Plasma and Urinary Amino Acid-Derived Catabolites as Potential Biomarkers of Protein and Amino Acid Deficiency in Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Pellet Preparation
2.3. Experimental Design
2.4. Urine Sampling
2.5. Metabolomics Sample Preparation and Liquid-Chromatography Mass Spectrometry (LC-MS) Analysis of Urine and Plasma
2.6. LC-MS Data Processing and Analysis
2.6.1. Data Pretreatment
2.6.2. Development of a Multivariate Model
Partial Least Square (PLS) Regression
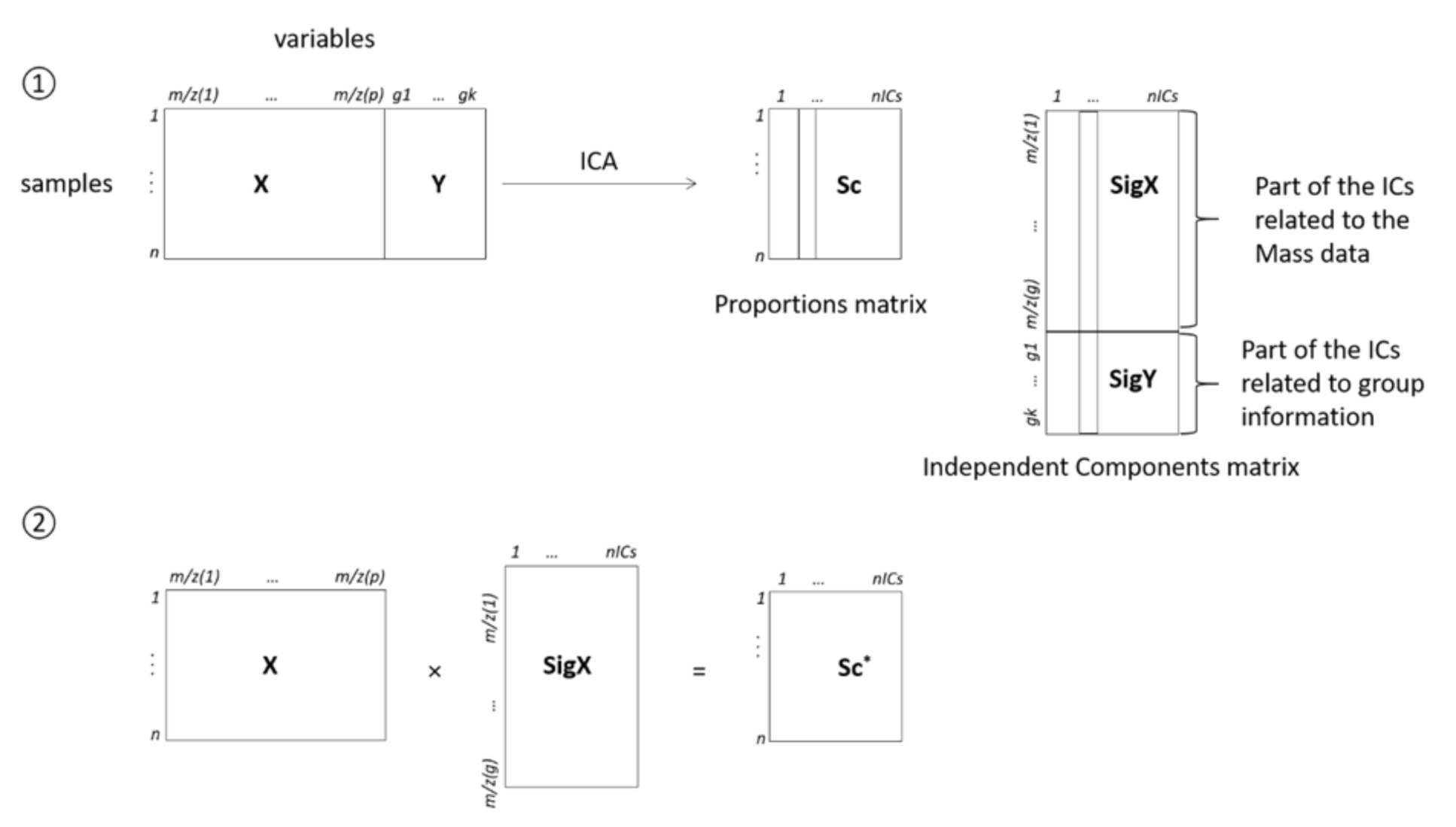
Independent Component Discriminant Analysis (ICDA)
2.6.3. Analysis by Univariate ANOVA Model for Discriminant Metabolites
3. Results
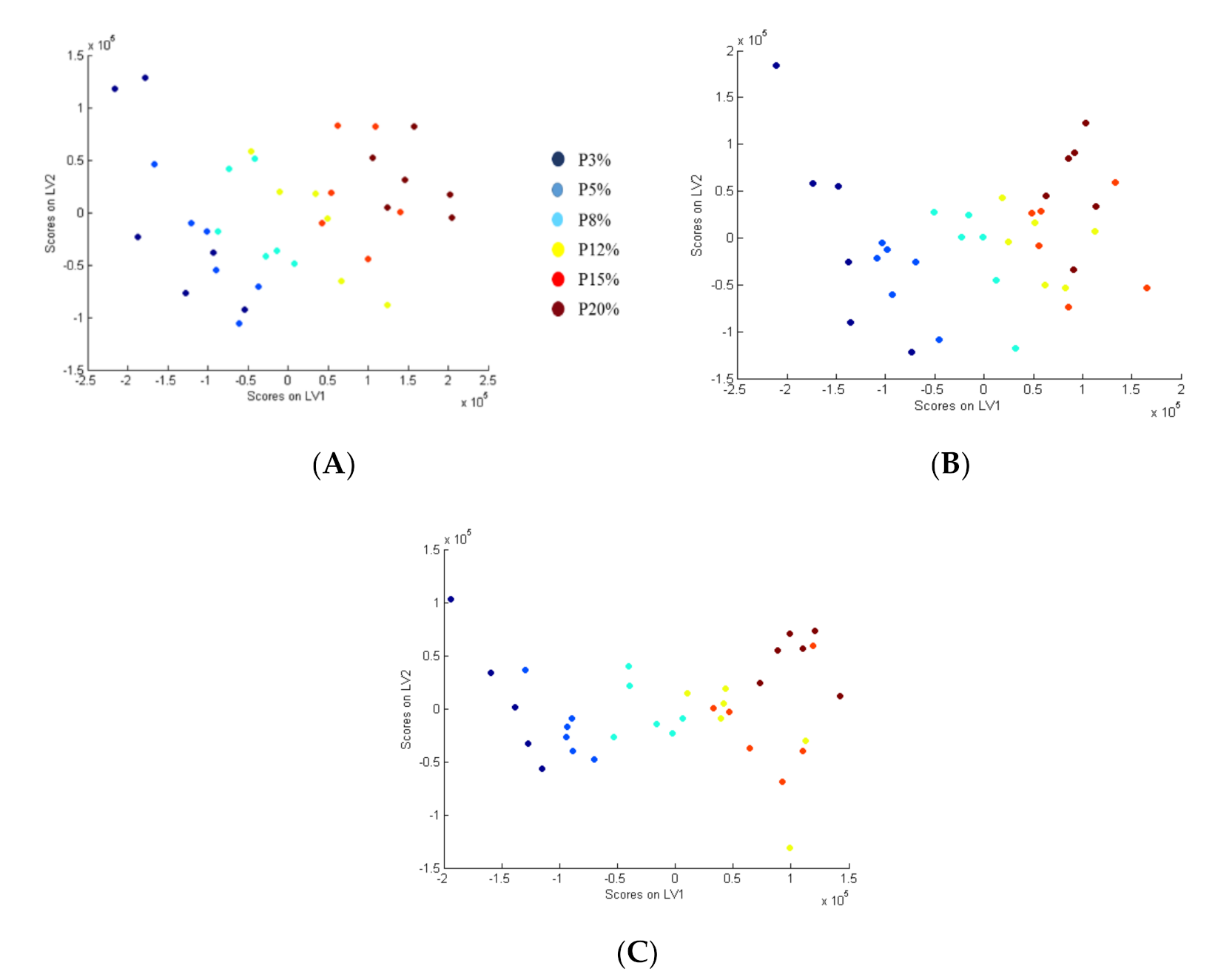
3.1. Urinary and Plasma Biomarkers Obtained with PLS Method
3.2. Urinary and Plasma Biomarkers Obtained with ICDA Method
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- ANSES. Apport en Protéines: Consommation, Qualité, Besoins et Recommandations; ANSES: Maisons-Alfort, France, 2007. [Google Scholar]
- Rand, W.M.; Young, V.R. Statistical analysis of nitrogen balance data with reference to the lysine requirement in adults. J. Nutr. 1999, 129, 1920–1926. [Google Scholar] [CrossRef] [PubMed]
- Young, V.R.; Marchini, J.S. Mechanisms and nutritional significance of metabolic responses to altered intakes of protein and amino acids, with reference to nutritional adaptation in humans. Am. J. Clin. Nutr. 1990, 51, 270–289. [Google Scholar] [CrossRef] [PubMed]
- Kurpad, A.V.; Thomas, T. Methods to assess amino acid requirements in humans. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Young, V.R.; Bier, D.M.; Pellett, P.L. A theoretical basis for increasing current estimates of the amino acid requirements in adult man, with experimental support. Am. J. Clin. Nutr. 1989, 50, 80–92. [Google Scholar] [CrossRef]
- Pencharz, P.B.; Ball, R.O. Different approaches to define individual amino acid requirements. Annu. Rev. Nutr. 2003, 23, 101–116. [Google Scholar] [CrossRef]
- WHO/FAO/UNU. Protein and Amino Acid Requirements in Human Nutrition; WHO: Geneva, Switzerland, 2007; pp. 1–265. [Google Scholar]
- FAO/WHO/UNU. Dietary Protein Quality Evaluation in Human Nutrition: Report of an FAO Expert Consultation; FAO: Rome, Italy, 2013. [Google Scholar]
- Rosique, C.; Lebsir, D.; Benatia, S.; Guigon, P.; Caire-Maurisier, F.; Benderitter, M.; Souidi, M.; Martin, J.-C. Metabolomics evaluation of repeated administration of potassium iodide on adult male rats. Arch. Toxicol. 2020, 94, 803–812. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Bouveresse, D.J.; Rutledge, D.N. Chapter 7-Independent Components Analysis: Theory and Applications. In Data Handling in Science and Technology no. 30; Ruckebusch, C., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 225–277. [Google Scholar]
- Habchi, B.; Alves, S.; Bouveresse, D.J.-R.; Moslah, B.; Paris, A.; Lécluse, Y.; Gauduchon, P.; LeBailly, P.; Rutledge, D.N.; Rathahao-Paris, E. An innovative chemometric method for processing direct introduction high resolution mass spectrometry metabolomic data: Independent component–discriminant analysis (IC–DA). Metabolomics 2017, 13, 45. [Google Scholar] [CrossRef]
- Khodorova, N.V.; Rutledge, D.N.; Oberli, M.; Mathiron, D.; Marcelo, P.; Benamouzig, R.; Tome, D.; Gaudichon, C.; Pilard, S. Urinary Metabolomics Profiles Associated to Bovine Meat Ingestion in Humans. Mol. Nutr. Food Res. 2019, 63, e1700834. [Google Scholar] [CrossRef]
- Horiuchi, M.; Takeda, T.; Takanashi, H.; Ozaki-Masuzawa, Y.; Taguchi, Y.; Toyoshima, Y.; Otani, L.; Kato, H.; Sone-Yonezawa, M.; Hakuno, F.; et al. Branched-chain amino acid supplementation restores reduced insulinotropic activity of a low-protein diet through the vagus nerve in rats. Nutr. Metab. 2017, 14. [Google Scholar] [CrossRef]
- Qiu, K.; Qin, C.F.; Luo, M.; Zhang, X.; Sun, W.J.; Jiao, N.; Li, D.F.; Yin, J.D. Protein Restriction with Amino Acid-Balanced Diets Shrinks Circulating Pool Size of Amino Acid by Decreasing Expression of Specific Transporters in the Small Intestine. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Torres, N.; Beristain, L.; Bourges, H.; Tovar, A.R. Histidine-imbalanced diets stimulate hepatic histidase gene expression in rats. J. Nutr. 1999, 129, 1979–1983. [Google Scholar] [CrossRef][Green Version]
- Torres, N.; Martínez, L.; Alemán, G.; Bourges, H.; Tovar, A.R. Histidase Expression Is Regulated by Dietary Protein at the Pretranslational Level in Rat Liver. J. Nutr. 1998, 128, 818–824. [Google Scholar] [CrossRef][Green Version]
- Mercer, L.P.; Dodds, S.J.; Schweisthal, M.R.; Dunn, J.D. Brain histidine and food intake in rats fed diets deficient in single amino acids. J. Nutr. 1989, 119, 66–74. [Google Scholar] [CrossRef]
- Adibi, S.A.; Modesto, T.A.; Morse, E.L.; Amin, P.M. Amino acid levels in plasma, liver, and skeletal muscle during protein deprivation. Am. J. Physiol. 1973, 225, 408–414. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Uppal, S.O.; Moorman, J.L.; Bennett, C.; Gruca, L.L.; Parimi, P.S.; Dasarathy, S.; Serre, D.; Hanson, R.W. Metabolic and Genomic Response to Dietary Isocaloric Protein Restriction in the Rat. J. Biol. Chem. 2011, 286, 5266–5277. [Google Scholar] [CrossRef]
- Nagao, K.; Bannai, M.; Seki, S.; Mori, M.; Takahashi, M. Adaptational modification of serine and threonine metabolism in the liver to essential amino acid deficiency in rats. Amino Acids 2009, 36, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Tominari, T.; Hirata, M.; Matsumoto, C.; Hirata, J.; Murphy, G.; Nagase, H.; Miyaura, C.; Inada, M. Indoxyl sulfate, a uremic toxin in chronic kidney disease, suppresses both bone formation and bone resorption. FEBS Open Bio. 2017, 7, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; A Soltow, Q.; E Mitchell, S.; Derous, D.; Wang, Y.; Chen, L.; Han, J.-D.J.; Promislow, D.E.L.; Lusseau, D.; Douglas, A.; et al. The Effects of Graded Levels of Calorie Restriction: XIII. Global Metabolomics Screen Reveals Graded Changes in Circulating Amino Acids, Vitamins, and Bile Acids in the Plasma of C57BL/6 Mice. J. Gerontol. A Biol. Sci. Med. Sci. 2019, 74, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Kimura, N.; Fukuwatari, T.; Sasaki, R.; Shibata, K. Comparison of Metabolic Fates of Nicotinamide, NAD+ and NADH Administered Orally and Intraperitoneally; Characterization of Oral NADH. J. Nutr. Sci. Vitaminol. 2006, 52, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Shibata, K.; Shiotani, M.; Onodera, M.; Suzuki, T. Effects of Protein-Free Diet Feeding or Starving on the Excretion Ratio of (N1-Methyl-2-pyridone-5-carboxamide + N1-methl-4-pyridone-3-carboxamideJ/N1-Methylnicotinamide. Agric. Biol. Chem. 1991, 55, 1483–1490. [Google Scholar] [CrossRef]
- Bortolotti, M.; Kreis, R.; Debard, C.; Cariou, B.; Faeh, D.; Chetiveaux, M.; Ith, M.; Vermathen, P.; Stefanoni, N.; Lê, K.-A.; et al. High protein intake reduces intrahepatocellular lipid deposition in humans. Am. J. Clin. Nutr. 2009, 90, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Morton, G.J.; Kaiyala, K.J.; Foster-Schubert, K.E.; Cummings, D.E.; Schwartz, M.W. Carbohydrate Feeding Dissociates the Postprandial FGF19 Response From Circulating Bile Acid Levels in Humans. J. Clin. Endocrinol. Metab. 2014, 99, E241–E245. [Google Scholar] [CrossRef]
- Bollard, M.E.; Stanley, E.G.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. Nmr. Biomed. 2005, 18, 143–162. [Google Scholar] [CrossRef]
- Wu, Z.; Li, M.; Zhao, C.; Zhou, J.; Chang, Y.; Li, X.; Gao, P.; Lü, X.; Li, Y.; Xu, G. Urinary metabonomics study in a rat model in response to protein-energy malnutrition by using gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry. Mol. Biosyst. 2010, 6, 2157–2163. [Google Scholar] [CrossRef]

| P3 | P5 | P8 | P12 | P15 | P20 | |
|---|---|---|---|---|---|---|
| Weight content (g/kg) | ||||||
| Milk proteins | 29 | 48 | 77 | 116 | 145 | 193.5 |
| Corn Starch | 717.9 | 701.5 | 676.6 | 643.1 | 618.1 | 576.4 |
| Sucrose | 115.8 | 113.2 | 109.1 | 103.6 | 99.6 | 92.8 |
| Soy Oil | 40 | 40 | 40 | 40 | 40 | 40 |
| Minerals | 35 | 35 | 35 | 35 | 35 | 35 |
| Vitamins | 10 | 10 | 10 | 10 | 10 | 10 |
| Cellulose | 50 | 50 | 50 | 50 | 50 | 50 |
| Choline | 2.3 | 2.3 | 2.3 | 2.3 | 2.3 | 2.3 |
| Energy content (%) | ||||||
| Protein | 3 | 5 | 8 | 12 | 15 | 20 |
| Carbohydrate | 86.6 | 84.6 | 81.6 | 77.6 | 74.6 | 69.5 |
| Fat | 9.3 | 9.3 | 9.3 | 9.3 | 9.3 | 9.3 |
| Energy density (kJ/g) | 14.54 | 14.55 | 14.55 | 14.56 | 14.56 | 14.57 |
| Urinary Discriminant Metabolites | Effect of Dietary Protein Content from P3 to P20 | |
|---|---|---|
| PLS | ICDA | |
| 3-methyl-2-oxovalerate | ↓ | ↓ |
| 4-methyl-2-oxovalerate | ↓ | ↓ |
| oxovaleric acid | ↓ | ↓ |
| azelate | ↓ | ↓ |
| creatine | ↓ | ↓ |
| d-mannose | ↓ | ↓ |
| d-raffinose | ↓ | ↓ |
| galactose | ↓ | ↓ |
| phosphoric acid | ↓ | ↓ |
| pyridoxine | ↓ | ↓ |
| raffinose | ↓ | ↓ |
| succinic acid | ↓ | ↓ |
| pantothenate | ↓ P5–P20 | ↓ |
| alpha d-glucose | ↓ | |
| galactitol | ↓ | |
| iso-maltose | ↓ | |
| l-carnitine | ↓ | |
| proline-leucine | ↓ | |
| sucrose | ↓ | |
| anthranilate | ↑ | ↑ |
| cadaverine | ↑ | ↑ |
| homogentisic acid | ↑ | ↑ |
| isovaleroylglycine | ↑ | ↑ |
| kynurenic acid | ↑ | ↑ |
| l-gulonolactone | ↑ | ↑ |
| n-methyl-2-pyridone-5carboxamide | ↑ | ↑ |
| pipecolate | ↑ | ↑ |
| uracil | ↑ | ↑ |
| xanthurenate | ↑ | ↑ |
| indoxyl sulfate | ↑ | ↑ |
| putrescine | ↑ | ↑ |
| 4-pyridoxate, | ↑ | |
| l-leucine | ↑ | |
| n-acetylputrescine | ↑ | |
| spermidine, | ↑ | |
| tyramine | ↓P3–P5↑P8–P12↓P15P20 | |
| Discriminant Plasma Metabolites | PLS | ICDA | ||
|---|---|---|---|---|
| PV | VC | PV | VC | |
| betaine | ↓ | ↓ | ↓ | ↓ |
| fucose | ↓ | ↓ | ↓ | ↓ |
| lysoPC (18:0) | ↓ | ↓ | ↓ | ↓ |
| l-carnitine | ↓ | ↓ | ||
| l-histidine | ↓ | ↓ | ↓ | ↓ |
| l-serine | ↓ | ↓ | ↓ | ↓ |
| malate | ↓ | ↓ | ↓ | |
| o-acetyl-carnitine | ↓ | ↓ | ||
| 2-hydroxyisocaproic acid | ↓ | ↓ | ||
| l-ornithine | ↓ | ↓ | ↓ | |
| pyroglutamate | ↓ | |||
| l-arginine | ↓ | ↓ | ||
| citrulline | ↓ | |||
| l-glutamine | ↓ | |||
| l-methionine | ↑ | ↑ | ↑ | ↑ |
| l-phenylalanine | ↑ | ↑ | ↑ | ↑ |
| l-threonine | ↑ | ↑ | ↑ | ↑ |
| l-tyrosine, | ↑ | ↑ | ↑ | ↑ |
| n-methyl-2-pyridone-5-carboxamide | ↑ | ↑ | ↑ | ↑ |
| taurocholic acid, | ↑ | ↑ | ||
| tryptophan | ↑ | ↑ | ↑ | |
| 3-isopropylmalic acid | ↑ | |||
| l-lysine | ↑ | ↑ | ↑ | |
| indoxyl sulfate | ↑ | |||
| l-valine | ↑ | |||
| l-leucine | ↑ | |||
| n-acetylserotonin | ↑ | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moro, J.; Khodorova, N.; Tomé, D.; Gaudichon, C.; Tardivel, C.; Berton, T.; Martin, J.-C.; Azzout-Marniche, D.; Jouan-Rimbaud Bouveresse, D. Plasma and Urinary Amino Acid-Derived Catabolites as Potential Biomarkers of Protein and Amino Acid Deficiency in Rats. Nutrients 2021, 13, 1567. https://doi.org/10.3390/nu13051567
Moro J, Khodorova N, Tomé D, Gaudichon C, Tardivel C, Berton T, Martin J-C, Azzout-Marniche D, Jouan-Rimbaud Bouveresse D. Plasma and Urinary Amino Acid-Derived Catabolites as Potential Biomarkers of Protein and Amino Acid Deficiency in Rats. Nutrients. 2021; 13(5):1567. https://doi.org/10.3390/nu13051567
Chicago/Turabian StyleMoro, Joanna, Nadezda Khodorova, Daniel Tomé, Claire Gaudichon, Catherine Tardivel, Thierry Berton, Jean-Charles Martin, Dalila Azzout-Marniche, and Delphine Jouan-Rimbaud Bouveresse. 2021. "Plasma and Urinary Amino Acid-Derived Catabolites as Potential Biomarkers of Protein and Amino Acid Deficiency in Rats" Nutrients 13, no. 5: 1567. https://doi.org/10.3390/nu13051567
APA StyleMoro, J., Khodorova, N., Tomé, D., Gaudichon, C., Tardivel, C., Berton, T., Martin, J.-C., Azzout-Marniche, D., & Jouan-Rimbaud Bouveresse, D. (2021). Plasma and Urinary Amino Acid-Derived Catabolites as Potential Biomarkers of Protein and Amino Acid Deficiency in Rats. Nutrients, 13(5), 1567. https://doi.org/10.3390/nu13051567









