Codium fragile Ameliorates High-Fat Diet-Induced Metabolism by Modulating the Gut Microbiota in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Codium fragile (CF) Extracts
2.2. Animal Experiment
2.3. Histological Analysis
2.4. Determination of Cholesterol Levels
2.5. Gut Microbiota Analysis
2.6. Statistical Analysis
3. Results
3.1. CF Extracts Reduce the High-Fat Diet (HF)-Induced Fat Deposition in Mice
3.2. CF Extracts Suppress the HF-Induced Lipid Level in Mice
3.3. Comparison Analysis of Mice Gut Microbiota According to the Types of Treatment
3.4. CF Extracts Ameliorate the HF-Induced Dysbiosis of the Gut Microbiota in Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Ferranti, S.; Mozaffarian, D. The perfect storm: Obesity, adipocyte dysfunction, and metabolic consequences. Clin. Chem. 2008, 54, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Peyton, K.J.; Liu, X.M.; Shebib, A.R.; Johnson, F.K.; Johnson, R.A.; Durante, W. Arginase inhibition prevents the development of hypertension and improves insulin resistance in obese rats. Amino Acids 2018, 50, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.J.; West, N.P.; Cripps, A.W. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015, 3, 207–215. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Y.; Zhang, G.; Zhang, T.; Lou, J.; Liu, J. L-Arabinose Elicits Gut-Derived Hydrogen Production and Ameliorates Metabolic Syndrome in C57BL/6J Mice on High-Fat-Diet. Nutrients 2019, 11, 3054. [Google Scholar] [CrossRef] [PubMed]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Jang, H.M.; Han, S.K.; Kim, J.K.; Oh, S.J.; Jang, H.B.; Kim, D.H. Lactobacillus sakei Alleviates High-Fat-Diet-Induced Obesity and Anxiety in Mice by Inducing AMPK Activation and SIRT1 Expression and Inhibiting Gut Microbiota-Mediated NF-kappaB Activation. Mol. Nutr. Food Res. 2019, 63, e1800978. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, L.; Li, Y.; Xia, G.; Chen, C.; Zhang, Y. Bamboo-shaving polysaccharide protects against high-diet induced obesity and modulates the gut microbiota of mice. J. Funct. Foods 2018, 49, 20–31. [Google Scholar] [CrossRef]
- Perez, M.J.; Falque, E.; Dominguez, H. Antimicrobial Action of Compounds from Marine Seaweed. Mar. Drugs 2016, 14, 52. [Google Scholar] [CrossRef]
- Su, J.; Guo, K.; Huang, M.; Liu, Y.; Zhang, J.; Sun, L.; Li, D.; Pang, K.L.; Wang, G.; Chen, L.; et al. Fucoxanthin, a Marine Xanthophyll Isolated From Conticribra weissflogii ND-8: Preventive Anti-Inflammatory Effect in a Mouse Model of Sepsis. Front. Pharmacol. 2019, 10, 906. [Google Scholar] [CrossRef]
- Yayeh, T.; Im, E.J.; Kwon, T.H.; Roh, S.S.; Kim, S.; Kim, J.H.; Hong, S.B.; Cho, J.Y.; Park, N.H.; Rhee, M.H. Hemeoxygenase 1 partly mediates the anti-inflammatory effect of dieckol in lipopolysaccharide stimulated murine macrophages. Int. Immunopharmacol. 2014, 22, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, S.A.M.; Elias, N.; Farag, M.A.; Chen, L.; Saeed, A.; Hegazy, M.F.; Moustafa, M.S.; Abd El-Wahed, A.; Al-Mousawi, S.M.; Musharraf, S.G.; et al. Marine Natural Products: A Source of Novel Anticancer Drugs. Mar. Drugs 2019, 17, 491. [Google Scholar] [CrossRef] [PubMed]
- Wan-Loy, C.; Siew-Moi, P. Marine Algae as a Potential Source for Anti-Obesity Agents. Mar. Drugs 2016, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- Sanjeewa, K.K.A.; Lee, W.; Jeon, Y.-J. Nutrients and bioactive potentials of edible green and red seaweed in Korea. Fish. Aquat. Sci. 2018, 21, 19. [Google Scholar] [CrossRef]
- Kimiya, T.; Ohtani, K.; Satoh, S.; Abe, Y.; Ogita, Y.; Kawakita, H.; Hamada, H.; Konishi, Y.; Kubota, S.; Tominaga, A. Inhibitory effects of edible marine algae extracts on degranulation of RBL-2H3 cells and mouse eosinophils. Fish. Sci. 2008, 74, 1157–1165. [Google Scholar] [CrossRef]
- Lee, S.A.; Moon, S.M.; Choi, Y.H.; Han, S.H.; Park, B.R.; Choi, M.S.; Kim, J.S.; Kim, Y.H.; Kim, D.K.; Kim, C.S. Aqueous extract of Codium fragile suppressed inflammatory responses in lipopolysaccharide-stimulated RAW264.7 cells and carrageenan-induced rats. Biomed. Pharmacother. 2017, 93, 1055–1064. [Google Scholar] [CrossRef]
- Kim, J.E.; Monmai, C.; Rod-In, W.; Jang, A.Y.; You, S.G.; Lee, S.M.; Park, W.J. Immune Enhancement Effects of Codium fragile Anionic Macromolecules Combined with Red Ginseng Extract in Immune-Suppressed Mice. J. Microbiol. Biotechnol. 2019, 29, 1361–1368. [Google Scholar] [CrossRef]
- Choi, J.H.; Sapkota, K.; Park, S.E.; Kim, S.; Kim, S.J. Thrombolytic, anticoagulant and antiplatelet activities of codiase, a bi-functional fibrinolytic enzyme from Codium fragile. Biochimie 2013, 95, 1266–1277. [Google Scholar] [CrossRef]
- Tabarsa, M.; Karnjanapratum, S.; Cho, M.; Kim, J.K.; You, S. Molecular characteristics and biological activities of anionic macromolecules from Codium fragile. Int. J. Biol. Macromol. 2013, 59, 1–12. [Google Scholar] [CrossRef]
- Kolsi, R.B.A.; Jardak, N.; Hajkacem, F.; Chaaben, R.; Jribi, I.; Feki, A.E.; Rebai, T.; Jamoussi, K.; Fki, L.; Belghith, H.; et al. Anti-obesity effect and protection of liver-kidney functions by Codium fragile sulphated polysaccharide on high fat diet induced obese rats. Int. J. Biol. Macromol. 2017, 102, 119–129. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Chai, B.; Farris, R.J.; Wang, Q.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Bandela, A.M.; Cardenas, E.; Garrity, G.M.; Tiedje, J.M. The ribosomal database project (RDP-II): Introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007, 35, D169–D172. [Google Scholar] [CrossRef] [PubMed]
- Westcott, S.L.; Schloss, P.D. OptiClust, an Improved Method for Assigning Amplicon-Based Sequence Data to Operational Taxonomic Units. MSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Chazdon, R.L.; Colwell, R.K.; Shen, T.-J. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 2004, 8, 148–159. [Google Scholar] [CrossRef]
- Shannon, C.E. The mathematical theory of communication 1963. MD Comput. 1997, 14, 306–317. [Google Scholar] [PubMed]
- Beals, E.W. Bray-Curtis Ordination: An Effective Strategy for Analysis of Multivariate Ecological Data. Adv. Ecol. Res. 1984, 14, 1–55. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and customizable approach for metagenome inference. bioRxiv 2020. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Vogel-Scheel, J.; Alpert, C.; Engst, W.; Loh, G.; Blaut, M. Requirement of purine and pyrimidine synthesis for colonization of the mouse intestine by Escherichia coli. Appl. Environ. Microbiol. 2010, 76, 5181–5187. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.S.; Jheng, H.F.; Iwase, M.; Kim, M.; Mohri, S.; Kwon, J.; Kawarasaki, S.; Li, Y.; Takahashi, H.; Ara, T.; et al. The Mevalonate Pathway Is Indispensable for Adipocyte Survival. iScience 2018, 9, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Alkhouri, N.; Eng, K.; Cikach, F.; Patel, N.; Yan, C.; Brindle, A.; Rome, E.; Hanouneh, I.; Grove, D.; Lopez, R.; et al. Breathprints of childhood obesity: Changes in volatile organic compounds in obese children compared with lean controls. Pediatr. Obes. 2015, 10, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Namkung, J.; Kim, H.; Park, S. Peripheral Serotonin: A New Player in Systemic Energy Homeostasis. Mol. Cells 2015, 38, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.F.; Li, J.; Zhang, Z.; Liu, J.; Sun, X.Y.; Feng, X.F.; Luo, H.H.; Yang, W.; Li, S.N.; Yang, X.; et al. Plasma Levels of Amino Acids Related to Urea Cycle and Risk of Type 2 Diabetes Mellitus in Chinese Adults. Front. Endocrinol. 2019, 10, 50. [Google Scholar] [CrossRef]
- Taira, R.; Yamaguchi, S.; Shimizu, K.; Nakamura, K.; Ayabe, T.; Taira, T. Bacterial cell wall components regulate adipokine secretion from visceral adipocytes. J. Clin. Biochem. Nutr. 2015, 56, 149–154. [Google Scholar] [CrossRef]
- Barathikannan, K.; Chelliah, R.; Rubab, M.; Daliri, E.B.; Elahi, F.; Kim, D.H.; Agastian, P.; Oh, S.Y.; Oh, D.H. Gut Microbiome Modulation Based on Probiotic Application for Anti-Obesity: A Review on Efficacy and Validation. Microorganisms 2019, 7, 456. [Google Scholar] [CrossRef]
- Lattimer, J.M.; Haub, M.D. Effects of dietary fiber and its components on metabolic health. Nutrients 2010, 2, 1266–1289. [Google Scholar] [CrossRef]
- Dreher, M.L. Whole Fruits and Fruit Fiber Emerging Health Effects. Nutrients 2018, 10, 1833. [Google Scholar] [CrossRef]
- Hervik, A.K.; Svihus, B. The Role of Fiber in Energy Balance. J. Nutr. Metab. 2019, 2019, 4983657. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, N.; Ma, Y.; Wen, D. Hydroxytyrosol Improves Obesity and Insulin Resistance by Modulating Gut Microbiota in High-Fat Diet-Induced Obese Mice. Front. Microbiol. 2019, 10, 390. [Google Scholar] [CrossRef]
- Sikalidis, A.K.; Maykish, A. The Gut Microbiome and Type 2 Diabetes Mellitus: Discussing a Complex Relationship. Biomedicines 2020, 8, 8. [Google Scholar] [CrossRef]
- Lee, S.; Keirsey, K.I.; Kirkland, R.; Grunewald, Z.I.; Fischer, J.G.; de La Serre, C.B. Blueberry Supplementation Influences the Gut Microbiota, Inflammation, and Insulin Resistance in High-Fat-Diet-Fed Rats. J. Nutr. 2018, 148, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Gerard, C.; Vidal, H. Impact of Gut Microbiota on Host Glycemic Control. Front. Endocrinol. 2019, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef]
- Lee, Y.S.; Park, M.S.; Choung, J.S.; Kim, S.S.; Oh, H.H.; Choi, C.S.; Ha, S.Y.; Kang, Y.; Kim, Y.; Jun, H.S. Glucagon-like peptide-1 inhibits adipose tissue macrophage infiltration and inflammation in an obese mouse model of diabetes. Diabetologia 2012, 55, 2456–2468. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Yabut, J.M.; Choo, J.M.; Page, A.J.; Sun, E.W.; Jessup, C.F.; Wesselingh, S.L.; Khan, W.I.; Rogers, G.B.; Steinberg, G.R.; et al. The gut microbiome regulates host glucose homeostasis via peripheral serotonin. Proc. Natl. Acad. Sci. USA 2019, 116, 19802–19804. [Google Scholar] [CrossRef]
- Pathak, P.; Xie, C.; Nichols, R.G.; Ferrell, J.M.; Boehme, S.; Krausz, K.W.; Patterson, A.D.; Gonzalez, F.J.; Chiang, J.Y.L. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology 2018, 68, 1574–1588. [Google Scholar] [CrossRef]
- Tung, Y.C.; Chang, W.T.; Li, S.; Wu, J.C.; Badmeav, V.; Ho, C.T.; Pan, M.H. Citrus peel extracts attenuated obesity and modulated gut microbiota in mice with high-fat diet-induced obesity. Food Funct. 2018, 9, 3363–3373. [Google Scholar] [CrossRef]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Geerlings, S.Y.; Kostopoulos, I.; de Vos, W.M.; Belzer, C. Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353. [Google Scholar] [CrossRef]
- Seregin, S.S.; Golovchenko, N.; Schaf, B.; Chen, J.; Pudlo, N.A.; Mitchell, J.; Baxter, N.T.; Zhao, L.; Schloss, P.D.; Martens, E.C.; et al. NLRP6 Protects Il10(-/-) Mice from Colitis by Limiting Colonization of Akkermansia muciniphila. Cell Rep. 2017, 19, 733–745. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Yan, X.; Yang, C.; Lin, G.; Chen, Y.; Miao, S.; Liu, B.; Zhao, C. Antidiabetic Potential of Green Seaweed Enteromorpha prolifera Flavonoids Regulating Insulin Signaling Pathway and Gut Microbiota in Type 2 Diabetic Mice. J. Food Sci. 2019, 84, 165–173. [Google Scholar] [CrossRef]
- Pfeiffer, N.; Desmarchelier, C.; Blaut, M.; Daniel, H.; Haller, D.; Clavel, T. Acetatifactor muris gen. nov., sp. nov., a novel bacterium isolated from the intestine of an obese mouse. Arch. Microbiol. 2012, 194, 901–907. [Google Scholar] [CrossRef]
- Feng, W.; Ao, H.; Peng, C. Gut Microbiota, Short-Chain Fatty Acids, and Herbal Medicines. Front. Pharmacol. 2018, 9, 1354. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.J.; et al. Short-Chain Fatty Acids Protect Against High-Fat Diet-Induced Obesity via a PPARgamma-Dependent Switch From Lipogenesis to Fat Oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Fan, C.; Li, P.; Lu, Y.; Chang, X.; Qi, K. Short Chain Fatty Acids Prevent High-fat-diet-induced Obesity in Mice by Regulating G Protein-coupled Receptors and Gut Microbiota. Sci. Rep. 2016, 6, 37589. [Google Scholar] [CrossRef] [PubMed]
- Chambers, E.S.; Morrison, D.J.; Frost, G. Control of appetite and energy intake by SCFA: What are the potential underlying mechanisms? Proc. Nutr. Soc. 2015, 74, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Weitkunat, K.; Stuhlmann, C.; Postel, A.; Rumberger, S.; Fankhanel, M.; Woting, A.; Petzke, K.J.; Gohlke, S.; Schulz, T.J.; Blaut, M.; et al. Short-chain fatty acids and inulin, but not guar gum, prevent diet-induced obesity and insulin resistance through differential mechanisms in mice. Sci. Rep. 2017, 7, 6109. [Google Scholar] [CrossRef]
- Dong, T.S.; Chang, H.H.; Hauer, M.; Lagishetty, V.; Katzka, W.; Rozengurt, E.; Jacobs, J.P.; Eibl, G. Metformin alters the duodenal microbiome and decreases the incidence of pancreatic ductal adenocarcinoma promoted by diet-induced obesity. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G763–G772. [Google Scholar] [CrossRef]
- Jeung, W.H.; Nam, W.; Kim, H.J.; Kim, J.Y.; Nam, B.; Jang, S.S.; Lee, J.L.; Sim, J.H.; Park, S.D. Oral Administration of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 with Cinnamomi Ramulus Extract Reduces Diet-Induced Obesity and Modulates Gut Microbiota. Prev. Nutr. Food Sci. 2019, 24, 136–143. [Google Scholar] [CrossRef]
- Kubeck, R.; Bonet-Ripoll, C.; Hoffmann, C.; Walker, A.; Muller, V.M.; Schuppel, V.L.; Lagkouvardos, I.; Scholz, B.; Engel, K.H.; Daniel, H.; et al. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol. Metab. 2016, 5, 1162–1174. [Google Scholar] [CrossRef]
- Shang, Q.; Shan, X.; Cai, C.; Hao, J.; Li, G.; Yu, G. Dietary fucoidan modulates the gut microbiota in mice by increasing the abundance of Lactobacillus and Ruminococcaceae. Food Funct. 2016, 7, 3224–3232. [Google Scholar] [CrossRef]
- Gao, F.; Lv, Y.W.; Long, J.; Chen, J.M.; He, J.M.; Ruan, X.Z.; Zhu, H.B. Butyrate Improves the Metabolic Disorder and Gut Microbiome Dysbiosis in Mice Induced by a High-Fat Diet. Front. Pharmacol. 2019, 10, 1040. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Takagaki, A.; Matsumoto, K.; Kato, Y.; Goto, K.; Benno, Y. Butyricimonas synergistica gen. nov., sp. nov. and Butyricimonas virosa sp. nov., butyric acid-producing bacteria in the family ’Porphyromonadaceae’ isolated from rat faeces. Int. J. Syst. Evol. Microbiol. 2009, 59, 1748–1753. [Google Scholar] [CrossRef] [PubMed]
- Shkoporov, A.N.; Khokhlova, E.V.; Chaplin, A.V.; Kafarskaia, L.I.; Nikolin, A.A.; Polyakov, V.Y.; Shcherbakova, V.A.; Chernaia, Z.A.; Efimov, B.A. Coprobacter fastidiosus gen. nov., sp. nov., a novel member of the family Porphyromonadaceae isolated from infant faeces. Int. J. Syst. Evol. Microbiol. 2013, 63, 4181–4188. [Google Scholar] [CrossRef] [PubMed]
- Jabari, L.; Gannoun, H.; Cayol, J.L.; Hedi, A.; Sakamoto, M.; Falsen, E.; Ohkuma, M.; Hamdi, M.; Fauque, G.; Ollivier, B.; et al. Macellibacteroides fermentans gen. nov., sp. nov., a member of the family Porphyromonadaceae isolated from an upflow anaerobic filter treating abattoir wastewaters. Int. J. Syst. Evol. Microbiol. 2012, 62, 2522–2527. [Google Scholar] [CrossRef] [PubMed]

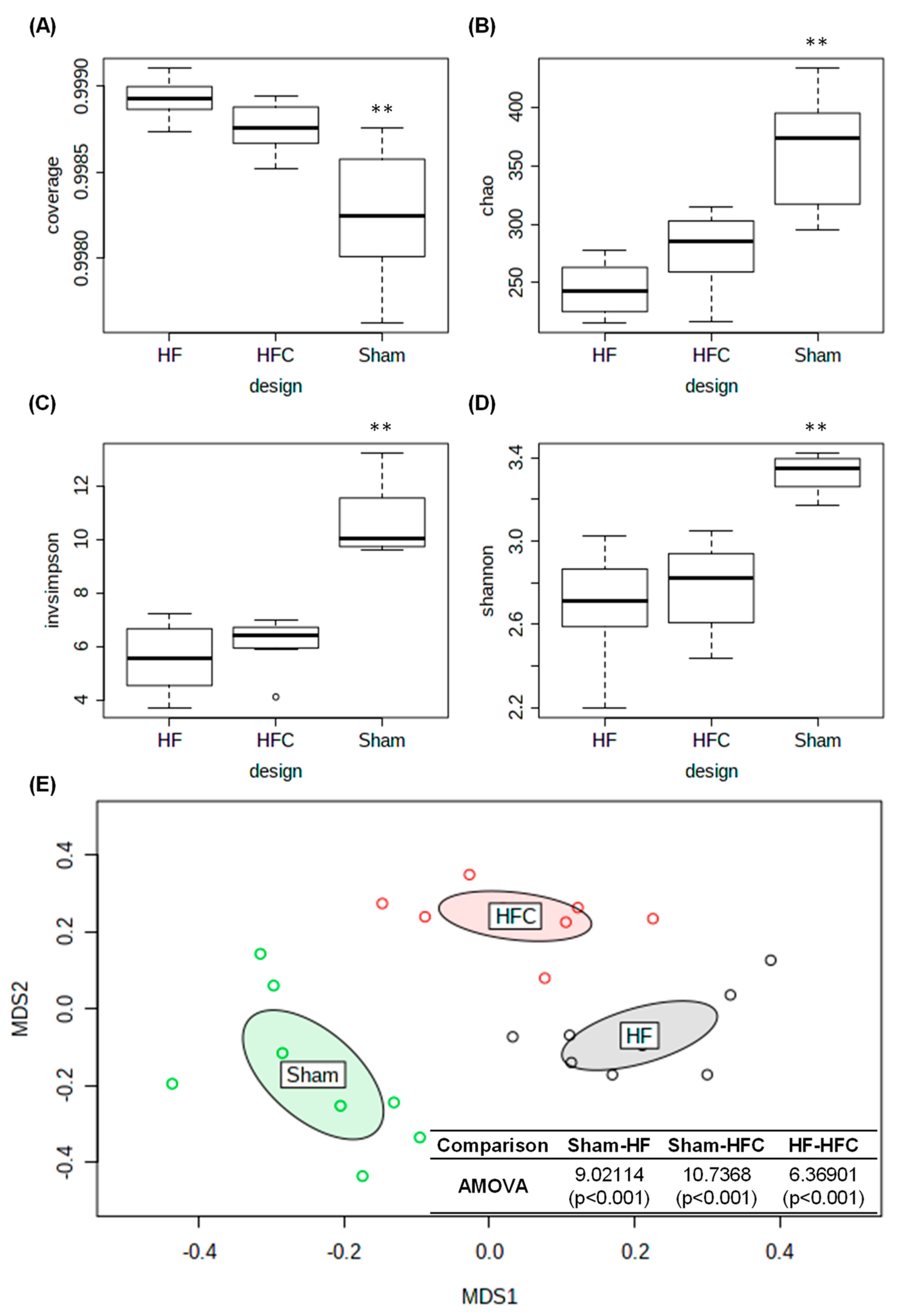
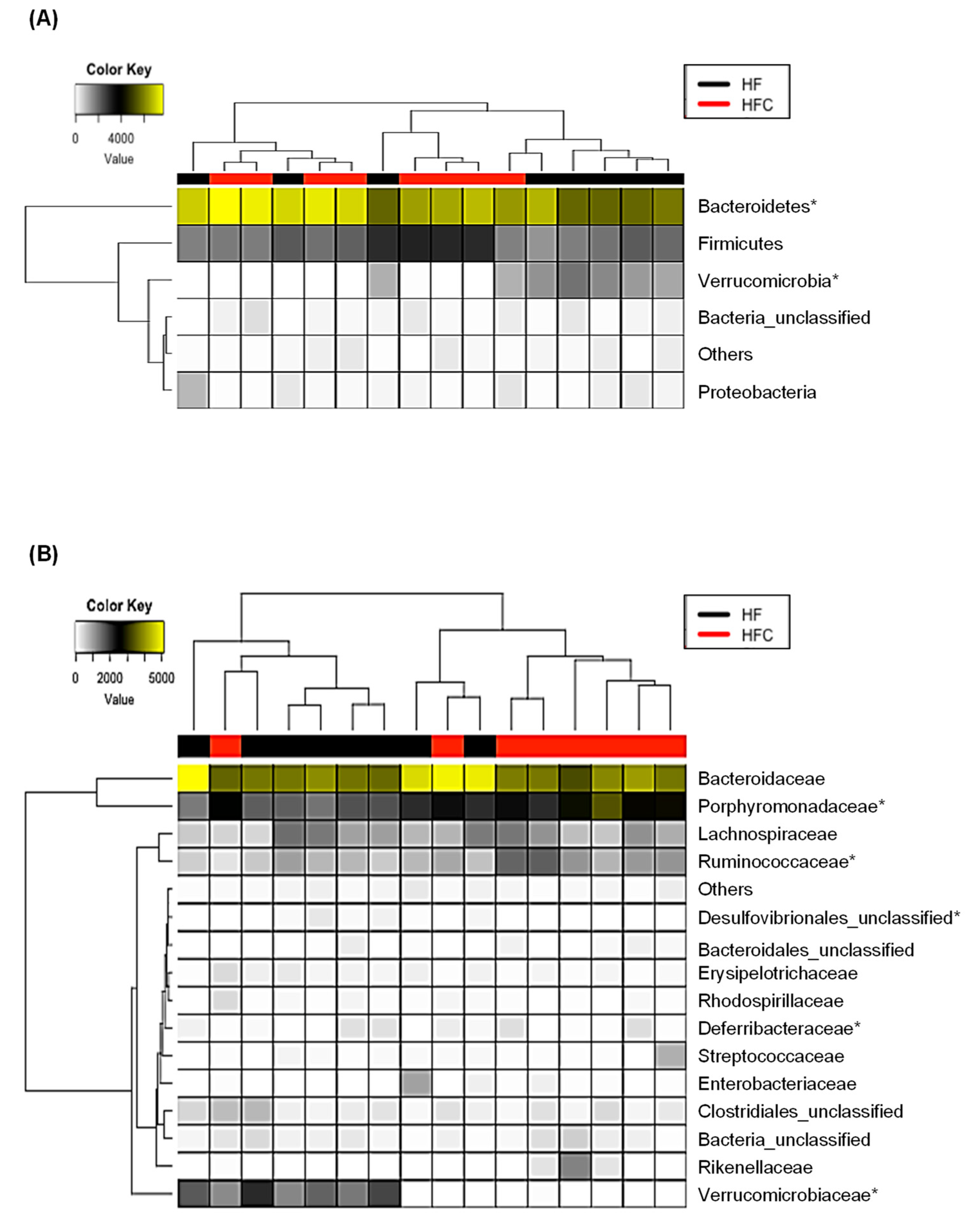
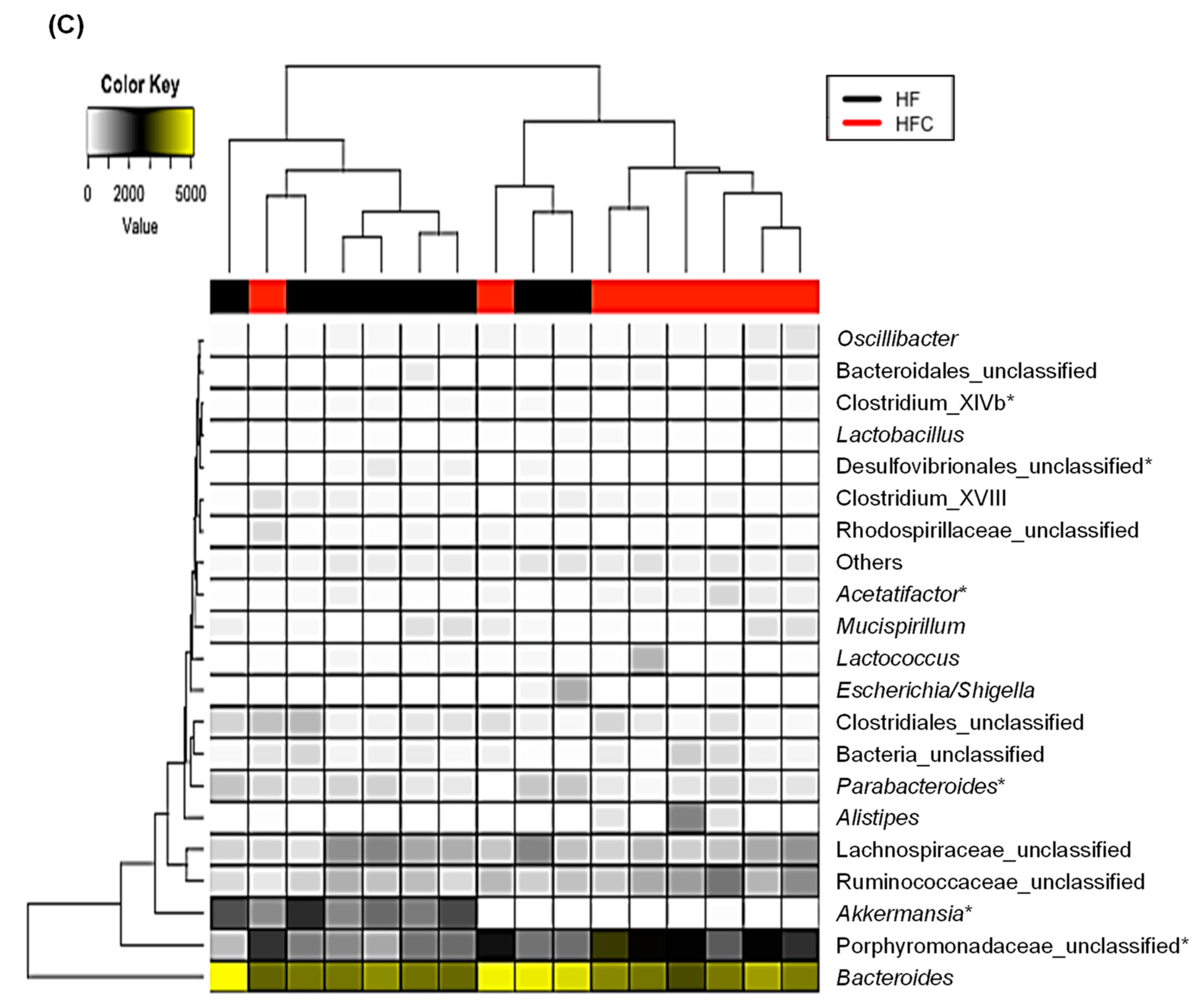
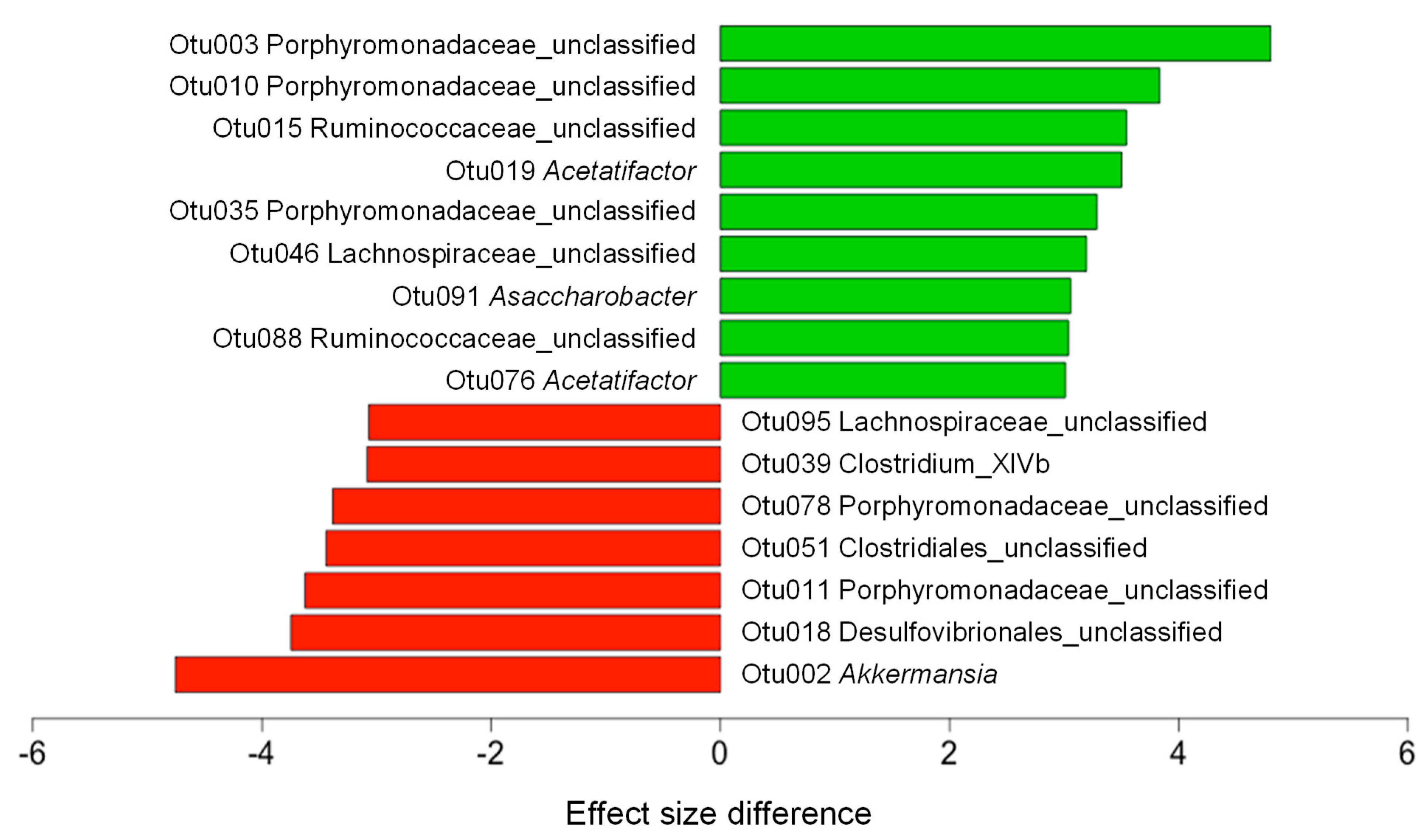
| Sham | HF | HFC | |
|---|---|---|---|
| Initial Body Weight (g) | 20.3 ± 0.81 | 20.0 ± 1.22 | 19.5 ± 1.53 |
| Final Body Weight (g) | 27.4 ± 1.47 | 36.2 ± 3.89 # | 30.8 ± 2.75 * |
| Body Weight Change (g) | 7.1 ± 0.77 | 16.2 ± 2.79 # | 11.3 ± 2.47 * |
| Sham | HF | HFC | |
|---|---|---|---|
| T-C (mg/dL) | 156.8 ± 15.43 | 208.3 ± 14.34 | 186.0 ± 21.62 |
| HDL-C (mg/dL) | 98.5 ± 6.26 | 100.6 ± 8.55 | 97.0 ± 8.92 |
| LDL-C (mg/dL) | 17.5 ± 2.50 | 28.8 ± 2.59 | 18.8 ± 3.96 * |
| Glucose (mg/dL) | 96.4 ± 11.48 | 128.4 ± 3.38 | 103.0 ± 7.77 * |
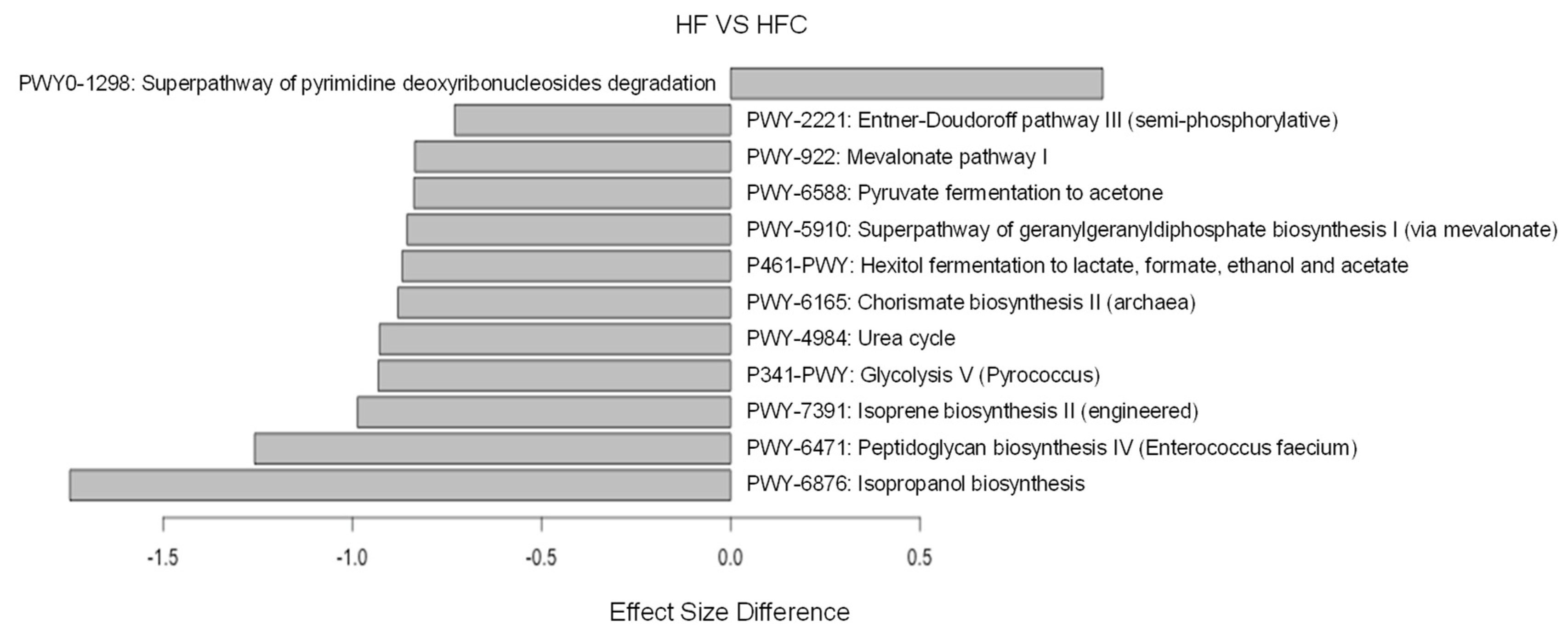
| Pathway | Increased/Decreased | Description | Associated Biomarker OTUs (Spearman Coef.) |
|---|---|---|---|
| PWY0-1298 | Increased | Pyrimidine deoxyribonucleosides degradation | Otu011: Porphyromonadaceae_unclassified (0.79) |
| PWY-6165 | Decreased | Chorismate biosynthesis II | Otu086: Acetatifactor (−0.76) Otu124 *: Ruminococceae_unclassified (0.73) |
| PWY-7391 | Decreased | Isoprene biosynthesis II | Otu157: Porphyromonadaceae_unclassified (−0.99) Otu120: Lachnospiraceae_unclassified (−0.81) Otu146: Acetatifactor (−0.81) Otu076: Acetatifactor (−0.71) Otu002 *: Akkermansia (0.77) |
| PWY-922 | Decreased | Mevalonate pathway I | Otu123: Ruminococcaceae_unclassified (−0.76) Otu124 *: Ruminococcaceae_unclassified (0.82) |
| PWY-5910 | Decreased | Geranylgeranyldiphosphate biosynthesis I | Otu136: Acetatifactor (−0.77) Otu175: Lachnospiraceae_unclassified (−0.73) Otu173: Acetatifactor (−0.71) Otu068 *: Lachnospiraceae_unclassified (0.73) Otu124 *: Ruminococcaceae_unclassified (0.82) |
| P461-PWY | Decreased | Hexitol fermentation to lactate, formate, ethanol and acetate | Otu039 *: Clostridium_XlVb (0.73) |
| PWY-2221 | Decreased | Enter-Doudoroff pathway III | Otu123: Ruminococcaceae_unclassified (−0.81) Otu124 *: Ruminococcaceae_unclassified (0.81) |
| PWY-6588 | Decreased | Pyruvate fermentation to acetone | Otu010: Porphyromonadaceae_unclassified (−0.77) Otu111: Lachnospiraceae_unclassified (−0.74) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Choi, J.H.; Oh, T.; Ahn, B.; Unno, T. Codium fragile Ameliorates High-Fat Diet-Induced Metabolism by Modulating the Gut Microbiota in Mice. Nutrients 2020, 12, 1848. https://doi.org/10.3390/nu12061848
Kim J, Choi JH, Oh T, Ahn B, Unno T. Codium fragile Ameliorates High-Fat Diet-Induced Metabolism by Modulating the Gut Microbiota in Mice. Nutrients. 2020; 12(6):1848. https://doi.org/10.3390/nu12061848
Chicago/Turabian StyleKim, Jungman, Jae Ho Choi, Taehwan Oh, Byungjae Ahn, and Tatsuya Unno. 2020. "Codium fragile Ameliorates High-Fat Diet-Induced Metabolism by Modulating the Gut Microbiota in Mice" Nutrients 12, no. 6: 1848. https://doi.org/10.3390/nu12061848
APA StyleKim, J., Choi, J. H., Oh, T., Ahn, B., & Unno, T. (2020). Codium fragile Ameliorates High-Fat Diet-Induced Metabolism by Modulating the Gut Microbiota in Mice. Nutrients, 12(6), 1848. https://doi.org/10.3390/nu12061848







