.R.; Kim, N.; Saccomano, N.A.; Wilcox, S.K.; Zichi, D.; et al. From SOMAmer-Based Biomarker Discovery to Diagnostic and Clinical Applications: A SOMAmer-Based, Streamlined Multiplex Proteomic Assay. PLoS ONE 2011, 6, e26332. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Smith, G.D.; Sheehan, N.; Thompson, J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I2 statistic. Int. J. Epidemiol. 2016, 45, 1961–1974. [Google Scholar] [CrossRef]
- Freeman, G.; Cowling, B.J.; Schooling, C.M. Power and sample size calculations for Mendelian randomization studies using one genetic instrument. Int. J. Epidemiol. 2013, 42, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, S.; Chen, C.-C.; Lan, L. GWAPower: A statistical power calculation software for genome-wide association studies with quantitative traits. BMC Genet. 2011, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian Randomization Analysis with Multiple Genetic Variants Using Summarized Data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Sedgwick, P. Multiple hypothesis testing and Bonferroni’s correction. BMJ 2014, 349, g6284. [Google Scholar] [CrossRef]
- Hemani, G.; Tilling, K.; Smith, G.D. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017, 13, e1007081. [Google Scholar] [CrossRef]
- Bowden, J.K.; Smith, G.D.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Smith, G.D.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Re: “Multivariable Mendelian Randomization: The Use of Pleiotropic Genetic Variants to Estimate Causal Effects”. Am. J. Epidemiol. 2015, 181, 290–291. [Google Scholar] [CrossRef]
- Rees, J.M.B.; Wood, A.M.; Burgess, S. Extending the MR-Egger method for multivariable Mendelian randomization to correct for both measured and unmeasured pleiotropy. Stat. Med. 2017, 36, 4705–4718. [Google Scholar] [CrossRef]
- Sanderson, E.; Smith, G.D.; Windmeijer, F.; Bowden, J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int. J. Epidemiol. 2019, 48, 713–727. [Google Scholar] [CrossRef]
- Mooijaart, S.P.; Berbée, J.F.P.; Van Heemst, D.; Havekes, L.M.; De Craen, A.J.M.; Slagboom, P.E.; Rensen, P.C.N.; Westendorp, R.G.J. ApoE Plasma Levels and Risk of Cardiovascular Mortality in Old Age. PLoS Med. 2006, 3, e176. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.L. Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: A review. Atherosclerosis 2016, 255, 145–155. [Google Scholar] [CrossRef]
- Lawlor, D.A. Commentary: Two-sample Mendelian randomization: Opportunities and challenges. Int. J. Epidemiol. 2016, 45, 908–915. [Google Scholar] [CrossRef]
- Deelen, J.; Evans, D.S.; Arking, D.E.; Tesi, N.; Nygaard, M.; Liu, X.; Wojczynski, M.K.; Biggs, M.L.; van der Spek, A.; Atzmon, G.; et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Schooling, C.M.; Lopez, P.M.; Yang, Z.; Zhao, J.V.; Yeung, S.L.A.; Huang, J.V. Use of Multivariable Mendelian Randomization to Address Biases Due to Competing Risk Before Recruitment. Front. Genet. 2021, 11. [Google Scholar] [CrossRef]
- Burgess, S.; Davies, N.; Thompson, S.G. Bias due to participant overlap in two-sample Mendelian randomization. Genet. Epidemiol. 2016, 40, 597–608. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.-Y.; Kwok, M.-K.; Schooling, C.M. Investigating Effects of Plasma Apolipoprotein E on Ischemic Heart Disease Using Mendelian Randomization Study. Nutrients 2021, 13, 2215. https://doi.org/10.3390/nu13072215
Li M-Y, Kwok M-K, Schooling CM. Investigating Effects of Plasma Apolipoprotein E on Ischemic Heart Disease Using Mendelian Randomization Study. Nutrients. 2021; 13(7):2215. https://doi.org/10.3390/nu13072215
Chicago/Turabian StyleLi, Meng-Yu, Man-Ki Kwok, and Catherine Mary Schooling. 2021. "Investigating Effects of Plasma Apolipoprotein E on Ischemic Heart Disease Using Mendelian Randomization Study" Nutrients 13, no. 7: 2215. https://doi.org/10.3390/nu13072215
APA StyleLi, M.-Y., Kwok, M.-K., & Schooling, C. M. (2021). Investigating Effects of Plasma Apolipoprotein E on Ischemic Heart Disease Using Mendelian Randomization Study. Nutrients, 13(7), 2215. https://doi.org/10.3390/nu13072215




