Tree Species Abundance Predictions in a Tropical Agricultural Landscape with a Supervised Classification Model and Imbalanced Data
Abstract
:1. Introduction
2. Materials and Methods
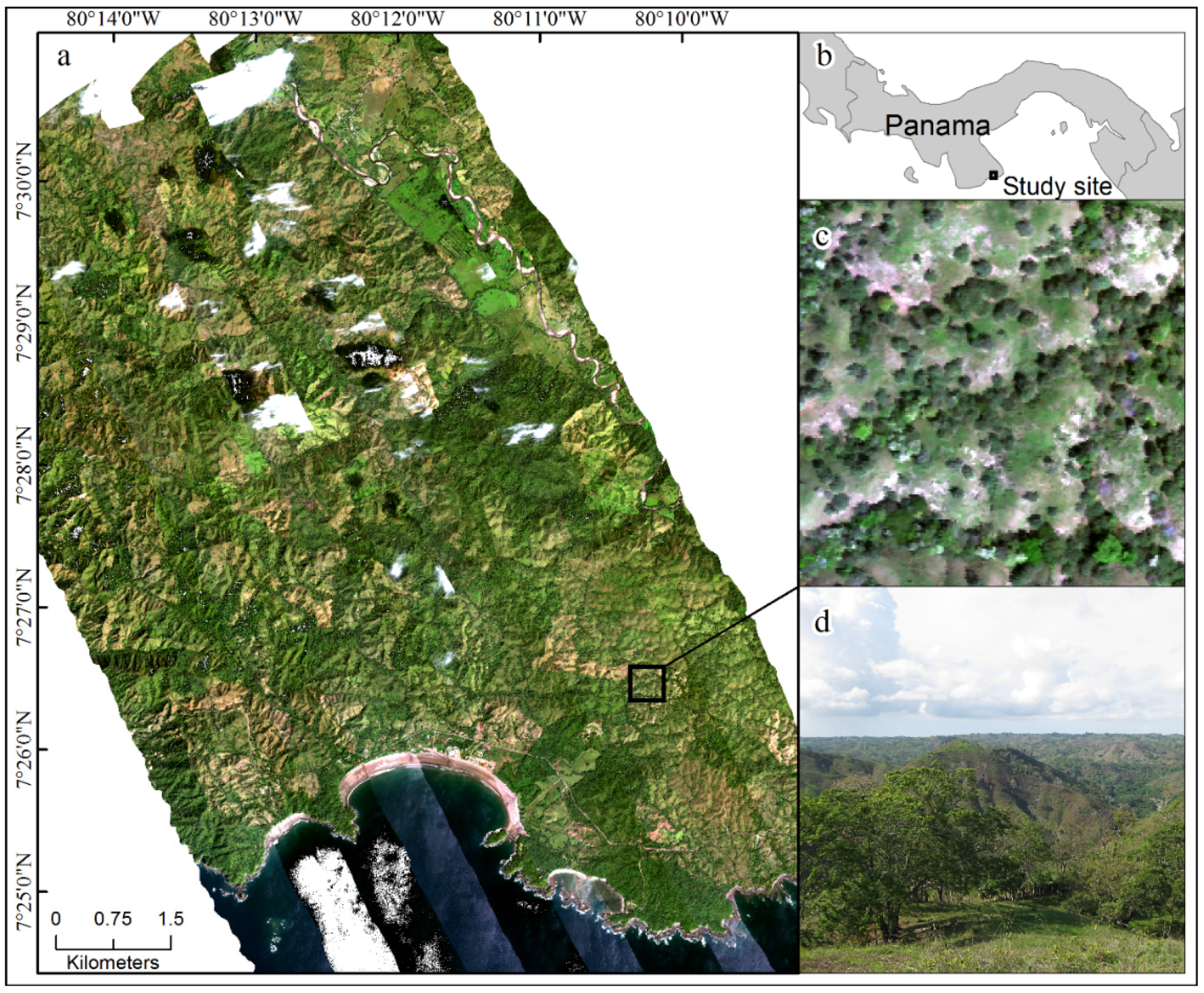
2.1. Study Site
2.2. Airborne and Field Data Collection
2.3. Training of an SVM Classifier
Parameter Optimization for an SVM Model
2.4. Assessment of SVM Classifier
Measures of Classification Performance
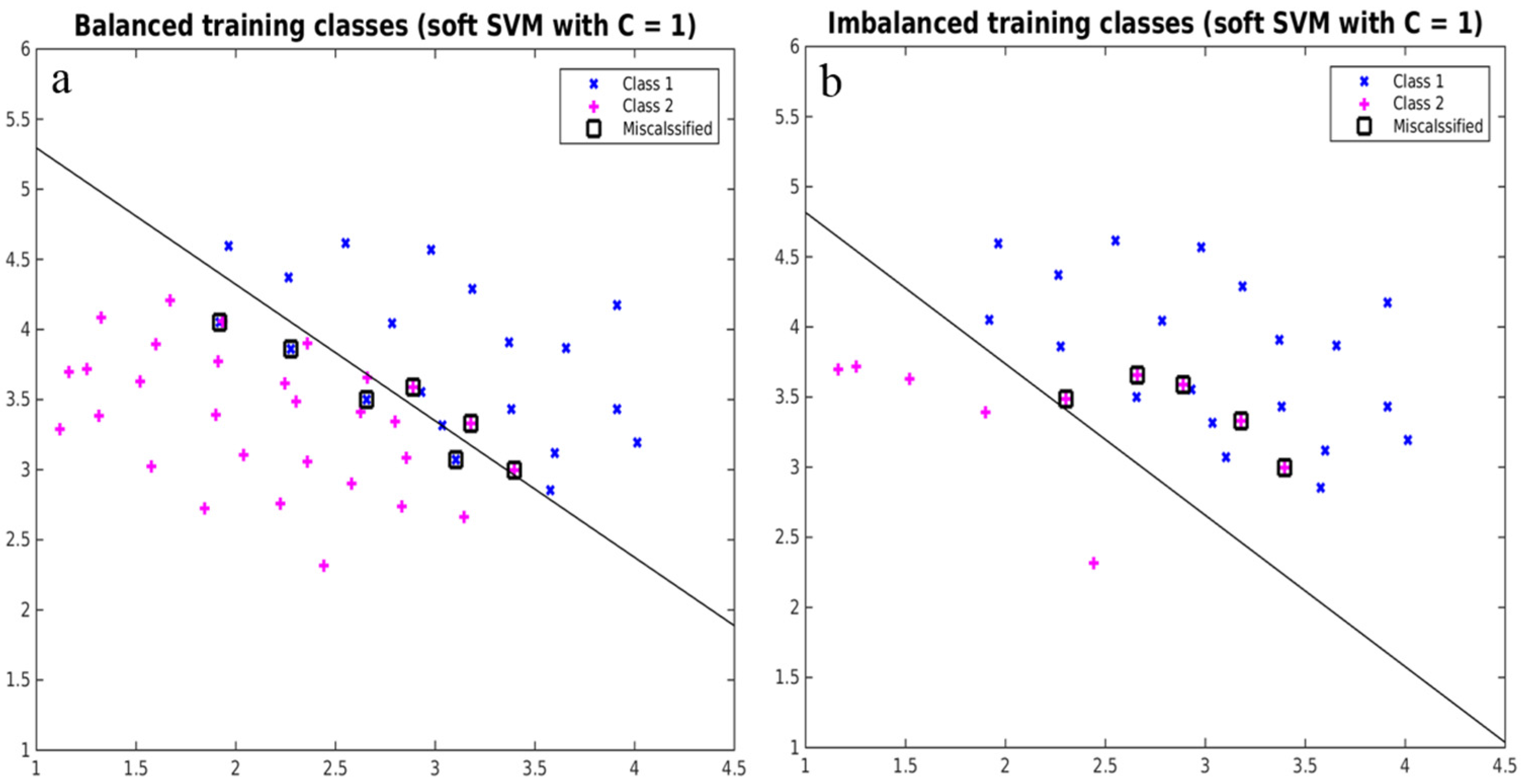
2.5. Implementing Strategies to Overcome Imbalance
2.6. Species Predictions across the Landscape
3. Results
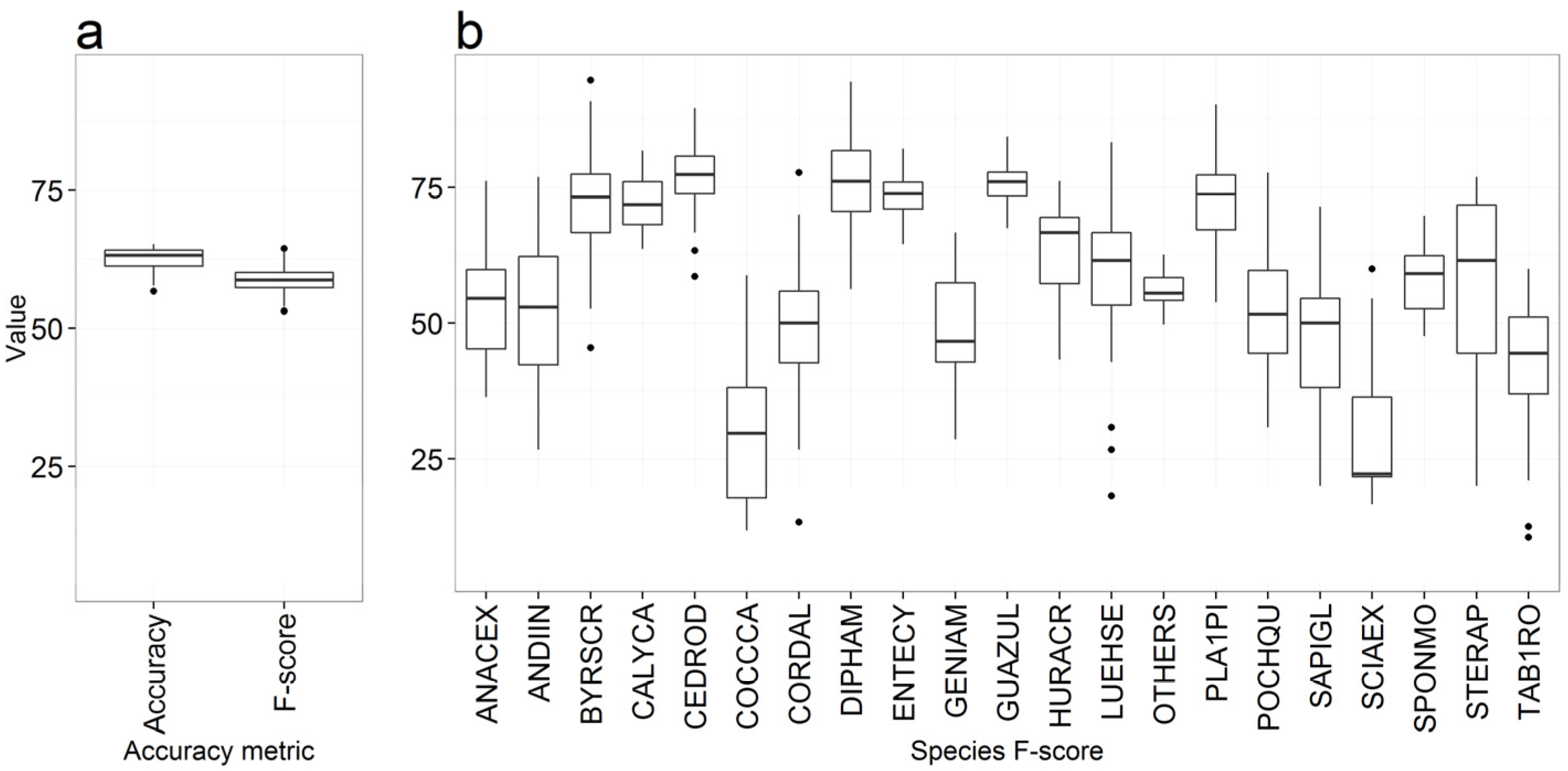
3.1. Overall Accuracy and F-score
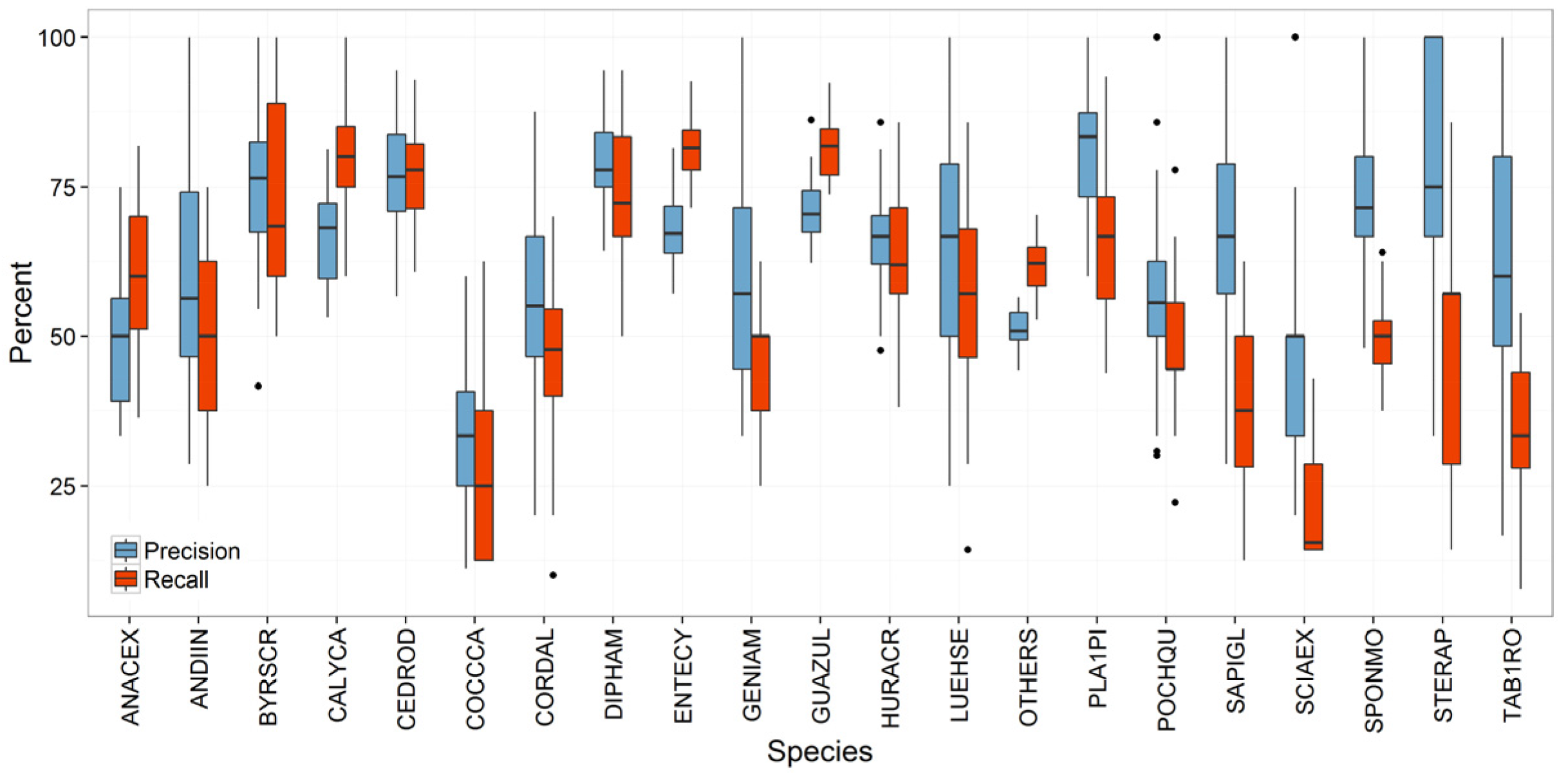
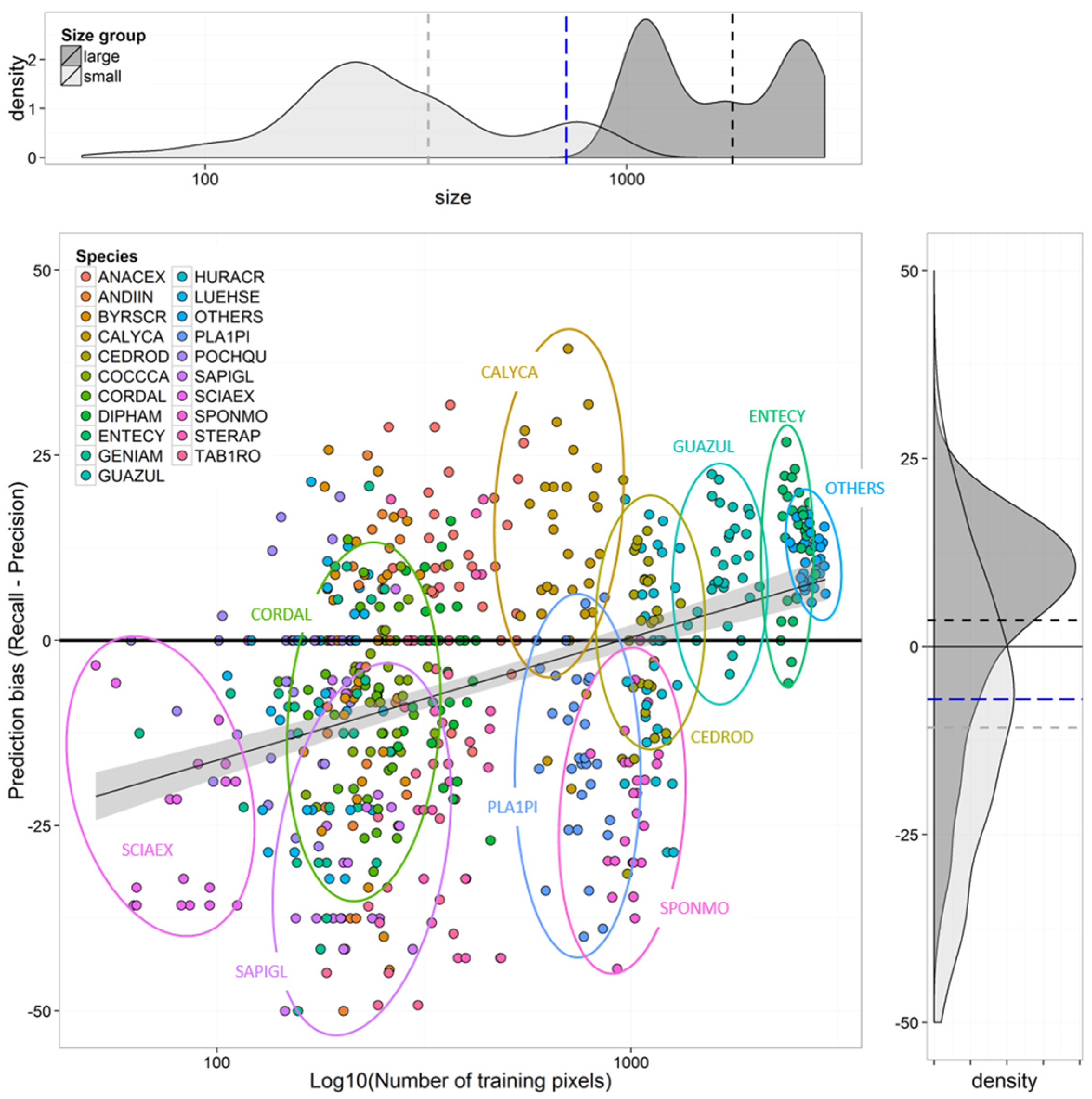
Species Prediction Errors
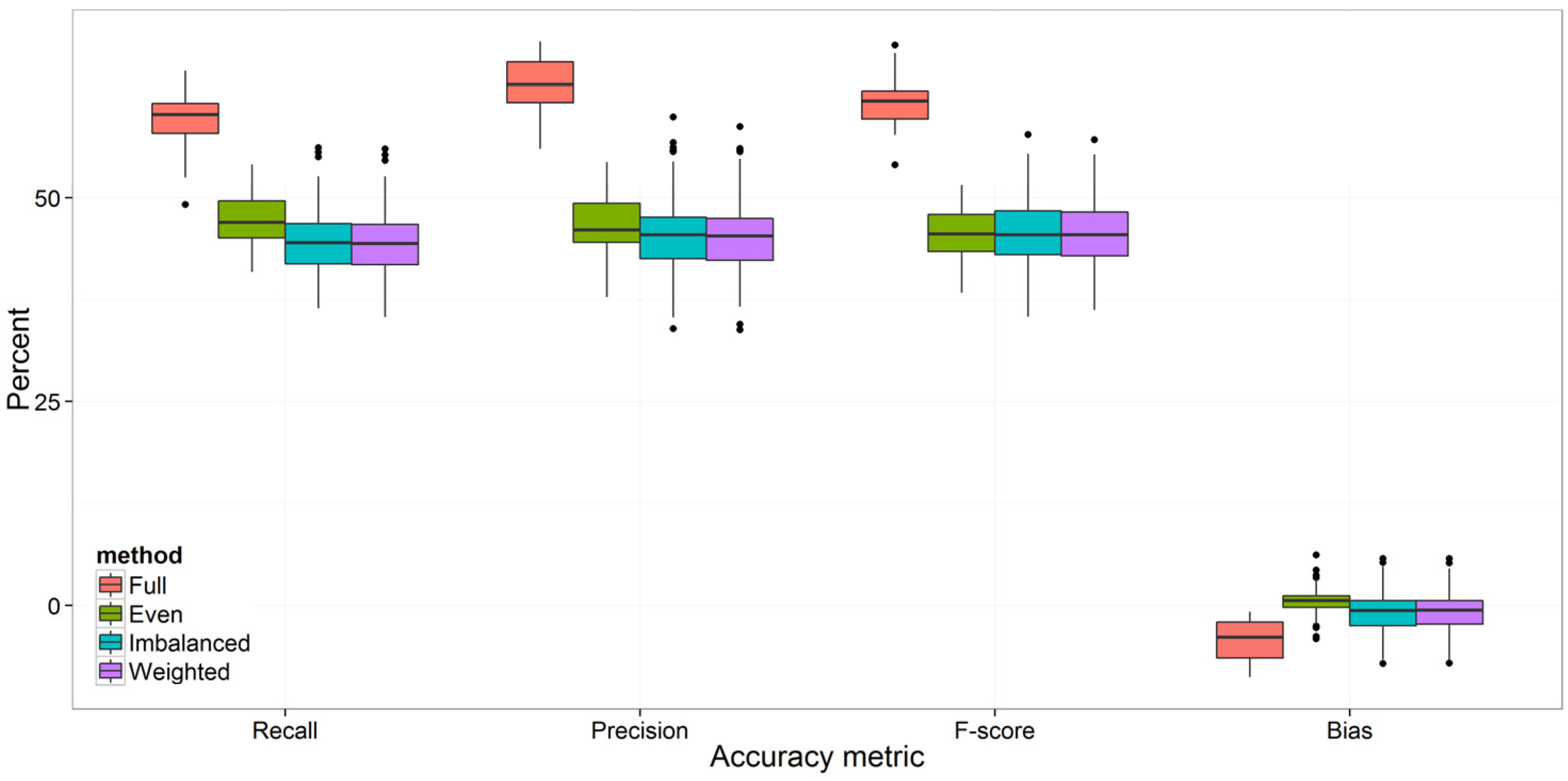
3.2. Strategies for Model Improvement
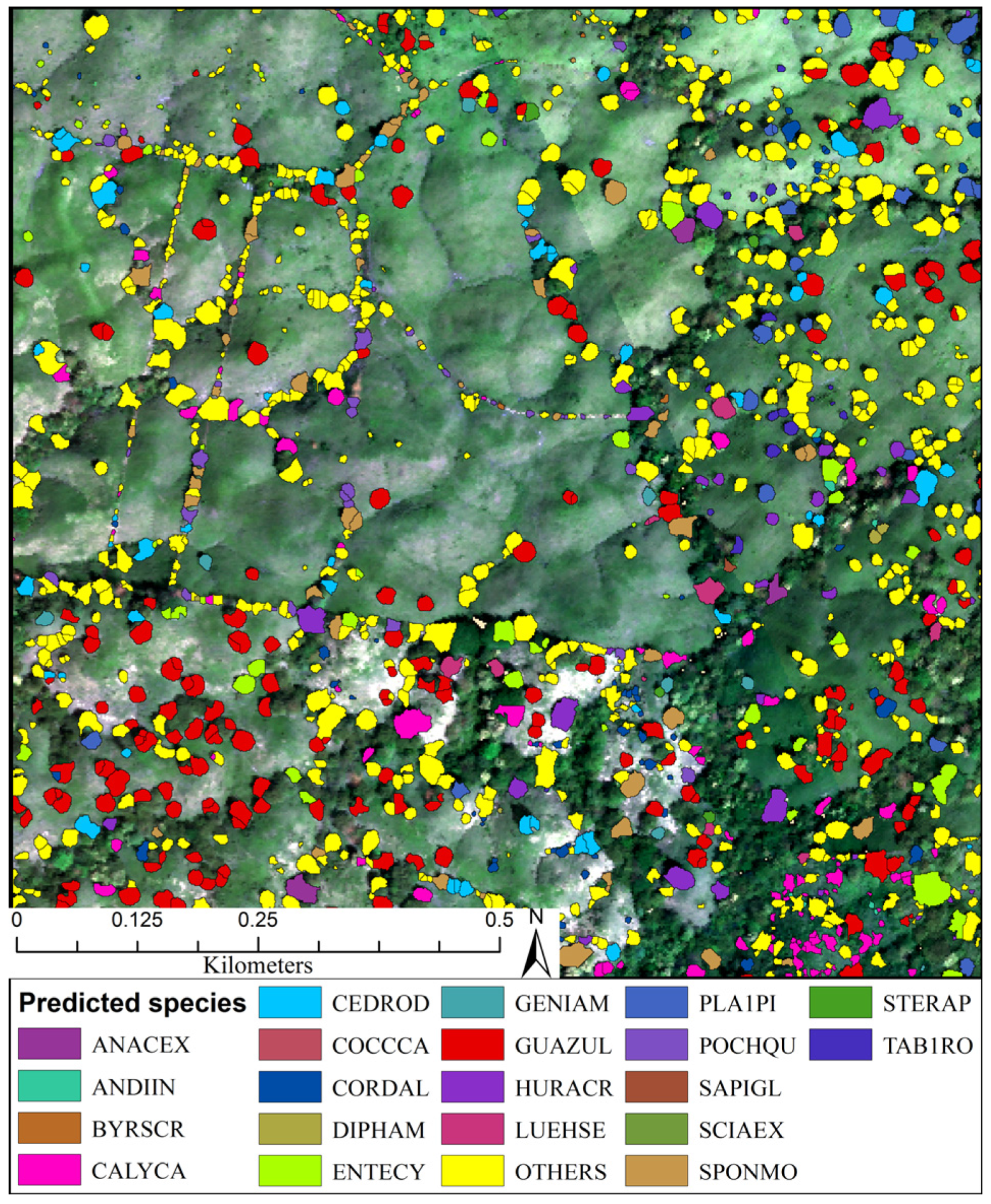
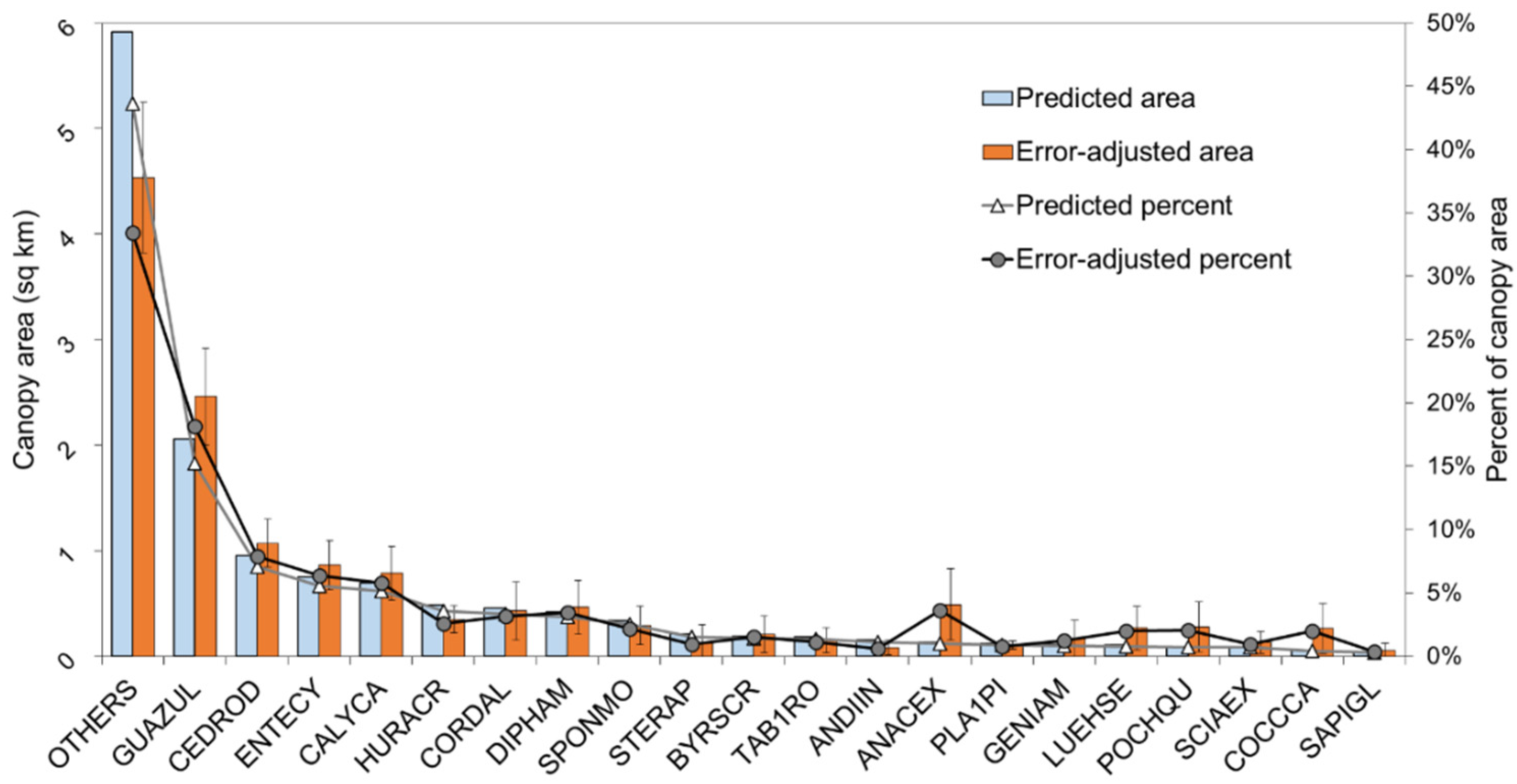
3.3. Predicted Landscape Species Distributions
4. Discussion
4.1. Effects of Imbalanced Data on Model Performance and Species Predictions
4.2. Selection of Training Data for Optimal Species-Level Accuracy
4.3. Minimum Sample Size Threshold
4.4. Operational Species Mapping in Species-Rich Landscapes
4.5. Operational Species Mapping Provides a View of the Diverse Tropical Landscape
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NIR | Near-infrared |
| NDVI | Normalized Difference Vegetation Index |
| SWIR | Shortwave-infrared |
| SVM | Support Vector Machine |
| VIS | Visible |
References
- Nagendra, H. Using remote sensing to assess biodiversity. Int. J. Remote Sens. 2001, 22, 2377–2400. [Google Scholar] [CrossRef]
- Turner, W.; Spector, S.; Gardiner, N.; Fladeland, M.; Sterling, E.; Steininger, M. Remote sensing for biodiversity science and conservation. Trends Ecol. Evol. 2003, 18, 306–314. [Google Scholar] [CrossRef]
- Colgan, M.S.; Asner, G.P. Coexistence and environmental filtering of species-specific biomass in an African savanna. Ecology 2014, 95, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Condit, R. Spatial patterns in the distribution of tropical tree species. Science 2000, 288, 1414–1418. [Google Scholar] [CrossRef] [PubMed]
- Lucas, R.; Bunting, P.; Paterson, M.; Chisholm, L. Classification of Australian forest communities using aerial photography, CASI and HyMap data. Remote Sens. Environ. 2008, 112, 2088–2103. [Google Scholar] [CrossRef]
- Ustin, S.L.; DiPietro, D.; Olmstead, K.; Underwood, E.; Scheer, G.J. Hyperspectral remote sensing for invasive species detection and mapping. IEEE Int. Geosci. Remote Sens. Sympos. 2002, 3, 1658–1660. [Google Scholar]
- Underwood, E.; Ustin, S.; DiPietro, D. Mapping nonnative plants using hyperspectral imagery. Remote Sens. Environ. 2003, 86, 150–161. [Google Scholar] [CrossRef]
- He, K.S.; Rocchini, D.; Neteler, M.; Nagendra, H. Benefits of hyperspectral remote sensing for tracking plant invasions. Divers. Distrib. 2011, 17, 381–392. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Nagendra, H.; Rocchini, D. High resolution satellite imagery for tropical biodiversity studies: The devil is in the detail. Biodivers. Conserv. 2008, 17, 3431–3442. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E.; Kellner, J.R.; Wright, S.J. Operational tree species mapping in a diverse tropical forest with airborne imaging spectroscopy. PLoS ONE 2015, 10, e0118403. [Google Scholar] [CrossRef] [PubMed]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Féret, J.B.; Asner, G.P. Semi-supervised methods to identify individual crowns of lowland tropical canopy species using imaging spectroscopy and lidar. Remote Sens. 2012, 4, 2457–2476. [Google Scholar] [CrossRef]
- Cochrane, M.A. Using vegetation reflectance variability for species level classification of hyperspectral data. Int. J. Remote Sens. 2000, 21, 2075–2087. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Asner, G.P. Improving remote species identification through efficient training data collection. Remote Sens. 2014, 6, 2682–2698. [Google Scholar] [CrossRef]
- Féret, J.B.; Asner, G.P. Tree species discrimination in tropical forests using airborne imaging spectroscopy. IEEE Trans. Geosci. Remote Sens. 2012, 51, 1–12. [Google Scholar] [CrossRef]
- Alonzo, M.; Roth, K.; Roberts, D. Identifying Santa Barbara’s urban tree species from AVIRIS imagery using canonical discriminant analysis. Remote Sens. Lett. 2013, 4, 513–521. [Google Scholar] [CrossRef]
- Pitman, N.C.A.; Terborgh, J.W.; Silman, M.R.; Núñez, P.V.; Neill, D.A.; Cerón, C.E.; Palacios, W.A.; Aulestia, M. Dominance and distribution of tree species in upper Amazonian terra firme forests. Ecology 2001, 82, 2101–2117. [Google Scholar] [CrossRef]
- Love, B.; Spaner, D. A survey of small-scale farmers using trees in pastures in Herrera Province, Panama. J. Sustain. For. 2005, 20, 37–65. [Google Scholar] [CrossRef]
- Griscom, H.P.; Connelly, A.B.; Ashton, M.S.; Wishnie, M.H.; Deago, J. The structure and composition of a tropical dry forest landscape after land clearance; Azuero peninsula, Panama. J. Sustain. For. 2011, 30, 37–41. [Google Scholar] [CrossRef]
- Sun, Y.; Wong, A.K.; Kamel, M.S. Classification of imbalanced data: A review. Int. J. Pattern Recognit. Artif. Intell. 2009, 23, 687–719. [Google Scholar] [CrossRef]
- Foody, G.M.; Mathur, A. Toward intelligent training of supervised image classifications: Directing training data acquisition for SVM classification. Remote Sens. Environ. 2004, 93, 107–117. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and lidar data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Cho, M.A.; Mathieu, R.; Asner, G.P.; Naidoo, L.; van Aardt, J.; Ramoelo, A.; Debba, P.; Wessels, K.; Main, R.; Smit, I.P.J.; et al. Mapping tree species composition in South African savannas using an integrated airborne spectral and lidar system. Remote Sens. Environ. 2012, 125, 214–226. [Google Scholar] [CrossRef]
- Knudby, A.; Nordlund, L.M.; Palmqvist, G.; Wikström, K.; Koliji, A.; Lindborg, R.; Gullström, M. Using multiple Landsat scenes in an ensemble classifier reduces classification error in a stable nearshore environment. Int. J. Appl. Earth Obs. Geoinf. 2014, 28, 90–101. [Google Scholar] [CrossRef]
- Roth, K.L.; Dennison, P.E.; Roberts, D.A. Comparing endmember selection techniques for accurate mapping of plant species and land cover using imaging spectrometer data. Remote Sens. Environ. 2012, 127, 139–152. [Google Scholar] [CrossRef]
- Jones, T.G.; Coops, N.C.; Sharma, T. Assessing the utility of airborne hyperspectral and lidar data for species distribution mapping in the coastal Pacific Northwest, Canada. Remote Sens. Environ. 2010, 114, 2841–2852. [Google Scholar] [CrossRef]
- Clark, M.L.; Roberts, D.A. Species-Level Differences in Hyperspectral Metrics among Tropical Rainforest Trees as Determined by a Tree-Based Classifier. Remote Sens. 2012, 4, 1820–1855. [Google Scholar] [CrossRef]
- Colgan, M.S.; Baldeck, C.A.; Féret, J.-B.; Asner, G.P. Mapping savanna tree species at ecosystem scales using support vector machine classification and BRDF correction on airborne hyperspectral and lidar data. Remote Sens. 2012, 4, 3462–3480. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. Support vector machines for classification in remote sensing. Int. J. Remote Sens. 2005, 26, 1007–1011. [Google Scholar] [CrossRef]
- Shao, Y.; Lunetta, R.S. Comparison of support vector machine, neural network, and CART algorithms for the land-cover classification using limited training data points. ISPRS J. Photogramm. Remote Sens. 2012, 70, 78–87. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Blagus, R.; Lusa, L. Class prediction for high-dimensional class-imbalanced data. BMC Bioinform. 2010, 11, 523. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.J.; Chen, J.J. Class-imbalanced classifiers for high-dimensional data. Brief. Bioinform. 2013, 14, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Shao, G.; Wu, J. On the accuracy of landscape pattern analysis using remote sensing data. Landsc. Ecol. 2008, 23, 505–511. [Google Scholar] [CrossRef]
- Peterson, C.J.; Dosch, J.J.; Carson, W.P. Pasture succession in the Neotropics: Extending the nucleation hypothesis into a matrix discontinuity hypothesis. Oecologia 2014, 175, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.M.; Dinerstein, E.; Wikramanayake, E.D.; Burgess, N.D.; Powell, G.V.N.; Underwood, E.C.; D’amico, J.A.; Itoua, I.; Strand, H.E.; Morrison, J.C.; et al. Terrestrial Ecoregions of the World: A New Map of Life on Earth. Bioscience 2001, 51, 933–938. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Boardman, J.; Green, R.O.; Kennedy-Bowdoin, T.; Eastwood, M.; Martin, R.E.; Anderson, C.; Field, C.B. Carnegie Airborne Observatory-2: Increasing science data dimensionality via high-fidelity multi-sensor fusion. Remote Sens. Environ. 2012, 124, 454–465. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying forest canopy traits: Imaging spectroscopy versus field survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Misc functions of the Department of Statistics, Probability Theory Group (Formally: E1071), TU Wein 2015. Available online: https://cran.r-project.org/web/packages/e1071/index.html (accessed on 1 December 2015).
- R Foundation for Statistical Computing: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org (accessed on 1 December 2015).
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Yang, J.-S.; Wang, J.-J. Margin calibration in SVM class-imbalanced learning. Neurocomputing 2009, 73, 397–411. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Colkesen, I. A kernel functions analysis for support vector machines for land cover classification. Int. J. Appl. Earth Obs. Geoinf. 2009, 11, 352–359. [Google Scholar] [CrossRef]
- Kim, J.-H. Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Comput. Stat. Data Anal. 2009, 53, 3735–3745. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, Y.Q.; Chawla, N.V.; Krasser, S. SVMs modeling for highly imbalanced classification. IEEE Trans. Syst. Man. Cybern. B. Cybern. 2009, 39, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Conrad, O. Module Watershed Segmentation. Available online: http://www.saga-gis.org/saga_module_doc/2.1.3/imagery_segmentation_0.html (accessed on 11 January 2016).
- Olofsson, P.; Foody, G.M.; Stehman, S.V.; Woodcock, C.E. Making better use of accuracy data in land change studies: Estimating accuracy and area and quantifying uncertainty using strati field estimation. Remote Sens. Environ. 2013, 129, 122–131. [Google Scholar] [CrossRef]
- Olofsson, P.; Foody, G.M.; Herold, M.; Stehman, S.V.; Woodcock, C.E.; Wulder, M.A. Good practices for estimating area and assessing accuracy of land change. Remote Sens. Environ. 2014, 148, 42–57. [Google Scholar] [CrossRef]
- Roth, K.L.; Roberts, D.A.; Dennison, P.E.; Alonzo, M.; Peterson, S.H.; Beland, M. Differentiating plant species within and across diverse ecosystems with imaging spectroscopy. Remote Sens. Environ. 2015, 167, 135–151. [Google Scholar] [CrossRef]
- Chen, X.; Gerlach, B.; Casasent, D. Pruning support vectors for imbalanced data classification. In Proceedings of International Joint Conference on Neural Networks, Montreal, QC, Canada, 31 July–4 August 2005; pp. 1883–1888.
- Baldeck, C.A.; Asner, G.P. Single-species detection with airborne imaging spectroscopy data: A comparison of support vector techniques. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 8, 2501–2512. [Google Scholar] [CrossRef]
- Platt, J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Adv. Large Margin Classif. 1999, 10, 61–74. [Google Scholar]
- Tipping, M.E. Sparse Bayesian learning and the relevance vector machine. J. Mach. Learn. Res. 2001, 1, 211–244. [Google Scholar]
- Rasmussen, C.E. Gaussian processes in machine learning. In Advanced Lectures on Machine Learning; Springer Berlin Heidelberg: Heidelberg, Germany, 2004; pp. 63–71. [Google Scholar]
- Stehman, S. V Estimating area from an accuracy assessment error matrix. Remote Sens. Environ. 2013, 132, 202–211. [Google Scholar] [CrossRef]
| Study | Sensor, Location | Spatial Resolution | Spectral Resolution | Number of Species | Imbalance | Accuracy (Classification Method) |
|---|---|---|---|---|---|---|
| Clark, Roberts, and Clark 2005 [9] | La Selva, Costa Rica; HYDICE | 1.6 m | VIS-SWIR (400–2500 nm; reduced to 30 bands selected) | 7 | I = 37 P = −0.956 | OA = 92% (LDA) |
| Jones et al. 2010 [27] | Gulf Islands, British Columbia; AISA Dual | 2 m | VIS-SWIR (429–2400 nm, reduced to 40 spectral bands) | 11 | I = 31 P = −0.896 | OA = 72% (SVM) |
| Dalponte et al. 2012 [23] | Val di Sella, Italy; AISA Eagle | 1 m | VIS-NIR (400–990 nm; 126 bands) | 7 species + 1 non-forest class | I = 35 P = −0.990 | OA = 74% (SVM) |
| Cho et al. 2012 [24] | Kruger National Park (KNP), South Africa; CAO Alpha | 1.1 m | VIS-NIR (384–1054 nm; 72 bands) | 6 | I = 31 P = −0.637 | OA = 65% (ML) |
| Clark and Roberts 2012 [28] | La Selva, Costa Rica; HYDICE | 1.6 m | VIS-SWIR (400–2500 nm; 210 bands) | Same as Clark et al. 2005 | Same as Clark et al. 2005 | OA = 87% (RF) |
| Colgan et al. 2012 [29] | KNP, South Africa; CAO Alpha | 1.1 m | VIS-NIR (385–1054 nm; 72 bands) | 15 species + 1 mixed species class | I = 15 P = −0.494 | OA = 76% (RBF-SVM) |
| Feret and Asner 2012 [16] | Hawaii, USA; CAO Alpha | 0.56 m | VIS-NIR (390–1044 nm; 24 bands) | 17 * | I = 25 P = −0.821 | OA = 73% (RBF-SVM) |
| Feret and Asner 2012 [13] | Hawaii, USA; CAO Alpha | 0.56 m | VIS-NIR (390–1044 nm; 24 bands) | 9* | I = 39 P = −1.161 | Balanced accuracy = 66% (SVM) |
| Alonzo et al. 2013 [17] | Santa Barbara, CA; AVIRIS | 3 m | VIS-SWIR (365–2500 nm, 178 bands) | 15 | Not applicable | OA = 86% (CDA, LDA) |
| Baldeck and Anser 2015 [11] | BCI, Panama; CAO AToMS | 2 m | VIS-SWIR (380–2512 nm) | 3 | Not applicable | Recall = 94-97% Prec. = 94-100% (Single-class SVM) |
| This study | Azuero Peninsula, Panama; CAO AToMS | 2 m | VIS-SWIR (380–2512 nm) | 20 + 1 mixed species class | I = 20 P = −0.756 | OA = 63% (RBF-SVM) |
| Species Code | Species | Family | Crowns | Pixels | Class Proportion |
|---|---|---|---|---|---|
| ANACEX | Anacardium excelsum | Anacardiaceae | 31 | 672 | 0.023 |
| ANDIIN | Andira inermis | Fabaceae | 24 | 618 | 0.025 |
| BYRSCR | Byrsonima crassifolia | Malpighiaceae | 29 | 402 | 0.014 |
| CALYCA | Calycophyllum candidissimum | Rubiaceae | 60 | 1163 | 0.040 |
| CEDROD | Cedrela odorata | Meliaceae | 83 | 1960 | 0.070 |
| COCCCA | Coccoloba caracasana | Polygonaceae | 24 | 422 | 0.014 |
| CORDAL | Cordia alliodora | Boraginaceae | 31 | 436 | 0.017 |
| DIPHAM | Diphysa americana | Fabaceae | 53 | 781 | 0.029 |
| ENTECY | Enterolobium cyclocarpum | Fabaceae | 82 | 4565 | 0.173 |
| GENIAM | Genipa americana | Rubiaceae | 24 | 315 | 0.013 |
| GUAZUL | Guazuma ulmifolia | Malvaceae | 116 | 2902 | 0.104 |
| HURACR | Hura crepitans | Euphorbiaceae | 62 | 2154 | 0.083 |
| LUEHSE | Luehea seemannii | Malvaceae | 21 | 510 | 0.020 |
| PLA1PI | Platymiscium pinnatum | Fabaceae | 47 | 1441 | 0.053 |
| POCHQU | Pachira quinata | Bombacaceae | 27 | 382 | 0.016 |
| SAPIGL | Sapium glandulosum | Euphorbiaceae | 24 | 416 | 0.018 |
| SCIAEX | Sciadodendron excelsum | Araliaceae | 20 | 131 | 0.004 |
| SPONMO | Spondias mombin | Anacardiaceae | 73 | 1651 | 0.064 |
| STERAP | Sterculia apetala | Malvaceae | 21 | 689 | 0.025 |
| TAB1RO | Tabebuia rosea | Bignoniaceae | 38 | 624 | 0.025 |
| OTHERS | 24 species | 222 | 4822 | 0.172 | |
| Total | 1112 | 27,056 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graves, S.J.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Colgan, M.S.; Kalantari, L.; Bohlman, S.A. Tree Species Abundance Predictions in a Tropical Agricultural Landscape with a Supervised Classification Model and Imbalanced Data. Remote Sens. 2016, 8, 161. https://doi.org/10.3390/rs8020161
Graves SJ, Asner GP, Martin RE, Anderson CB, Colgan MS, Kalantari L, Bohlman SA. Tree Species Abundance Predictions in a Tropical Agricultural Landscape with a Supervised Classification Model and Imbalanced Data. Remote Sensing. 2016; 8(2):161. https://doi.org/10.3390/rs8020161
Chicago/Turabian StyleGraves, Sarah J., Gregory P. Asner, Roberta E. Martin, Christopher B. Anderson, Matthew S. Colgan, Leila Kalantari, and Stephanie A. Bohlman. 2016. "Tree Species Abundance Predictions in a Tropical Agricultural Landscape with a Supervised Classification Model and Imbalanced Data" Remote Sensing 8, no. 2: 161. https://doi.org/10.3390/rs8020161
APA StyleGraves, S. J., Asner, G. P., Martin, R. E., Anderson, C. B., Colgan, M. S., Kalantari, L., & Bohlman, S. A. (2016). Tree Species Abundance Predictions in a Tropical Agricultural Landscape with a Supervised Classification Model and Imbalanced Data. Remote Sensing, 8(2), 161. https://doi.org/10.3390/rs8020161







