Evaluating Different Methods for Grass Nutrient Estimation from Canopy Hyperspectral Reflectance
Abstract
:1. Introduction
2. Material and Methods
2.1. Sampling Design and Canopy Spectral Measurements
2.2. Chemical Analysis
2.3. Spectral Pre-Processing
2.4. Modeling Methods
2.4.1. Univariate Linear Regression with VIs
| VIs | Formula | Plant Species | R2Val | Literature |
|---|---|---|---|---|
| SRI1 | Wheat | 0.847 | Yao et al. [23] | |
| SRI2 | Sugarcane | 0.760 | Abdel-Rahman et al. [12] | |
| SRI3 | Corn | 0.706 | Corp et al. [22] | |
| NDI1 | Wheat | 0.836 | Yao et al. [23] | |
| NDI2 | Corn | 0.672 | Corp et al. [22] | |
| TBI1 | Rice | 0.830 | Tian et al. [24] | |
| TBI2 | Rice | 0.866 | Wang et al. [25] | |
| Wheat | 0.883 | |||
| TBI3 | Holm oak | 0.760 | Pacheco-Labrador et al. [15] | |
| REP | Maize Rye | 0.860 | Cho and Skidmore [21] | |
| 0.820 |
2.4.2. Classical Multivariate Regression Methods
| Method | Reflectance Spectra Used | N (R2Val | RMSEVal) | P (R2Val | RMSEVal) | Literature |
|---|---|---|---|---|
| SMLR | Canopy, continuum-removed | 0.700–0.760 | 0.310–0.460 | Mutanga et al. [6] |
| SMLR | Leaf, absorbance | 0.232 | 0.087% | Bogrekci and Lee [5] | |
| SMLR | Image, continuum-removed | 0.210 | Ullah et al. [8] | |
| SMLR | Canopy, 1st derivative | 0.590 | 0.450% | 0.250 | 0.080% | Ramoelo et al. [27] |
| SMLR | Canopy, water-removed | 0.870 | 0.250% | 0.640 | 0.060% | Ramoelo et al. [27] |
| PLSR | Canopy, 1st derivative | 0.590 | 0.450% | 0.470 | 0.070% | Ramoelo et al. [27] |
| PLSR | Canopy, water-removed | 0.840 | 0.280% | 0.430 | 0.070% | Ramoelo et al. [27] |
| PLSR | Leaf, absorbance | 0.425 | 0.073% | Bogrekci and Lee [5] | |
| PLSR | Canopy, 1st derivative | 0.830 | 0.210% | 0.770 | 0.050% | Sanches et al. [9] |
| SVR | Image, original | 0.030–0.673 | Karimi et al. [28] | |
| SVR | Leaf, 1st derivative | 0.706 | 0.521% | 0.722 | 0.073% | Zhai et al. [14] |
| SVR | Leaf, original | 0.197 | 0.946% | 0.458 | 0.097% | Zhai et al. [14] |
2.4.3. SPA-MLR
2.5. Model Development
| Nutrient | Dataset | n | Range (%) | Mean (%) | CV (%) | |
|---|---|---|---|---|---|---|
| Original data | N | December 2012 | 66 | 1.73−3.08 | 2.44 | 14.56 |
| April 2013 | 71 | 1.00−2.07 | 1.59 | 12.98 | ||
| P | December 2012 | 66 | 0.17−0.41 | 0.28 | 17.51 | |
| April 2013 | 71 | 0.08−0.20 | 0.16 | 13.57 | ||
| Data partitioning | N | Training set | 69 | 1.00−3.08 | 2.00 | 26.03 |
| Test set | 68 | 1.12−3.06 | 2.00 | 25.38 | ||
| P | Training set | 69 | 0.08−0.41 | 0.22 | 33.06 | |
| Test set | 68 | 0.18−0.37 | 0.22 | 31.68 |
2.6. Model Comparison
3. Results
3.1. Comparison of the Modeling Methods for N Estimation
| Modeling Methods | Model Equation | Cross-Validation (n = 69) | Validation (n = 68) | ||||
|---|---|---|---|---|---|---|---|
| R2CV | RMSECV | R2Val | RMSEVal | bias | RPD | ||
| SRI1 | 0.649 | 0.306 | 0.639 | 0.303 | −0.028 | 1.66 | |
| SRI2 | 0.653 | 0.304 | 0.619 | 0.315 | −0.031 | 1.60 | |
| SRI3 | 0.709 | 0.278 | 0.705 | 0.275 | −0.032 | 1.83 | |
| NDI1 | 0.675 | 0.294 | 0.674 | 0.289 | −0.033 | 1.74 | |
| NDI2 | 0.671 | 0.296 | 0.650 | 0.301 | −0.044 | 1.67 | |
| TBI1 | 1 | 0.580 | 0.335 | 0.480 | 0.367 | −0.042 | 1.37 |
| TBI2 | 0.668 | 0.298 | 0.599 | 0.320 | −0.026 | 1.57 | |
| TBI3 | 0.260 | 0.447 | 0.354 | 0.406 | −0.018 | 1.24 | |
| REP | 0.699 | 0.283 | 0.692 | 0.282 | −0.039 | 1.78 | |
| SMLR | 0.708 | 0.279 | 0.670 | 0.291 | 0.031 | 1.73 | |
| PLSR | 4 LVs | 0.538 | 0.363 | 0.747 | 0.248 | 0.048 | 2.03 |
| SVR | 69 SVs | 0.594 | 0.334 | 0.740 | 0.254 | 0.051 | 1.98 |
| SPA-MLR | 0.710 | 0.278 | 0.738 | 0.258 | −0.007 | 1.95 | |
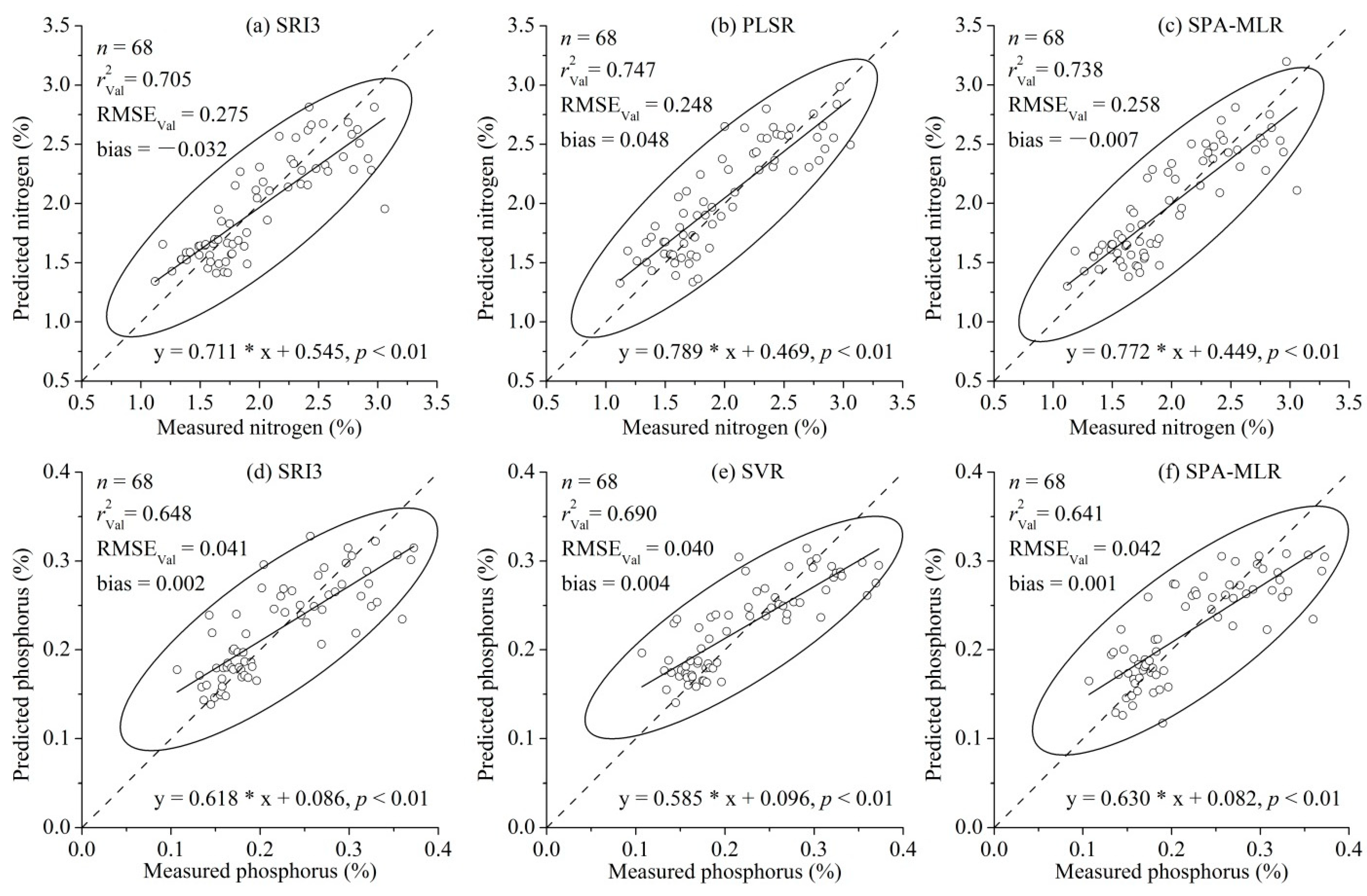
3.2. Comparison of the Modeling Methods for P Estimation
| Modeling Methods | Model Equation | Cross-Validation (n = 69) | Validation (n = 68) | ||||
|---|---|---|---|---|---|---|---|
| R2CV | RMSECV | R2Val | RMSEVal | bias | RPD | ||
| SRI1 | 0.646 | 0.043 | 0.605 | 0.044 | 0.004 | 1.58 | |
| SRI2 | 0.545 | 0.049 | 0.601 | 0.046 | −0.113 | 1.55 | |
| SRI3 | 0.666 | 0.042 | 0.648 | 0.041 | 0.002 | 1.70 | |
| NDI1 | 0.663 | 0.042 | 0.609 | 0.044 | 0.005 | 1.58 | |
| NDI2 | 0.608 | 0.046 | 0.591 | 0.045 | 0.003 | 1.54 | |
| TBI1 | 1 | 0.417 | 0.056 | 0.423 | 0.053 | 0.001 | 1.31 |
| TBI2 | 0.568 | 0.048 | 0.557 | 0.046 | 0.002 | 1.51 | |
| TBI3 | 0.403 | 0.056 | 0.307 | 0.062 | 0.012 | 1.12 | |
| REP | 0.637 | 0.044 | 0.618 | 0.043 | 0.004 | 1.62 | |
| SMLR | 0.621 | 0.045 | 0.583 | 0.045 | 0.006 | 1.53 | |
| PLSR | 2 LVs | 0.559 | 0.048 | 0.547 | 0.047 | 0.003 | 1.48 |
| SVR | 60 SVs | 0.599 | 0.046 | 0.690 | 0.040 | 0.004 | 1.77 |
| SPA-MLR | 0.620 | 0.045 | 0.641 | 0.042 | 0.001 | 1.66 | |
3.3. Differences in Model Accuracy
4. Discussion
4.1. Comparison of Model Performance
| Model Sequence Number | Mean Difference (I−J) | p-Value | 95% Confidence Interval | ||
|---|---|---|---|---|---|
| I | J | Lower Bound | Upper Bound | ||
| 13 | 1 | 0.068 | 0.283 | −0.060 | 0.195 |
| 2 | 0.080 | 0.208 | −0.048 | 0.207 | |
| 3 | 0.013 | 0.834 | −0.115 | 0.141 | |
| 4 | 0.048 | 0.441 | −0.080 | 0.176 | |
| 5 | 0.069 | 0.272 | −0.059 | 0.197 | |
| 6 | 0.238 | 0.001 | 0.110 | 0.366 | |
| 7 | 0.112 | 0.083 | −0.016 | 0.239 | |
| 8 | 0.359 | 0.000 | 0.231 | 0.487 | |
| 9 | 0.035 | 0.579 | −0.093 | 0.162 | |
| 10 | 0.043 | 0.495 | −0.085 | 0.170 | |
| 11 | −0.026 | 0.681 | −0.153 | 0.102 | |
| 12 | 0.025 | 0.693 | −0.103 | 0.152 | |
4.2. Interpretation of the SPA-MLR Model
5. Conclusions
- The SPA-MLR method had the optimal performance in both N and P estimations synthetically considering model accuracy, simplicity, robustness and interpretation, and the SRI3 model had comparable performance to the SPA-MLR model in P estimation
- The sensitive spectral bands employed by SPA-MLR method for N (715 and 731 nm) and P (714 and 729 nm) estimations were indirectly related to foliar chlorophyll content.
- Apart from two VIs (TBI1 and TBI3) that provided unsuccessful N and P estimations (R2Val < 0.50), the other seven VIs (SRI1, SRI2, SRI3, NDI1, NDI2, TBI2 and REP) had the potential to be applied to grass species (e.g., C. cinerascens).
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ingestad, T.; Lund, A.B. Theory and techniques for steady state mineral nutrition and growth of plants. Scand. J. For. Res. 1986, 1, 439–453. [Google Scholar] [CrossRef]
- Van der Graaf, A.; Stahl, J.; Veen, G.; Havinga, R.; Drent, R. Patch choice of avian herbivores along a migration trajectory—From Temperate to Arctic. Basic Appl. Ecol. 2007, 8, 354–363. [Google Scholar]
- Van Gils, J.A.; Dekinga, A.; van den Hout, P.J.; Spaans, B.; Piersma, T. Digestive organ size and behavior of red knots (Calidris canutus) indicate the quality of their benthic food stocks. Isr. J. Ecol. Evol. 2007, 53, 329–346. [Google Scholar]
- Yoder, B.J.; Pettigrew-Crosby, R.E. Predicting nitrogen and chlorophyll content and concentrations from reflectance spectra (400–2500 nm) at leaf and canopy scales. Remote Sens. Environ. 1995, 53, 199–211. [Google Scholar] [CrossRef]
- Bogrekci, I.; Lee, W. Spectral phosphorus mapping using diffuse reflectance of soils and grass. Biosyst. Eng. 2005, 91, 305–312. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.; Kumar, L.; Ferwerda, J. Estimating tropical pasture quality at canopy level using band depth analysis with continuum removal in the visible domain. Int. J. Remote Sens. 2005, 26, 1093–1108. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C.; Corsi, F.; Cho, M. LAI and chlorophyll estimation for a heterogeneous grassland using hyperspectral measurements. ISPRS J. Photogramm. Remote Sens. 2008, 63, 409–426. [Google Scholar] [CrossRef]
- Ullah, S.; Si, Y.; Schlerf, M.; Skidmore, A.K.; Shafique, M.; Iqbal, I.A. Estimation of grassland biomass and nitrogen using meris data. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 196–204. [Google Scholar] [CrossRef]
- Sanches, I.; Tuohy, M.; Hedley, M.; Mackay, A. Seasonal prediction of in situ pasture macronutrients in New Zealand pastoral systems using hyperspectral data. Int. J. Remote Sens. 2013, 34, 276–302. [Google Scholar] [CrossRef]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Smith, R.B.; De Pauw, E. Hyperspectral vegetation indices and their relationships with agricultural crop characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Abdel-Rahman, E.M.; Ahmed, F.B.; van den Berg, M. Estimation of sugarcane leaf nitrogen concentration using in situ spectroscopy. Int. J. Appl. Earth Obs. Geoinf. 2010, 12, S52–S57. [Google Scholar] [CrossRef]
- Yu, K.; Li, F.; Gnyp, M.L.; Miao, Y.; Bareth, G.; Chen, X. Remotely detecting canopy nitrogen concentration and uptake of paddy rice in the Northeast China Plain. ISPRS J. Photogramm. Remote Sens. 2013, 78, 102–115. [Google Scholar] [CrossRef]
- Zhai, Y.; Cui, L.; Zhou, X.; Gao, Y.; Fei, T.; Gao, W. Estimation of nitrogen, phosphorus, and potassium contents in the leaves of different plants using laboratory-based visible and near-infrared reflectance spectroscopy: Comparison of partial least-square regression and support vector machine regression methods. Int. J. Remote Sens. 2013, 34, 2502–2518. [Google Scholar] [CrossRef]
- Pacheco-Labrador, J.; González-Cascón, R.; Martín, M.P.; Riaño, D. Understanding the optical responses of leaf nitrogen in Mediterranean holm oak (Quercus ilex) using field spectroscopy. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 105–118. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Pearson, R.L.; Miller, L.D. Remote Mapping of Standing Crop Biomass for Estimation of the Productivity of the Shortgrass Prairie; Eighth International Symposium on Remote Sensing of Environment; Willow Run Laboratories: Ann Arbor, MI, USA, 1972; p. 1355. [Google Scholar]
- Jackson, R.D.; Huete, A.R. Interpreting vegetation indices. Prev. Vet. Med. 1991, 11, 185–200. [Google Scholar] [CrossRef]
- Vogelmann, J.; Rock, B.; Moss, D. Red edge spectral measurements from sugar maple leaves. Int. J. Remote Sens. 1993, 14, 1563–1575. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Cho, M.A.; Skidmore, A.K. A new technique for extracting the red edge position from hyperspectral data: The linear extrapolation method. Remote Sens. Environ. 2006, 101, 181–193. [Google Scholar] [CrossRef]
- Corp, L.A.; Middleton, E.M.; Campbell, P.E.; Daughtry, C.S.; Russ, A.; Cheng, Y.-B.; Huemmrich, K.F. Spectral indices to monitor nitrogen-driven carbon uptake in field corn. J. Appl. Remote Sens. 2010, 4, 043555. [Google Scholar]
- Yao, X.; Zhu, Y.; Tian, Y.; Feng, W.; Cao, W. Exploring hyperspectral bands and estimation indices for leaf nitrogen accumulation in wheat. Int. J. Appl. Earth Obs. Geoinf. 2010, 12, 89–100. [Google Scholar] [CrossRef]
- Tian, Y.; Yao, X.; Yang, J.; Cao, W.; Hannaway, D.; Zhu, Y. Assessing newly developed and published vegetation indices for estimating rice leaf nitrogen concentration with ground-and space-based hyperspectral reflectance. Field Crop. Res. 2011, 120, 299–310. [Google Scholar] [CrossRef]
- Wang, W.; Yao, X.; Yao, X.; Tian, Y.; Liu, X.; Ni, J.; Cao, W.; Zhu, Y. Estimating leaf nitrogen concentration with three-band vegetation indices in rice and wheat. Field Crop. Res. 2012, 129, 90–98. [Google Scholar] [CrossRef]
- Thomas, J.; Oerther, G. Estimating nitrogen content of sweet pepper leaves by reflectance measurements. Agron. J. 1972, 64, 11–13. [Google Scholar] [CrossRef]
- Ramoelo, A.; Skidmore, A.K.; Schlerf, M.; Mathieu, R.; Heitkönig, I. Water-removed spectra increase the retrieval accuracy when estimating savanna grass nitrogen and phosphorus concentrations. ISPRS J. Photogramm. Remote Sens. 2011, 66, 408–417. [Google Scholar] [CrossRef]
- Karimi, Y.; Prasher, S.; Madani, A.; Kim, S. Application of support vector machine technology for the estimation of crop biophysical parameters using aerial hyperspectral observations. Can. Biosyst. Eng. 2008, 50, 13–20. [Google Scholar]
- Wise, B.; Gallagher, N.; Bro, R.; Shaver, J.; Windig, W.; Koch, R. Chemometrics Tutorial for PLS_Toolbox and Solo; Eigenvector Research: Wenatchee, WA, USA, 2006. [Google Scholar]
- Arkan, E.; Shahlaei, M.; Pourhossein, A.; Fakhri, K.; Fassihi, A. Validated QSAR analysis of some diaryl substituted pyrazoles as CCR2 inhibitors by various linear and nonlinear multivariate chemometrics methods. Eur. J. Med. Chem. 2010, 45, 3394–3406. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Fei, T.; Qi, Q.; Liu, Y.; Wu, G. Estimating carex quality with laboratory-based hyperspectral measurements. Int. J. Remote Sens. 2013, 34, 1866–1878. [Google Scholar] [CrossRef]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvão, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Di Nezio, M.S.; Pistonesi, M.F.; Fragoso, W.D.; Pontes, M.J.; Goicoechea, H.C.; Araujo, M.C. Successive projections algorithm improving the multivariate simultaneous direct spectrophotometric determination of five phenolic compounds in sea water. Microchem. J. 2007, 85, 194–200. [Google Scholar] [CrossRef]
- Liu, F.; He, Y. Application of successive projections algorithm for variable selection to determine organic acids of plum vinegar. Food Chem. 2009, 115, 1430–1436. [Google Scholar] [CrossRef]
- Kemper, T.; Sommer, S. Estimate of heavy metal contamination in soils after a mining accident using reflectance spectroscopy. Environ. Sci. Technol. 2002, 36, 2742–2747. [Google Scholar] [CrossRef] [PubMed]
- Saeys, W.; Mouazen, A.M.; Ramon, H. Potential for onsite and online analysis of pig manure using visible and near infrared reflectance spectroscopy. Biosyst. Eng. 2005, 91, 393–402. [Google Scholar] [CrossRef]
- Myung, I.J.; Pitt, M.A. Applying occam’s razor in modeling cognition: A Bayesian approach. Psychon. Bull. Rev. 1997, 4, 79–95. [Google Scholar] [CrossRef]
- Zeaiter, M.; Roger, J.-M.; Bellon-Maurel, V.; Rutledge, D. Robustness of models developed by multivariate calibration. Part I: The assessment of robustness. TrAC Trends Anal. Chem. 2004, 23, 157–170. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, J. Feeding ecology of two wintering geese species at Poyang Lake, China. J. Freshw. Ecol. 1999, 14, 439–445. [Google Scholar] [CrossRef]
- Bradstreet, R. Kjeldahl method for organic nitrogen. Anal. Chem. 1954, 26, 185–187. [Google Scholar] [CrossRef]
- Yuan, G.; Lavkulich, L. Colorimetric determination of phosphorus in citrate-bicarbonate-dithionite extracts of soils. Commun. Soil Sci. Plant Anal. 1995, 26, 1979–1988. [Google Scholar] [CrossRef]
- Schmidt, K.; Skidmore, A. Smoothing vegetation spectra with wavelets. Int. J. Remote Sens. 2004, 25, 1167–1184. [Google Scholar] [CrossRef]
- Mirik, M.; Norland, J.E.; Crabtree, R.L.; Biondini, M.E. Hyperspectral one-meter-resolution remote sensing in Yellowstone National Park, Wyoming: I. Forage nutritional values. Rangel. Ecol. Manag. 2005, 58, 452–458. [Google Scholar] [CrossRef]
- Pimstein, A.; Karnieli, A.; Bansal, S.K.; Bonfil, D.J. Exploring remotely sensed technologies for monitoring wheat potassium and phosphorus using field spectroscopy. Field Crop. Res. 2011, 121, 125–135. [Google Scholar] [CrossRef]
- Neter, J.; Kutner, M.H.; Wasserman, W.; Nachtsheim, C. Applied Linear Regression Models, 4th ed.; McGraw-Hill/Irwin: New York, NY, USA, 1996. [Google Scholar]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Kooistra, L.; Salas, E.; Clevers, J.; Wehrens, R.; Leuven, R.; Nienhuis, P.; Buydens, L. Exploring field vegetation reflectance as an indicator of soil contamination in river floodplains. Environ. Pollut. 2004, 127, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef]
- Basak, D.; Pal, S.; Patranabis, D.C. Support vector regression. Neural Inf. Process.-Lett. Rev. 2007, 11, 203–224. [Google Scholar]
- Ye, S.; Wang, D.; Min, S. Successive projections algorithm combined with uninformative variable elimination for spectral variable selection. Chemom. Intell. Lab. Syst. 2008, 91, 194–199. [Google Scholar] [CrossRef]
- Williams, P. Near-Infrared Technology: Getting the Best Out of Light: A Short Course in the Practical Implementation of Near-Infrared Spectroscopy for the User, 2nd ed.; Value Added Wheat CRC: Sydney, NSW, Australia, 2004. [Google Scholar]
- Bian, M.; Skidmore, A.K.; Schlerf, M.; Wang, T.; Liu, Y.; Zeng, R.; Fei, T. Predicting foliar biochemistry of tea (Camellia sinensis) using reflectance spectra measured at powder, leaf and canopy levels. ISPRS J. Photogramm. Remote Sens. 2013, 78, 148–156. [Google Scholar] [CrossRef]
- Kawamura, K.; Mackay, A.; Tuohy, M.; Betteridge, K.; Sanches, I.; Inoue, Y. Potential for spectral indices to remotely sense phosphorus and potassium content of legume-based pasture as a means of assessing soil phosphorus and potassium fertility status. Int. J. Remote Sens. 2011, 32, 103–124. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Wang, T.; Skidmore, A.K.; Shi, T.; Wu, G. Evaluating Different Methods for Grass Nutrient Estimation from Canopy Hyperspectral Reflectance. Remote Sens. 2015, 7, 5901-5917. https://doi.org/10.3390/rs70505901
Wang J, Wang T, Skidmore AK, Shi T, Wu G. Evaluating Different Methods for Grass Nutrient Estimation from Canopy Hyperspectral Reflectance. Remote Sensing. 2015; 7(5):5901-5917. https://doi.org/10.3390/rs70505901
Chicago/Turabian StyleWang, Junjie, Tiejun Wang, Andrew K. Skidmore, Tiezhu Shi, and Guofeng Wu. 2015. "Evaluating Different Methods for Grass Nutrient Estimation from Canopy Hyperspectral Reflectance" Remote Sensing 7, no. 5: 5901-5917. https://doi.org/10.3390/rs70505901
APA StyleWang, J., Wang, T., Skidmore, A. K., Shi, T., & Wu, G. (2015). Evaluating Different Methods for Grass Nutrient Estimation from Canopy Hyperspectral Reflectance. Remote Sensing, 7(5), 5901-5917. https://doi.org/10.3390/rs70505901







