Ileal Lactase Expression Associates with Lactase Persistence Genotypes
Abstract
1. Introduction
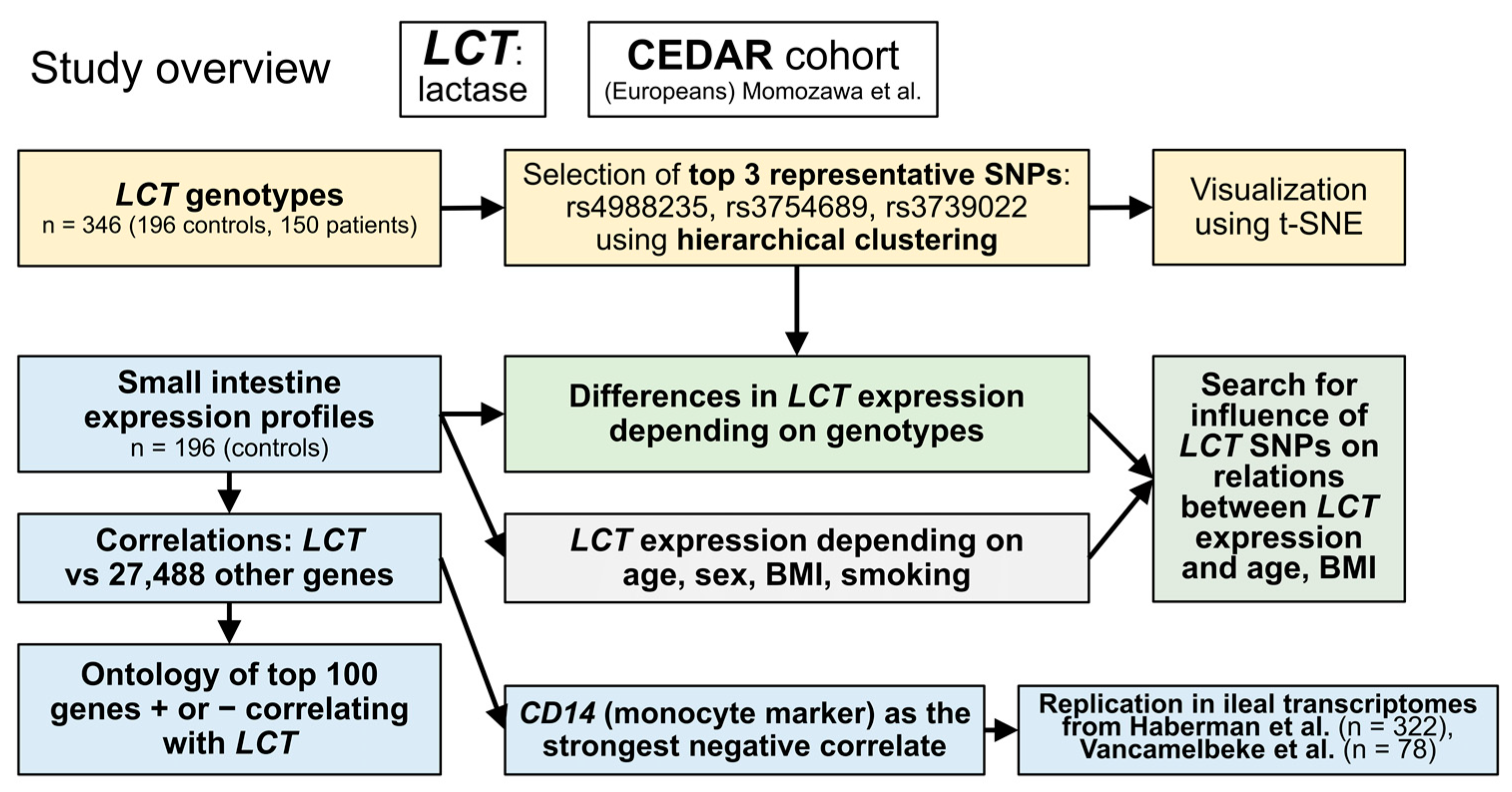
2. Materials and Methods
2.1. Genotypes
2.2. Transcriptomic Data
3. Results
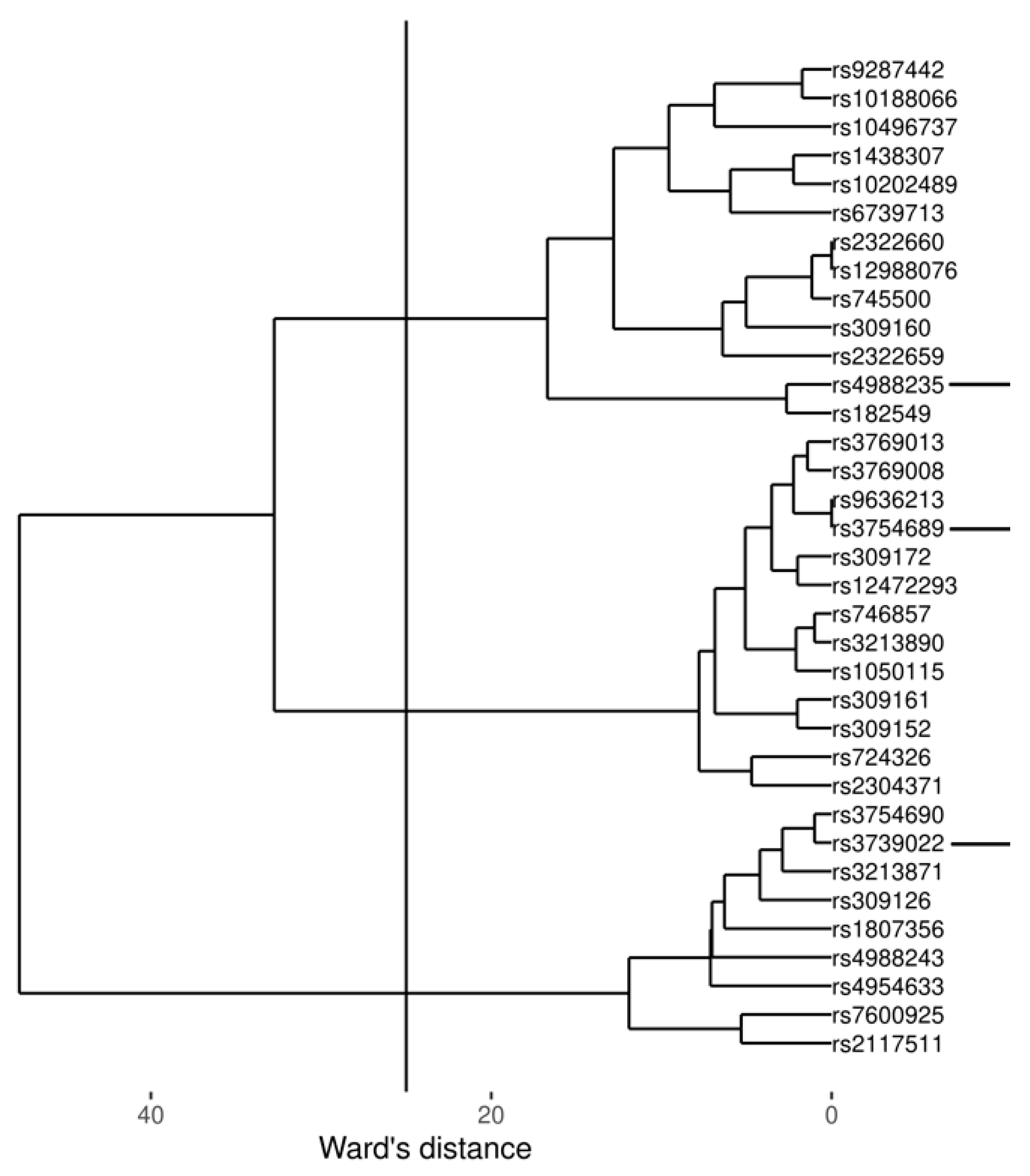
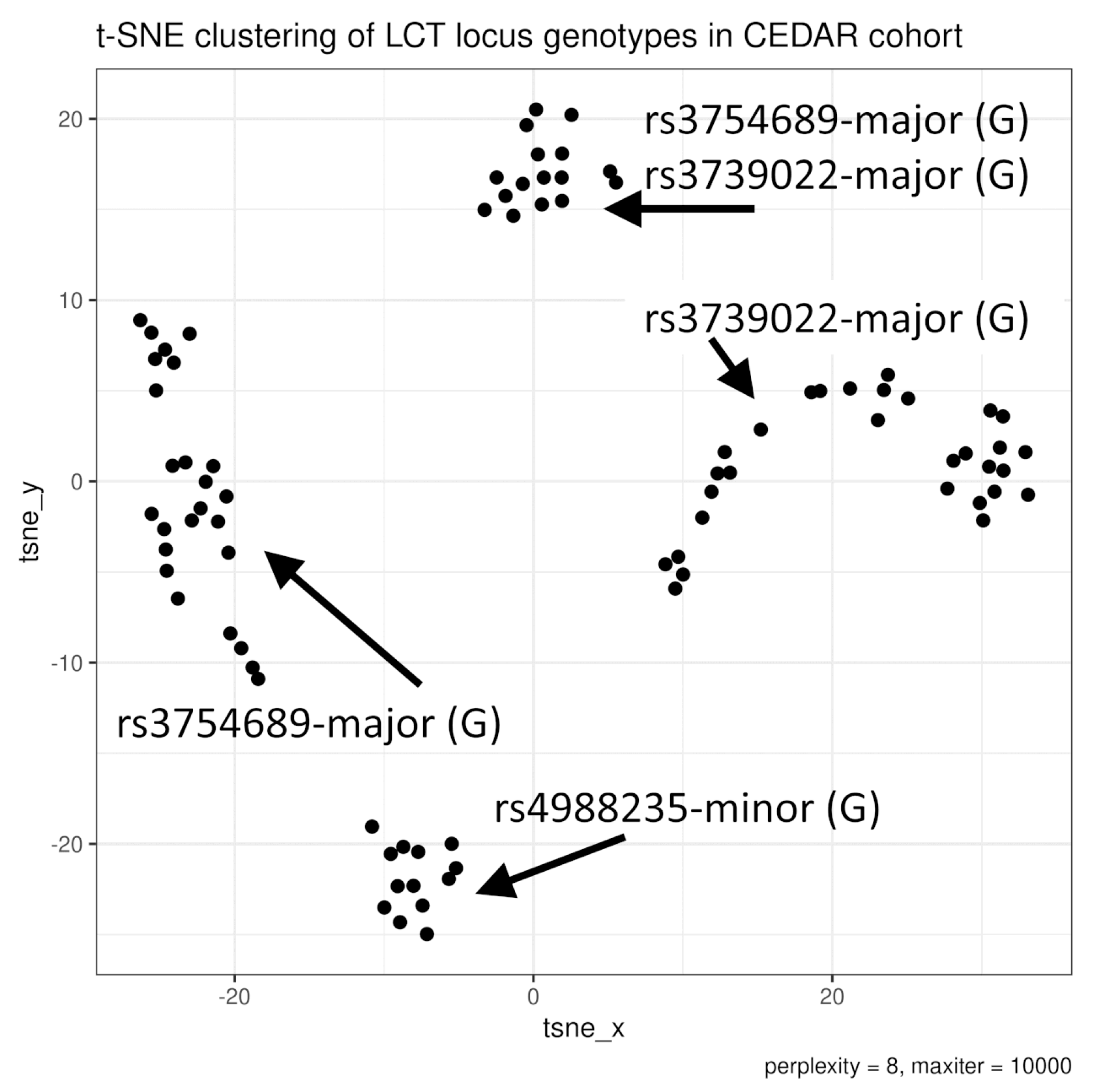
3.1. Genotypes
3.2. Expression
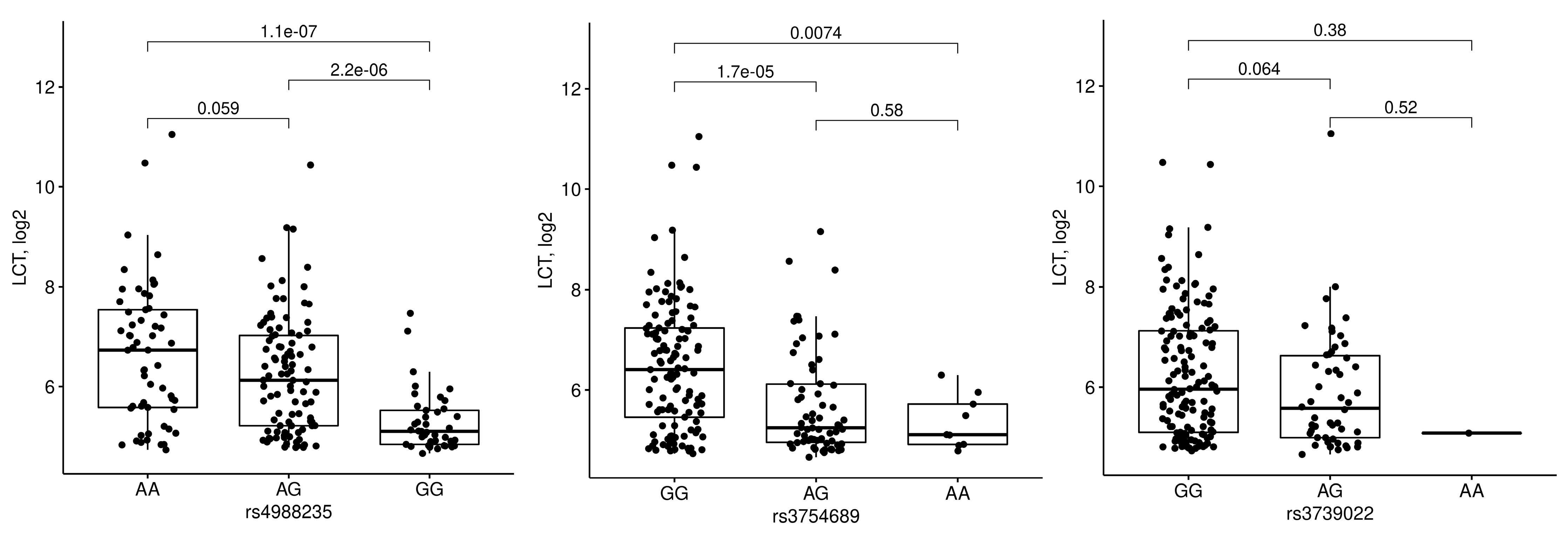
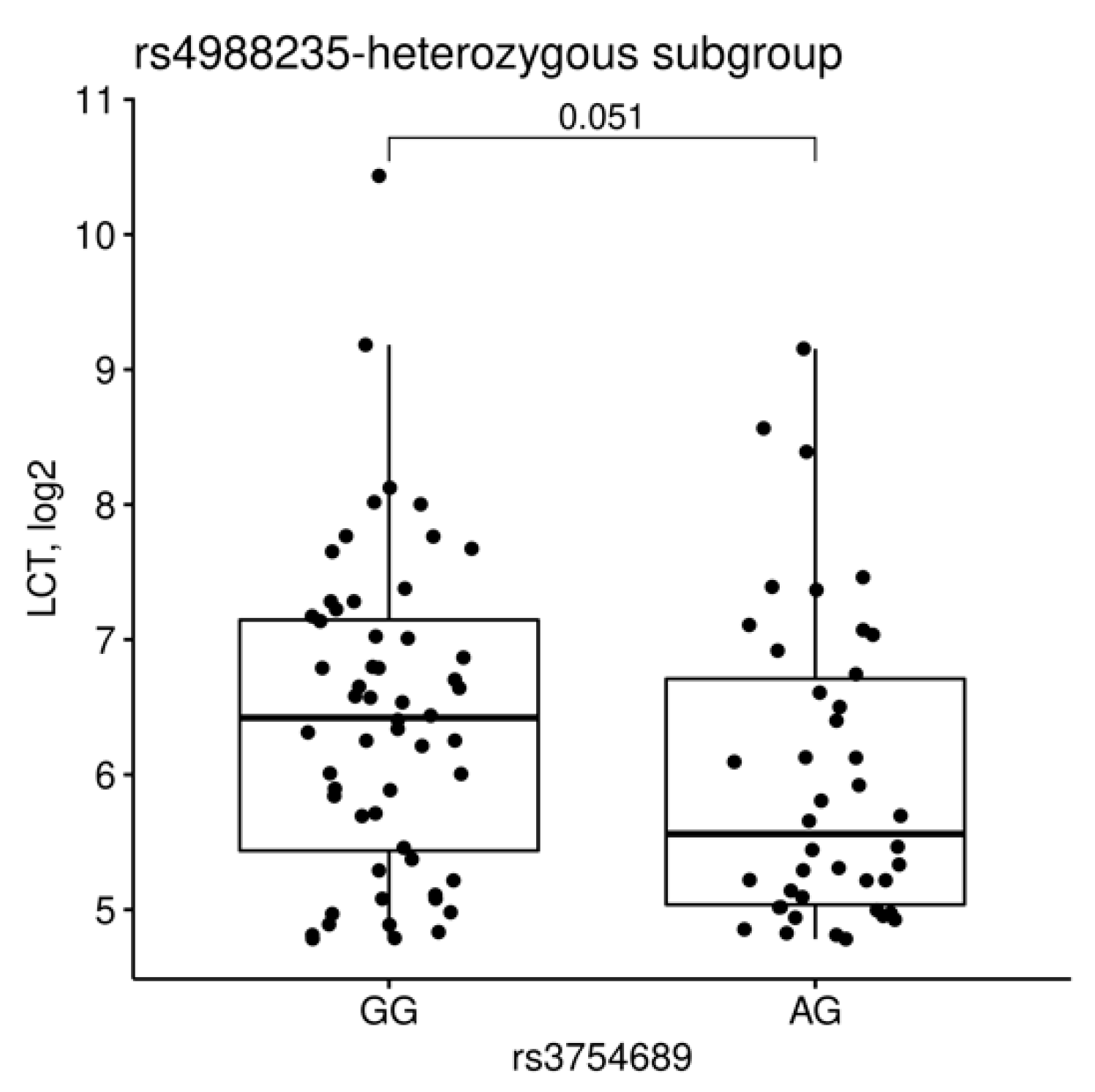
3.3. Relations between LCT Genotype and Expression
4. Discussion
4.1. The Role of rs4988235 and rs3754689
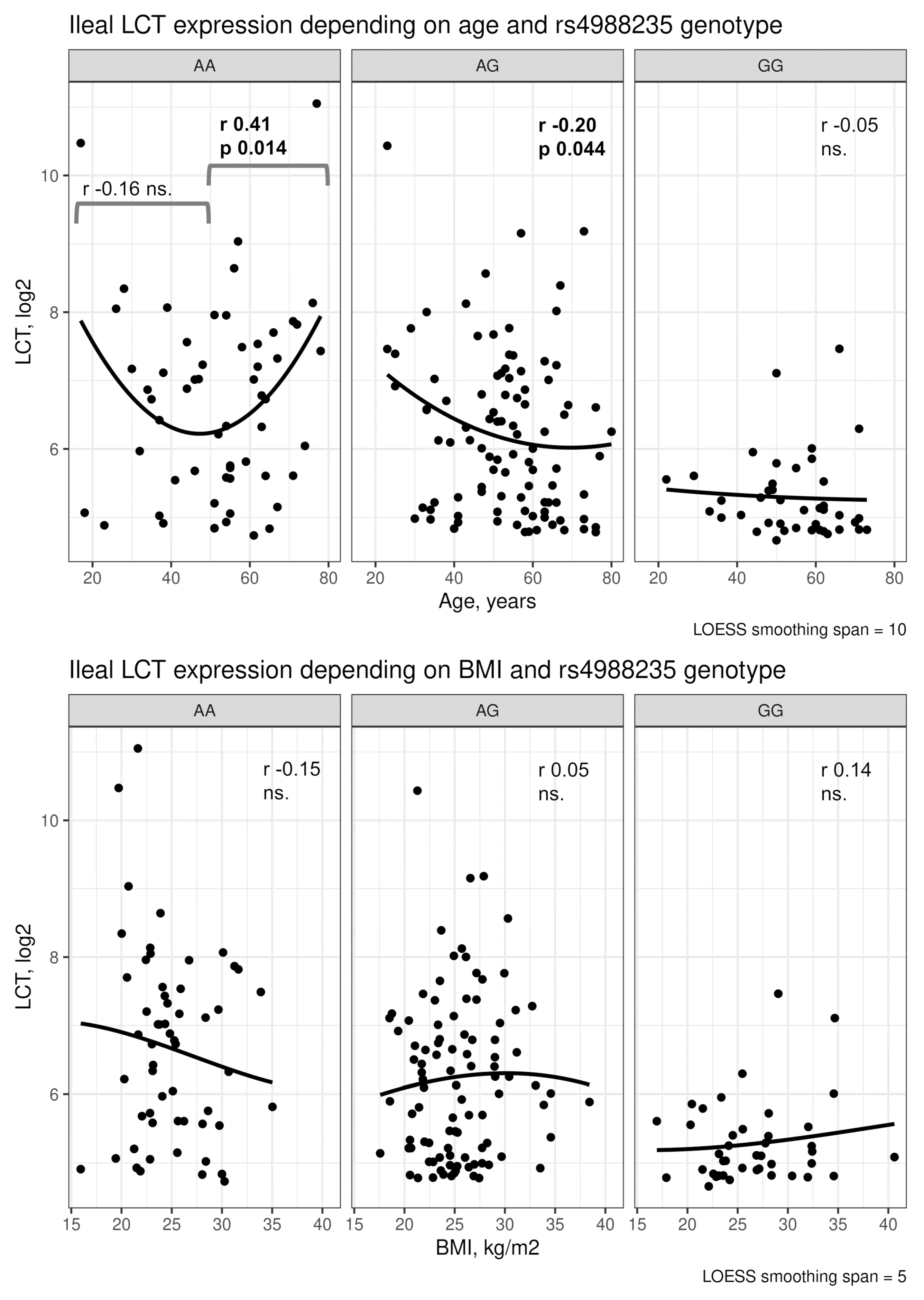
4.2. Ileal LCT Expression and Clinical Characteristics
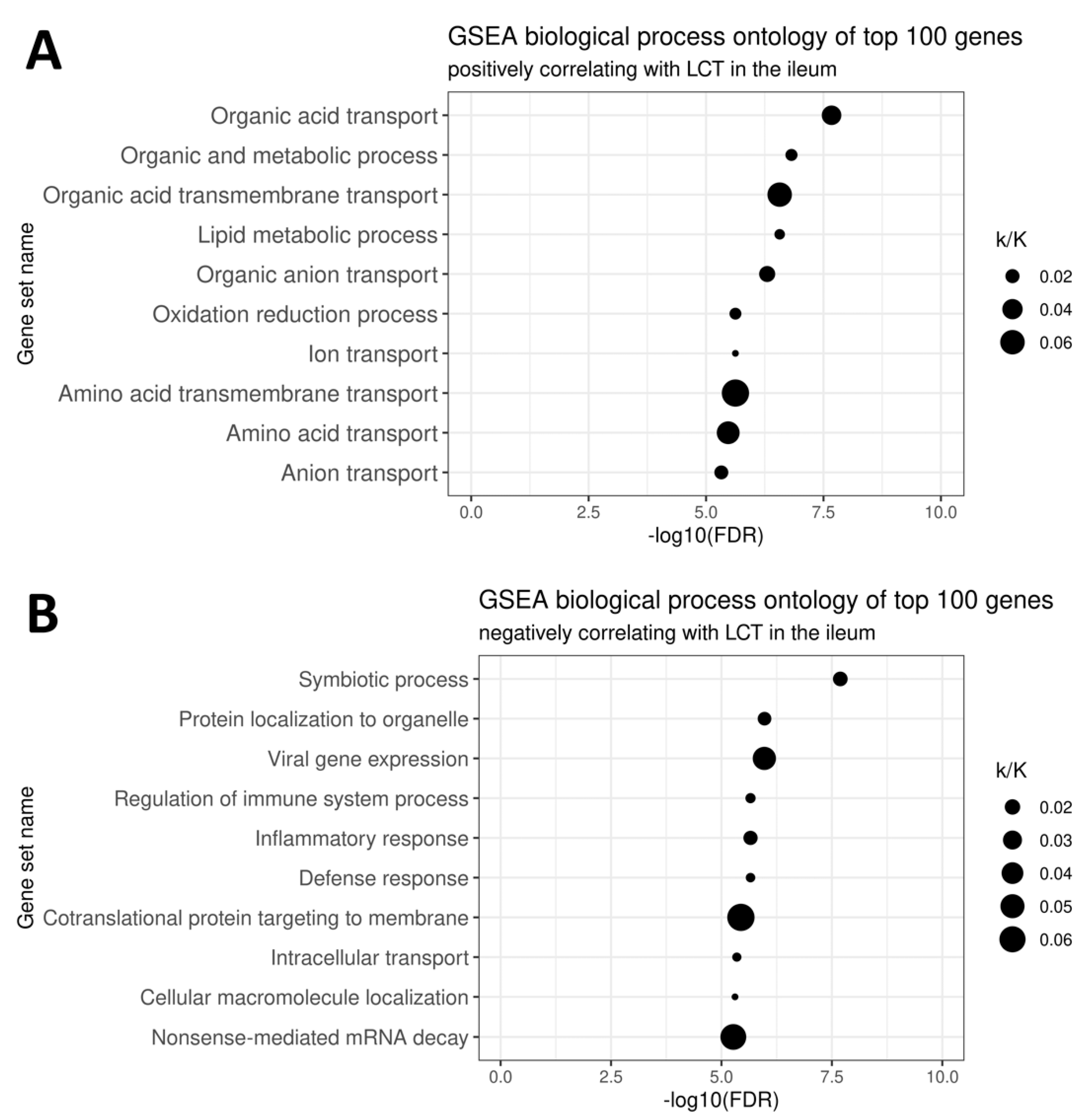
4.3. Transcriptomic Correlates of Ileal LCT Expression Including CD14
4.4. Generalization and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mądry, E.; Fidler, E.; Walkowiak, J. Lactose Intolerance—Current State of Knowledge. Acta Sci. Pol. Technol. Aliment. 2010, 9, 343–350. [Google Scholar]
- Misselwitz, B.; Butter, M.; Verbeke, K.; Fox, M.R. Update on Lactose Malabsorption and Intolerance: Pathogenesis, Diagnosis and Clinical Management. Gut 2019, 68, 2080–2091. [Google Scholar] [CrossRef] [PubMed]
- Couce, M.L.; Sánchez-Pintos, P.; González-Vioque, E.; Leis, R. Clinical Utility of LCT Genotyping in Children with Suspected Functional Gastrointestinal Disorder. Nutrients 2020, 12, 3017. [Google Scholar] [CrossRef]
- Forsgård, R.A. Lactose Digestion in Humans: Intestinal Lactase Appears to Be Constitutive Whereas the Colonic Microbiome Is Adaptable. Am. J. Clin. Nutr. 2019, 110, 273–279. [Google Scholar] [CrossRef]
- Rasinperä, H.; Kuokkanen, M.; Kolho, K.-L.; Lindahl, H.; Enattah, N.S.; Savilahti, E.; Orpana, A.; Järvelä, I. Transcriptional Downregulation of the Lactase (LCT) Gene during Childhood. Gut 2005, 54, 1660–1661. [Google Scholar] [CrossRef] [PubMed]
- Mądry, E.; Lisowska, A.; Kwiecień, J.; Marciniak, R.; Korzon-Burakowska, A.; Drzymała-Czyż, S.; Mojs, E.; Walkowiak, J. Adult-Type Hypolactasia and Lactose Malabsorption in Poland. Acta Biochim. Pol. 2010, 57, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Tishkoff, S.A.; Reed, F.A.; Ranciaro, A.; Voight, B.F.; Babbitt, C.C.; Silverman, J.S.; Powell, K.; Mortensen, H.M.; Hirbo, J.B.; Osman, M.; et al. Convergent Adaptation of Human Lactase Persistence in Africa and Europe. Nat. Genet 2007, 39, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Schirru, E.; Corona, V.; Usai-Satta, P.; Scarpa, M.; Oppia, F.; Loriga, F.; Cucca, F.; De Virgiliis, S.; Rossino, R.; Macis, M.D.; et al. Genetic Testing Improves the Diagnosis of Adult Type Hypolactasia in the Mediterranean Population of Sardinia. Eur. J. Clin. Nutr. 2007, 61, 1220–1225. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sibley, E.; Ahn, J.K.; Theodore, E. Woodward Award: Lactase Persistence SNPs in African Populations Regulate Promoter Activity in Intestinal Cell Culture. Trans. Am. Clin. Clim. Assoc. 2011, 122, 155–165. [Google Scholar]
- Schlebusch, C.M.; Sjödin, P.; Skoglund, P.; Jakobsson, M. Stronger Signal of Recent Selection for Lactase Persistence in Maasai than in Europeans. Eur. J. Hum. Genet. 2013, 21, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Anguita-Ruiz, A.; Aguilera, C.M.; Gil, Á. Genetics of Lactose Intolerance: An Updated Review and Online Interactive World Maps of Phenotype and Genotype Frequencies. Nutrients 2020, 12, 2689. [Google Scholar] [CrossRef] [PubMed]
- Enattah, N.S.; Sahi, T.; Savilahti, E.; Terwilliger, J.D.; Peltonen, L.; Järvelä, I. Identification of a Variant Associated with Adult-Type Hypolactasia. Nat. Genet. 2002, 30, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Leseva, M.N.; Grand, R.J.; Klett, H.; Boerries, M.; Busch, H.; Binder, A.M.; Michels, K.B. Differences in DNA Methylation and Functional Expression in Lactase Persistent and Non-Persistent Individuals. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Olsen, W.A.; Li, B.U.K.; Lloyd, M.; Korsmo, H. Heterogeneity of Intestinal Lactase Activity in Children: Relationship to Lactase-Phlorizin Hydrolase Messenger RNA Abundance. Pediatr. Res. 1996, 39, 877–881. [Google Scholar] [CrossRef]
- Momozawa, Y.; Dmitrieva, J.; Théâtre, E.; Deffontaine, V.; Rahmouni, S.; Charloteaux, B.; Crins, F.; Docampo, E.; Elansary, M.; Gori, A.-S.; et al. IBD Risk Loci Are Enriched in Multigenic Regulatory Modules Encompassing Putative Causative Genes. Nat. Commun. 2018, 9, 2427. [Google Scholar] [CrossRef]
- Haberman, Y.; Tickle, T.L.; Dexheimer, P.J.; Kim, M.-O.; Tang, D.; Karns, R.; Baldassano, R.N.; Noe, J.D.; Rosh, J.; Markowitz, J.; et al. Pediatric Crohn Disease Patients Exhibit Specific Ileal Transcriptome and Microbiome Signature. J. Clin. Investig. 2014, 124, 3617–3633. [Google Scholar] [CrossRef]
- Vancamelbeke, M.; Vanuytsel, T.; Farré, R.; Verstockt, S.; Ferrante, M.; Van Assche, G.; Rutgeerts, P.; Schuit, F.; Vermeire, S.; Arijs, I.; et al. Genetic and Transcriptomic Bases of Intestinal Epithelial Barrier Dysfunction in Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2017, 23, 1718–1729. [Google Scholar] [CrossRef]
- Heianza, Y.; Sun, D.; Ma, W.; Zheng, Y.; Champagne, C.M.; Bray, G.A.; Sacks, F.M.; Qi, L. Gut-microbiome-related LCT genotype and 2-year changes in body composition and fat distribution: The POUNDS Lost Trial. Int. J. Obes. 2018, 42, 1565–1573. [Google Scholar] [CrossRef]
- Nowak, J.K.; Lindstrøm, J.C.; Kalla, R.; Ricanek, P.; Halfvarson, J.; Satsangi, J. Age, Inflammation, and Disease Location Are Critical Determinants of Intestinal Expression of SARS-CoV-2 Receptor ACE2 and TMPRSS2 in Inflammatory Bowel Disease. Gastroenterology 2020, 159, 1151–1154.e2. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Xiang, L.; Chen, W.; Li, S.; Huang, S.; Li, J.; Zhuge, L.; Jin, L.; Feng, W.; Chen, Y.; et al. LncRNA GAS5 Enhanced the Killing Effect of NK Cell on Liver Cancer through Regulating MiR-544/RUNX3. Innate Immun. 2019, 25, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Kuokkanen, M.; Enattah, N.S.; Oksanen, A.; Savilahti, E.; Orpana, A.; Järvelä, I. Transcriptional Regulation of the Lactase-Phlorizin Hydrolase Gene by Polymorphisms Associated with Adult-Type Hypolactasia. Gut 2003, 52, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Araujo, E.M.; Dos Santos, L.; Coutinho, R.; Assis, V.; Brandão, N.; Almeida, D.; Conceição, G.; Figueredo, C.; Fonseca, H.; Lima, M.L.; et al. Genetic and Oral Tests for the Diagnosis of Lactose Intolerance in Mixed-Ancestry Brazilians with Metabolic Syndrome. Lifestyle Genom. 2019, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gorski, M.M.; Blighe, K.; Lotta, L.A.; Pappalardo, E.; Garagiola, I.; Mancini, I.; Mancuso, M.E.; Fasulo, M.R.; Santagostino, E.; Peyvandi, F. Whole-Exome Sequencing to Identify Genetic Risk Variants Underlying Inhibitor Development in Severe Hemophilia A Patients. Blood 2016, 127, 2924–2933. [Google Scholar] [CrossRef] [PubMed]
- Tomczonek-Moruś, J.; Wojtasik, A.; Zeman, K.; Smolarz, B.; Bąk-Romaniszyn, L. 13910C>T and 22018G>A LCT Gene Polymorphisms in Diagnosing Hypolactasia in Children. United Eur. Gastroenterol. J. 2019, 7, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Labrie, V.; Buske, O.J.; Oh, E.; Jeremian, R.; Ptak, C.; Gasiūnas, G.; Maleckas, A.; Petereit, R.; Žvirbliene, A.; Adamonis, K.; et al. Lactase Nonpersistence Is Directed by DNA-Variation-Dependent Epigenetic Aging. Nat. Struct. Mol. Biol. 2016, 23, 566–573. [Google Scholar] [CrossRef]
- Oh, E.; Jeremian, R.; Oh, G.; Groot, D.; Susic, M.; Lee, K.; Foy, K.; Laird, P.W.; Petronis, A.; Labrie, V. Transcriptional Heterogeneity in the Lactase Gene within Cell-Type Is Linked to the Epigenome. Sci. Rep. 2017, 7, 41843. [Google Scholar] [CrossRef]
- Kettunen, J.; Silander, K.; Saarela, O.; Amin, N.; Müller, M.; Timpson, N.; Surakka, I.; Ripatti, S.; Laitinen, J.; Hartikainen, A.-L.; et al. European Lactase Persistence Genotype Shows Evidence of Association with Increase in Body Mass Index. Hum. Mol. Genet 2010, 19, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Corella, D.; Arregui, M.; Coltell, O.; Portolés, O.; Guillem-Sáiz, P.; Carrasco, P.; Sorlí, J.V.; Ortega-Azorín, C.; González, J.I.; Ordovás, J.M. Association of the LCT-13910C>T Polymorphism with Obesity and Its Modulation by Dairy Products in a Mediterranean Population. Obes. Silver Spring 2011, 19, 1707–1714. [Google Scholar] [CrossRef]
- Almon, R.; Álvarez-León, E.E.; Serra-Majem, L. Association of the European Lactase Persistence Variant (LCT-13910 C>T Polymorphism) with Obesity in the Canary Islands. PLoS ONE 2012, 7, e43978. [Google Scholar] [CrossRef]
- Chen, M.-H.; Raffield, L.M.; Mousas, A.; Sakaue, S.; Huffman, J.E.; Moscati, A.; Trivedi, B.; Jiang, T.; Akbari, P.; Vuckovic, D.; et al. Trans-Ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations. Cell 2020, 182, 1198–1213.e14. [Google Scholar] [CrossRef]
- Ng, T.B.; Cheung, R.C.F.; Wong, J.H.; Wang, Y.; Ip, D.T.M.; Wan, D.C.C.; Xia, J. Antiviral Activities of Whey Proteins. Appl. Microbiol. Biotechnol. 2015, 99, 6997–7008. [Google Scholar] [CrossRef] [PubMed]
- Cederlund, A.; Kai-Larsen, Y.; Printz, G.; Yoshio, H.; Alvelius, G.; Lagercrantz, H.; Strömberg, R.; Jörnvall, H.; Gudmundsson, G.H.; Agerberth, B. Lactose in Human Breast Milk an Inducer of Innate Immunity with Implications for a Role in Intestinal Homeostasis. PLoS ONE 2013, 8, e53876. [Google Scholar] [CrossRef] [PubMed]
- Dharuri, H.; ’t Hoen, P.A.C.; van Klinken, J.B.; Henneman, P.; Laros, J.F.J.; Lips, M.A.; el Bouazzaoui, F.; van Ommen, G.-J.B.; Janssen, I.; van Ramshorst, B.; et al. Downregulation of the Acetyl-CoA Metabolic Network in Adipose Tissue of Obese Diabetic Individuals and Recovery after Weight Loss. Diabetologia 2014, 57, 2384–2392. [Google Scholar] [CrossRef] [PubMed]
- Taminelli, G.L.; Sotomayor, V.; Valdivieso, Á.G.; Teiber, M.L.; Marín, M.C.; Santa-Coloma, T.A. CISD1 Codifies a Mitochondrial Protein Upregulated by the CFTR Channel. Biochem. Biophys. Res. Commun. 2008, 365, 856–862. [Google Scholar] [CrossRef] [PubMed]
| SNP Pair | Pearson’s r | P |
|---|---|---|
| rs4988235–rs3754689 | 0.60 | 2.2 × 10−16 |
| rs4988235–rs3739022 | 0.38 | 2.6 × 10−13 |
| rs3754689–rs3739022 | −0.18 | 6.8 × 10−4 |
| Polymorphism | Homozygous Major | Heterozygous | Homozygous Minor |
|---|---|---|---|
| rs4988235 | AA 57 (29.1%) | AG 98 (50.0%) | GG 41 (20.9%) |
| rs3754689 | GG 121 (61.7%) | AG 66 (33.7%) | AA 9 (4.6%) |
| rs3739022 | GG 141 (72.3%) | AG 50 (25.6%) | AA 1 (0.5%) |
| Gene | r | pFDR |
|---|---|---|
| Positively correlating with LCT expression | ||
| ALDH6A1—Aldehyde Dehydrogenase 6 Family Member A1 | 0.56 | 2.4 × 10−14 |
| LOC441442 | 0.56 | 8.9 × 10−14 |
| C18orf18—Long Intergenic Non-Protein Coding RNA 526 | 0.55 | 2.5 × 10−13 |
| XYLB—Xylulokinase | 0.55 | 2.8 × 10−13 |
| CISD1—CDGSH Iron Sulfur Domain 1 | 0.54 | 6.3 × 10−13 |
| BCL2L15—BCL2 Like 15 | 0.54 | 6.5 × 10−13 |
| LOC100128907 | 0.54 | 8.3 × 10−13 |
| CCDC25—Coiled-Coil Domain Containing 25 | 0.54 | 8.9 × 10−13 |
| BEND7—BEN Domain Containing 7 | 0.53 | 2.3 × 10−12 |
| APOM—Apolipoprotein M | 0.53 | 3.2 × 10−12 |
| Negatively correlating with LCT expression | ||
| CD14—Monocyte Differentiation Antigen CD14 | −0.57 | 1.1 × 10−14 |
| RNASET2—Ribonuclease T2 | −0.50 | 2.9 × 10−11 |
| CDC37—Cell Division Cycle 37, HSP90 Cochaperone | −0.50 | 3.6 × 10−11 |
| C7orf50—Chromosome 7 Open Reading Frame 50 | −0.50 | 5.4 × 10−11 |
| LOC730187 | −0.49 | 6.3 × 10−11 |
| UBE2I—Ubiquitin Conjugating Enzyme E2 I | −0.49 | 1.0 × 10−10 |
| LOC650369 | −0.49 | 1.0 × 10−10 |
| RCC2—Regulator of Chromosome Condensation 2 | −0.49 | 1.0 × 10−10 |
| HK1—Hexokinase 1 | −0.49 | 1.1 × 10−10 |
| RPL27—Ribosomal Protein L27 | −0.48 | 2.3 × 10−10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowak, J.K.; Dybska, E.; Dworacka, M.; Tsikhan, N.; Kononets, V.; Bermagambetova, S.; Walkowiak, J. Ileal Lactase Expression Associates with Lactase Persistence Genotypes. Nutrients 2021, 13, 1340. https://doi.org/10.3390/nu13041340
Nowak JK, Dybska E, Dworacka M, Tsikhan N, Kononets V, Bermagambetova S, Walkowiak J. Ileal Lactase Expression Associates with Lactase Persistence Genotypes. Nutrients. 2021; 13(4):1340. https://doi.org/10.3390/nu13041340
Chicago/Turabian StyleNowak, Jan Krzysztof, Emilia Dybska, Marzena Dworacka, Natallia Tsikhan, Victoria Kononets, Saule Bermagambetova, and Jarosław Walkowiak. 2021. "Ileal Lactase Expression Associates with Lactase Persistence Genotypes" Nutrients 13, no. 4: 1340. https://doi.org/10.3390/nu13041340
APA StyleNowak, J. K., Dybska, E., Dworacka, M., Tsikhan, N., Kononets, V., Bermagambetova, S., & Walkowiak, J. (2021). Ileal Lactase Expression Associates with Lactase Persistence Genotypes. Nutrients, 13(4), 1340. https://doi.org/10.3390/nu13041340








