Individual Tree Detection in a Eucalyptus Plantation Using Unmanned Aerial Vehicle (UAV)-LiDAR
Abstract
1. Introduction
2. Materials and Methods
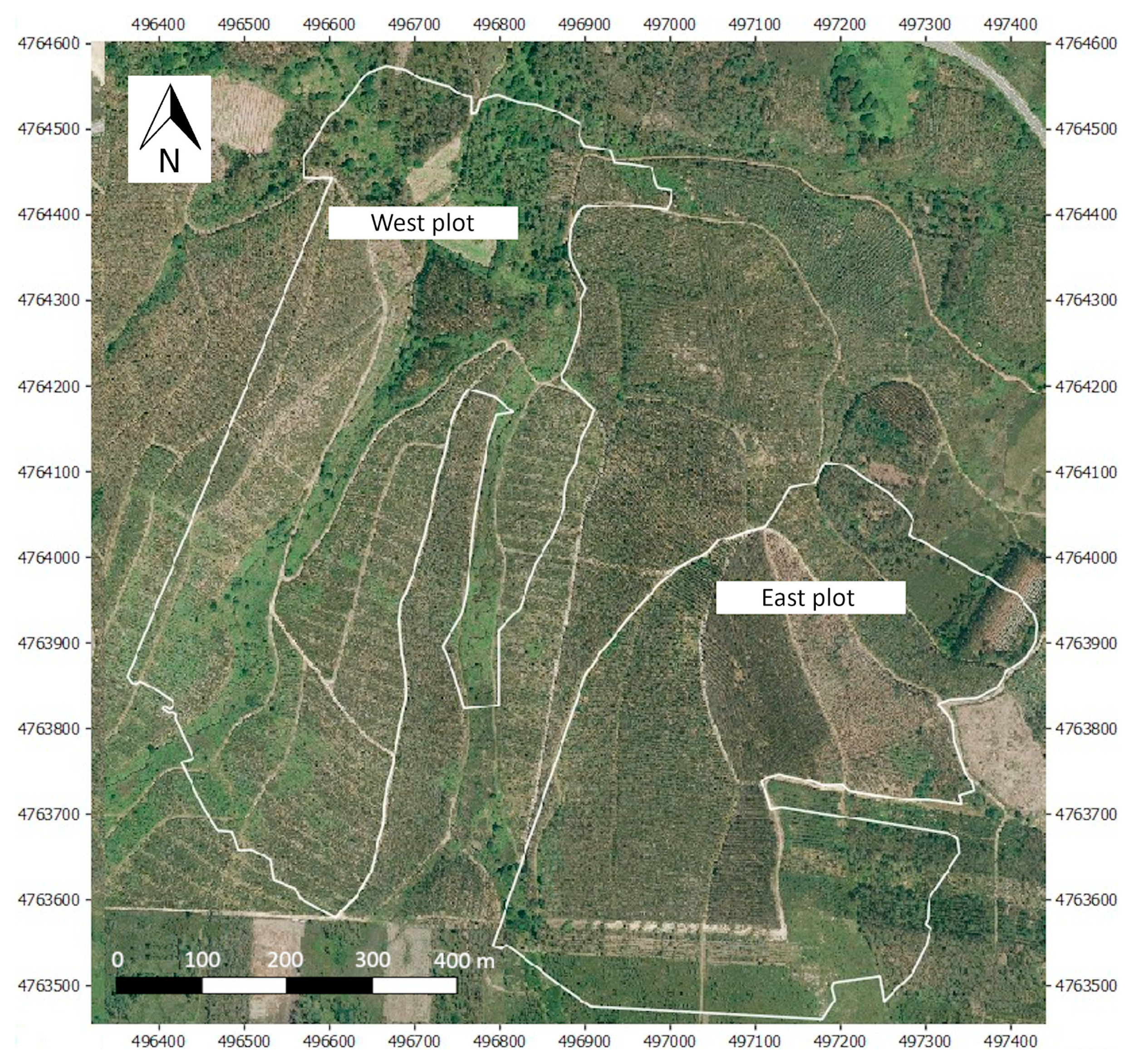
2.1. Study Area
2.2. Data Acquisition and Pre-Processing
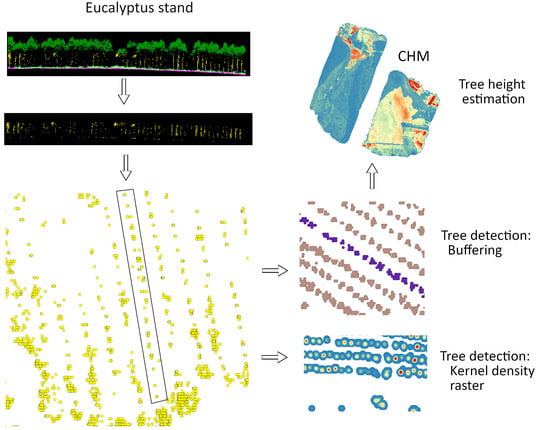
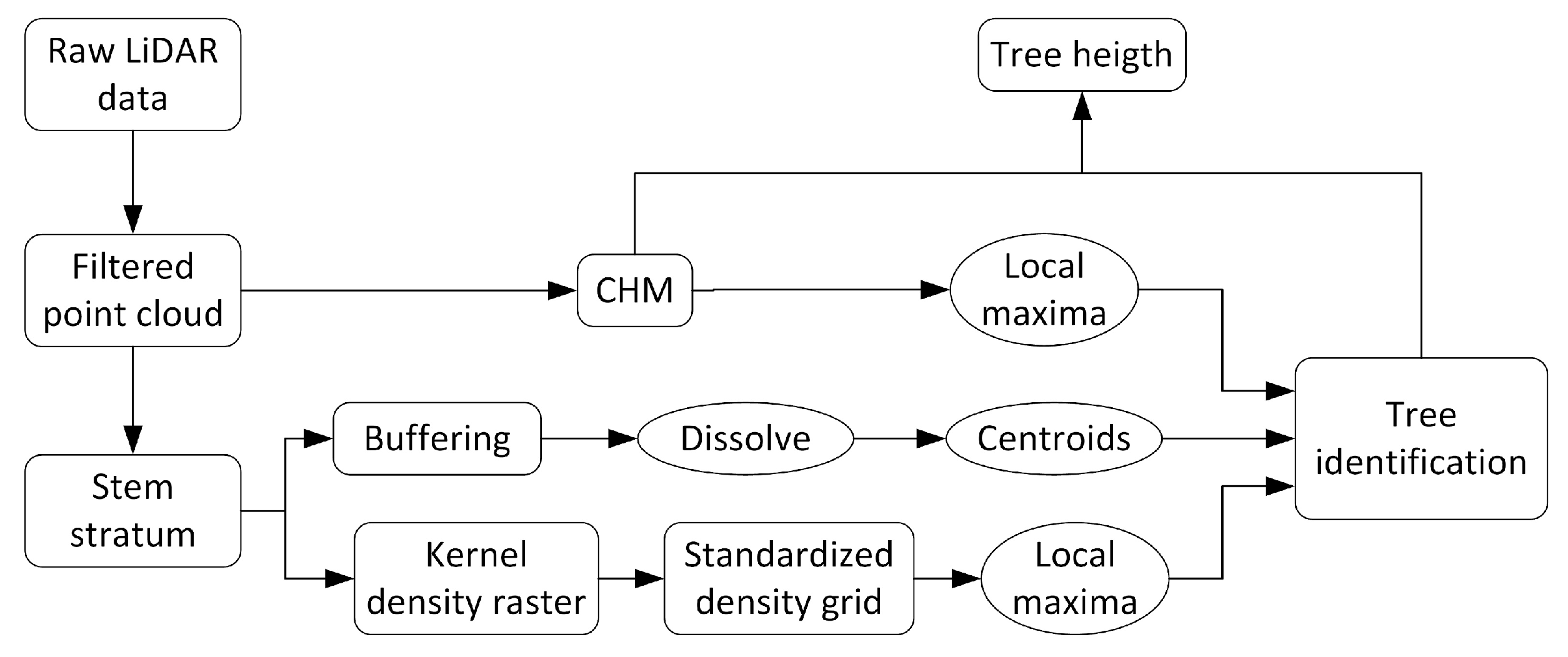
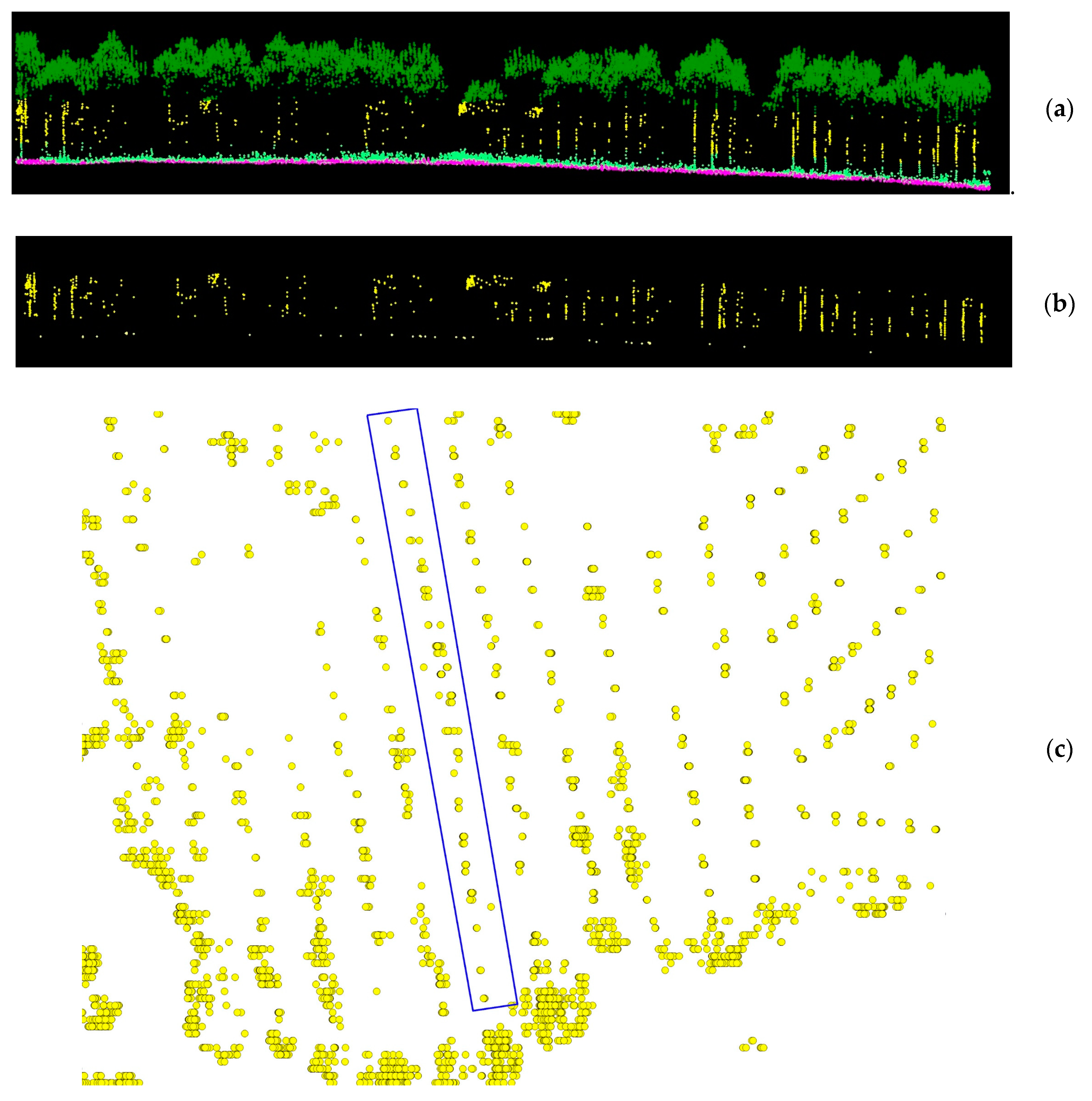
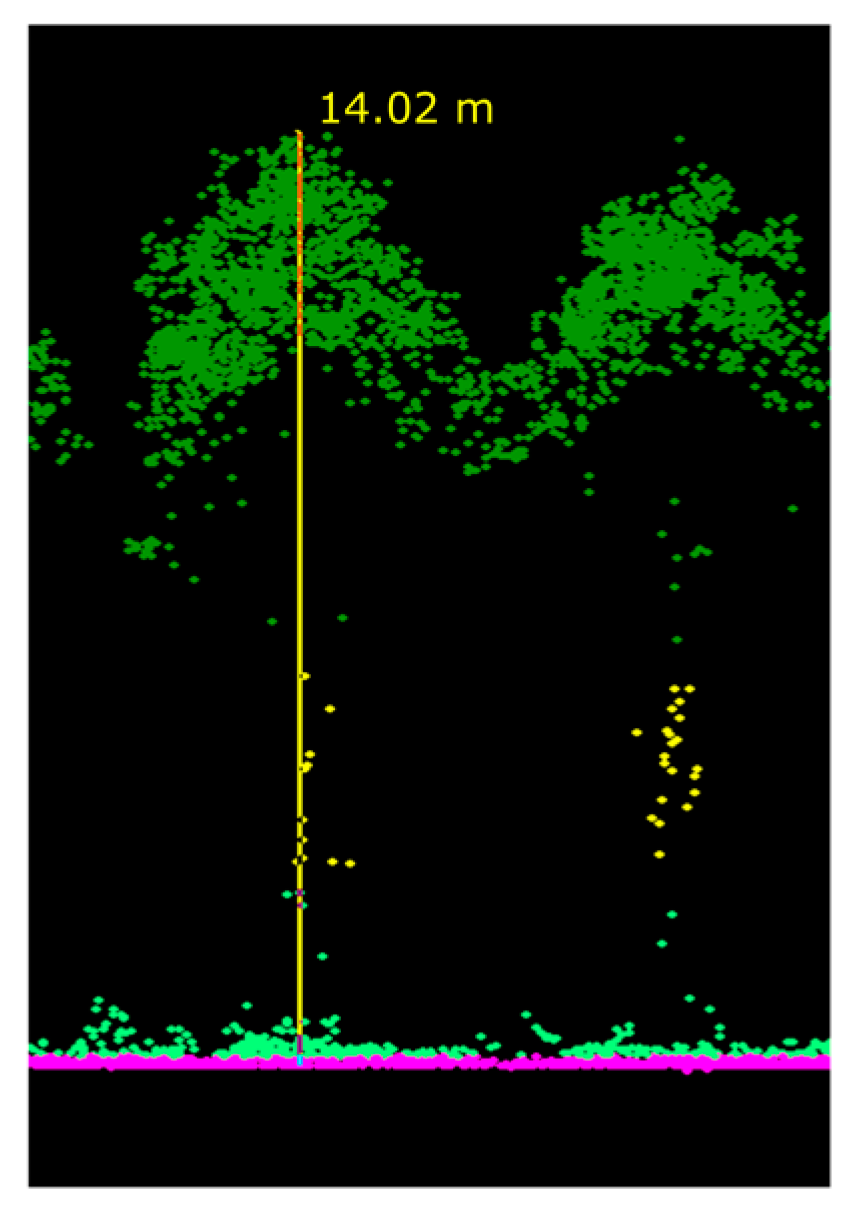
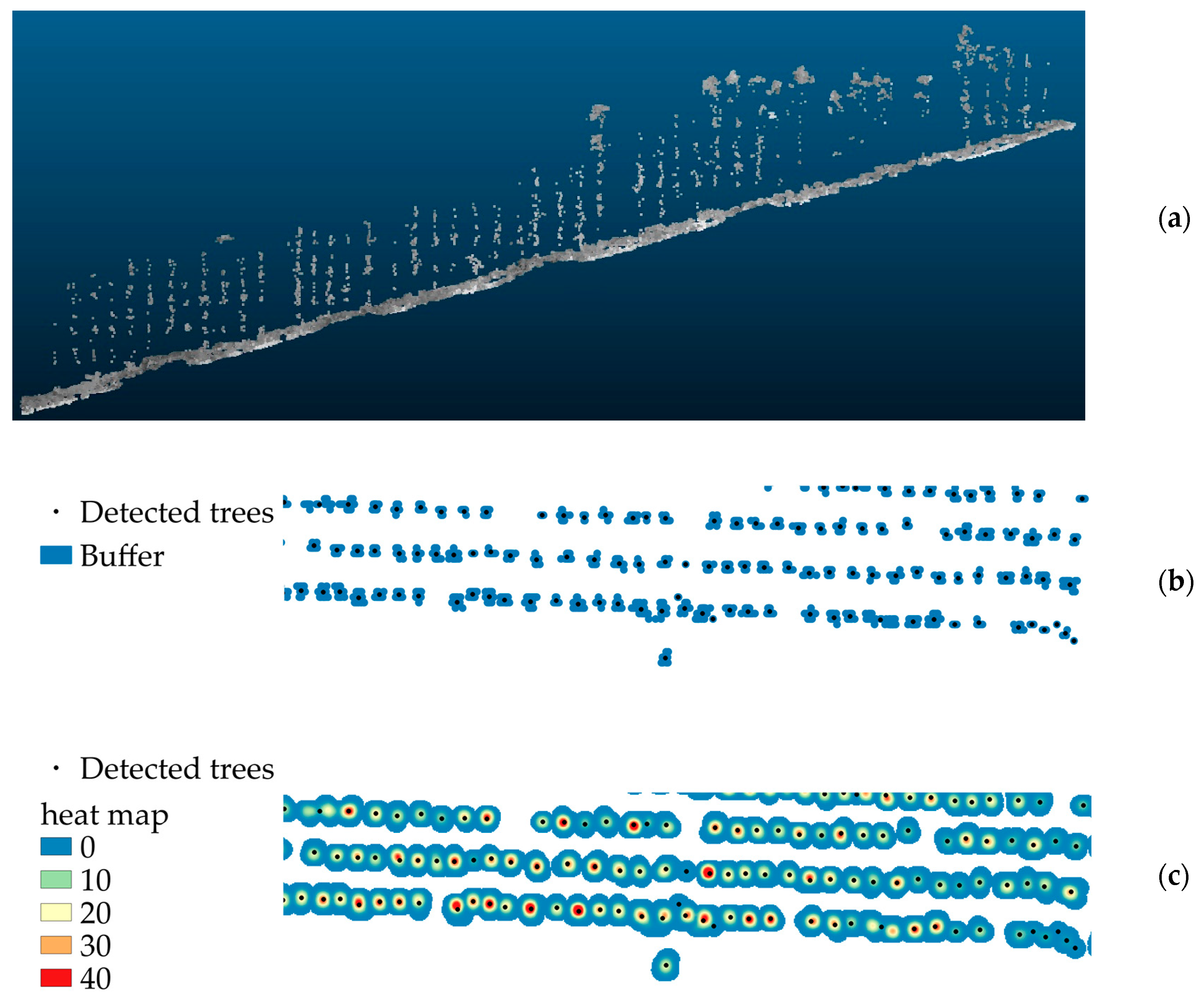
2.3. Tree Detection and Tree Height Determination
2.4. Assessment of Methods
3. Results
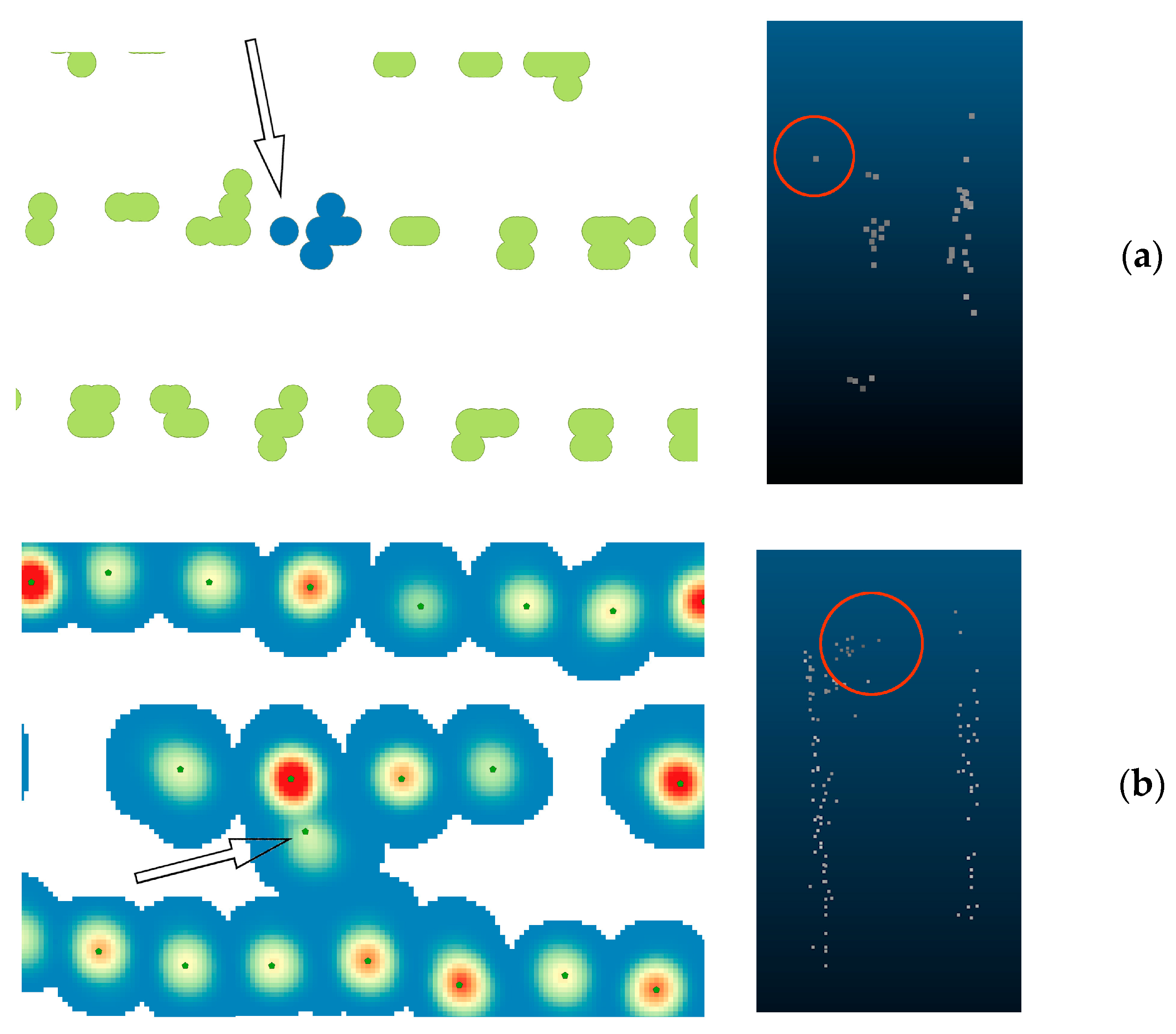
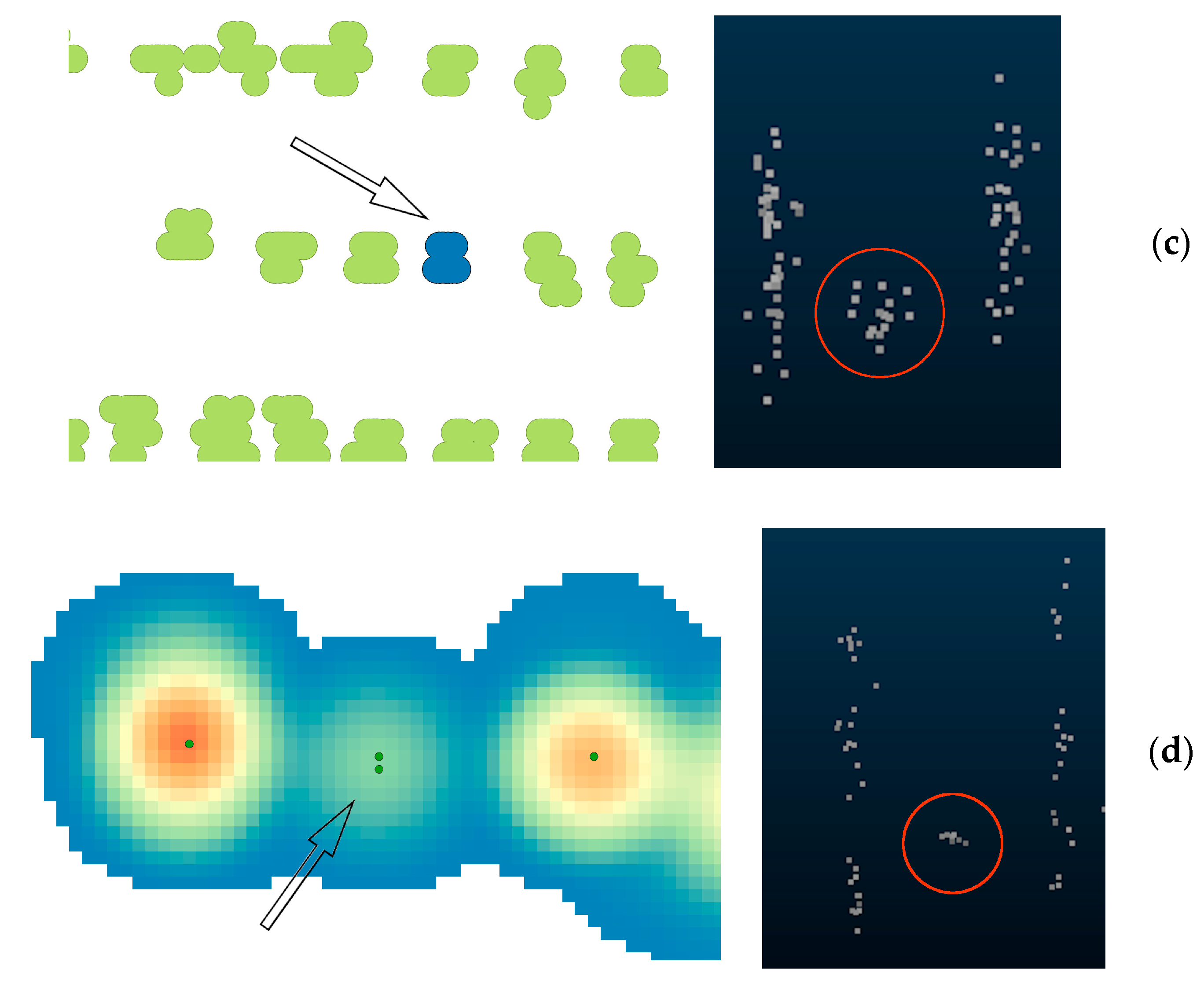
3.1. Tree detection
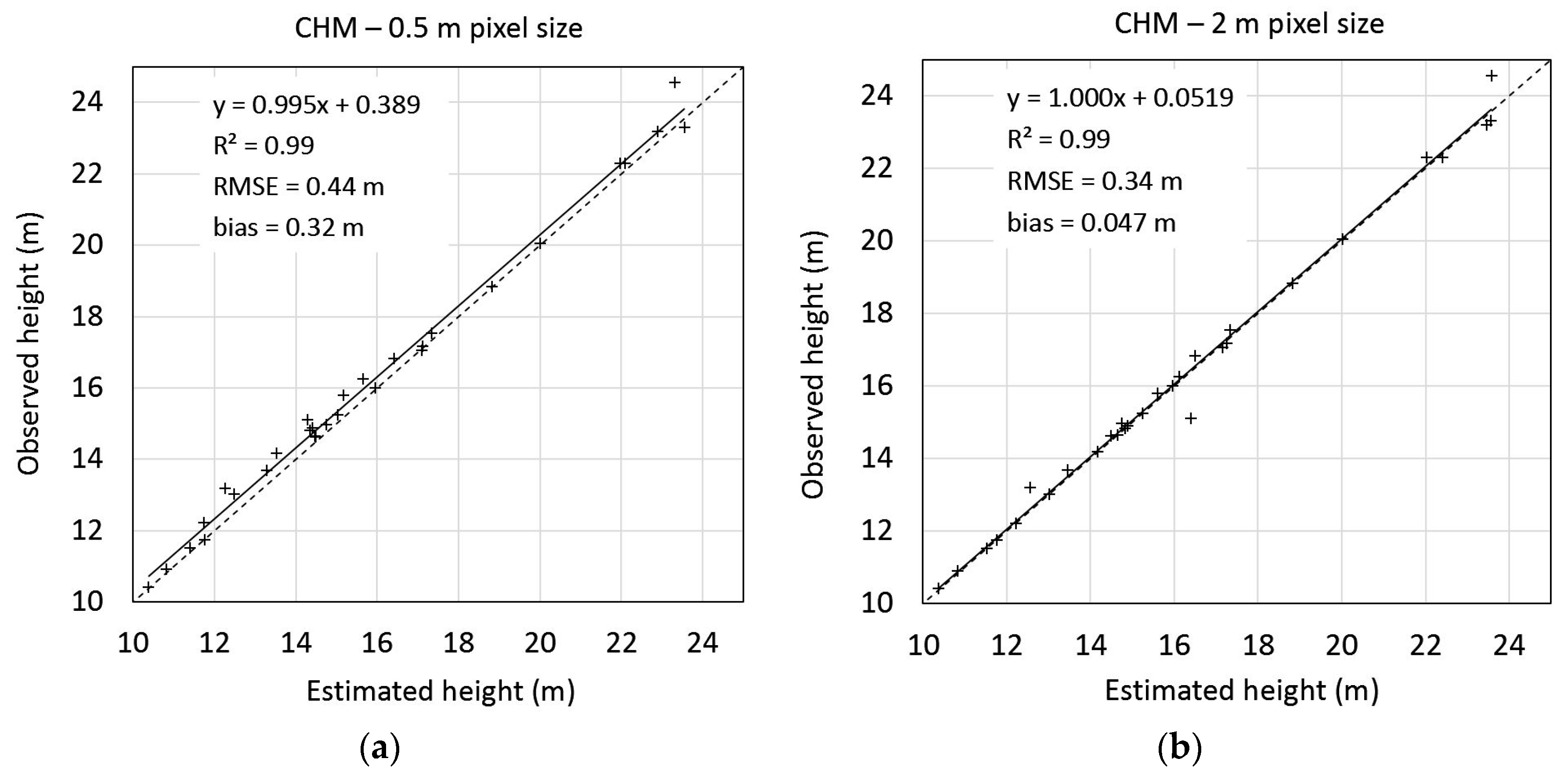
3.2. Tree Height Determination
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, J.; Hu, B.; Noland, T.L. Classification of tree species based on structural features derived from high density LiDAR data. Agric. For. Meteorol. 2013, 171, 104–114. [Google Scholar] [CrossRef]
- Edson, C.; Wing, M.G. Airborne Light Detection and Ranging (LiDAR) for individual tree stem location, height, and biomass measurements. Remote Sens. 2011, 3, 2494–2528. [Google Scholar] [CrossRef]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.-P. Generalizing predictive models of forest inventory attributes using an area-based approach with airborne LiDAR data. Remote Sens. Environ. 2015, 156, 322–334. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Næsset, E.; Ørka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef]
- Maltamo, M.; Næsset, E.; Vauhkonen, J. Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Springer: Dordrecht, The Netherlands, 2014. [Google Scholar] [CrossRef]
- Hyyppä, J.; Hyyppä, H.; Leckie, D.; Gougeon, F.; Yu, X.; Maltamo, M. Review of methods of small-footprint airborne laser scanning for extracting forest inventory data in boreal forests. Int. J. Remote Sens. 2008, 29, 1339–1366. [Google Scholar] [CrossRef]
- Budei, B.C.; St-Onge, B.; Hopkinson, C.; Audet, F.-A. Identifying the genus or species of individual trees using a three-wavelength airborne lidar system. Remote Sens. Environ. 2018, 204, 632–647. [Google Scholar] [CrossRef]
- Stone, C.; Webster, M.; Osborn, J.; Iqbal, I. Alternatives to LiDAR-derived canopy height models for softwood plantations: A review and example using photogrammetry. Aust. For. 2016, 79, 271–282. [Google Scholar] [CrossRef]
- Shinzato, E.T.; Shimabukuro, Y.E.; Coops, N.C.; Tompalski, P.; Gasparoto, E.A. Integrating area-based and individual tree detection approaches for estimating tree volume in plantation inventory using aerial image and airborne laser scanning data. iFor. Biogeosci. For. 2017, 10, 296–302. [Google Scholar] [CrossRef]
- Jeronimo, S.M.A.; Kane, V.R.; Churchill, D.J.; McGaughey, R.J.; Franklin, J.F. Applying LiDAR individual tree detection to management of structurally diverse forest landscapes. J. For. 2018, 116, 336–346. [Google Scholar] [CrossRef]
- Næsset, E. Estimating timber volume of forest stands using airborne laser scanner data. Remote Sens. Environ. 1997, 61, 246–253. [Google Scholar] [CrossRef]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Cosenza, D.N.; Soares, V.P.; Leite, H.G.; Gleriani, J.M.; Amaral, C.H.d.; GrippJúnior, J.; Silva, A.A.L.d.; Soares, P.; Tomé, M. Airborne laser scanning applied to eucalyptus stand inventory at individual tree level. Pesqui. Agropecu. Bras. 2018, 53, 1373–1382. [Google Scholar] [CrossRef]
- Chen, Q.; Baldocchi, D.; Gong, P.; Kelly, M. Isolating individual trees in a savanna woodland using small footprint lidar data. Photogramm. Eng. RemoteSens. 2006, 72, 923–932. [Google Scholar] [CrossRef]
- Wulder, M.A.; Bater, C.W.; Coops, N.C.; Hilker, T.; White, J.C. The role of LiDAR in sustainable forest management. For. Chron. 2008, 84, 807–826. [Google Scholar] [CrossRef]
- Liu, Q.; Li, Z.; Chen, E.; Pang, Y.; Tian, X.; Cao, C. Estimating biomass of individual trees using point cloud data of airborne LIDAR. Chin. High. Technol. Lett. 2010, 20, 765–770. [Google Scholar]
- Morsdorf, F.; Meier, E.; Kötz, B.; Itten, K.I.; Dobbertin, M.; Allgöwer, B. LIDAR-based geometric reconstruction of boreal type forest stands at singletree level for forest and wildland fire management. Remote Sens. Environ. 2004, 92, 353–362. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M. Generating spike-free digital surface models using LiDAR raw point clouds: A new approach for forestry applications. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 104–114. [Google Scholar] [CrossRef]
- Iglhaut, J.; Cabo, C.; Puliti, S.; Piermattei, L.; O’Connor, J.; Rosette, J. Structure from Motion Photogrammetry in Forestry: A review. Curr. For. Reports. 2019, 5, 155–168. [Google Scholar] [CrossRef]
- Goodbody, T.R.H.; Coops, N.C.; White, J.C. Digital Aerial Photogrammetry for Updating Area-Based Forest Inventories: A Review of Opportunities, Challenges, and Future Directions. Curr. For. Reports. 2019, 5, 55–75. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Malenovský, Z.; Turner, D.; Vopěnka, P. Assessment of Forest Structure Using Two UAV Techniques: A Comparison of Airborne Laser Scanning and Structure from Motion (SfM) Point Clouds. Forests 2016, 7, 62. [Google Scholar] [CrossRef]
- Zhao, K.; Popescu, S. Hierarchical watershed segmentation of canopy height model for multi-scale forest inventory. In Proceedings of the ISPRS Workshop on Laser Scanning 2007 and SilviLaser 2007, Espoo, Finland, 12–14 September 2007; Volume XXXVI. Part3/W52. [Google Scholar]
- Bortolot, Z.J.; Wynne, R.H. Estimating forest biomass using small footprint LiDAR data: An individual tree-based approach that incorporates training data. ISPRS J. Photogramm. Remote Sens. 2005, 59, 342–360. [Google Scholar] [CrossRef]
- Mohan, M.; Silva, A.C.; Klauberg, C.; Jat, P.; Catts, G.; Cardil, A.; Hudak, T.A.; Dia, M. Individual tree detection from Unmanned Aerial Vehicle (UAV) derived Canopy Height Model in an open canopy mixed conifer forest. Forests 2017, 8, 340. [Google Scholar] [CrossRef]
- Harikumar, A.; Bovolo, F.; Bruzzone, L. A local projection-based approach to individual tree detection and 3-D crown delineation in multistoried coniferous forests using high-density airborne LiDAR data. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1168–1182. [Google Scholar] [CrossRef]
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A new method for segmenting individual trees from the Lidar point cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef]
- Chen, Q.; Gong, P.; Baldocchi, D.; Tian, Y.Q. Estimating basal area and stem volume for individual trees from Lidar data. Photogramm. Eng. Remote Sens. 2007, 73, 1355–1365. [Google Scholar] [CrossRef]
- Korpela, I.; Dahlin, B.; Schäfer, H.; Bruun, E.; Haapaniemi, F.; Honkasalo, J.; Ilvesniemi, S.; Kuutti, V.; Linkosalmi, M.; Mustonen, J.; et al. Single-tree forest inventory using LIDAR and aerial images for 3D treetop positioning, species recognition, height and crown width estimation. Proc. IAPRS 2007, 36, 227–233. [Google Scholar]
- Yu, X.; Hyyppä, J.; Vastaranta, M.; Holopainen, M.; Viitala, R. Predicting individual tree attributes from airborne laser point clouds based on the random forests technique. ISPRS J. Photogramm. Remote Sens. 2011, 66, 28–37. [Google Scholar] [CrossRef]
- Gleason, C.J.; Im, J. A review of remote sensing of forest biomass and biofuel: Options for small-area applications. GISci. Remote Sens. 2011, 48, 141–170. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Smith, A.M.S.; Hudak, A.T.; Gessler, P.E.; Vierling, L.A.; Crookston, N.L. Automated estimation of individual conifer tree height and crown diameter via two-dimensional spatial wavelet analysis of Lidar data. Can. J. Remote Sens. 2006, 32, 153–161. [Google Scholar] [CrossRef]
- Goerndt, M.E.; Monleon, V.J.; Temesgen, H. Relating forest attributes with area- and tree-based light detection and ranging metrics for Western Oregon. West. J. Appl. For. 2010, 25, 105–111. [Google Scholar] [CrossRef]
- Véga, C.; Durrieu, S. Multi-level filtering segmentation to measure individual tree parameters based on Lidar data: Application to a mountainous forest with heterogeneous stands. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 646–656. [Google Scholar] [CrossRef]
- Sothe, C.; Dalponte, M.; Almeida, M.C.; Schimalski, B.M.; Lima, L.C.; Liesenberg, V.; Miyoshi, T.G.; Tommaselli, M.A. Tree species classification in a highly diverse subtropical forest integrating UAV-based photogrammetric point cloud and hyperspectral data. Remote Sens. 2019, 11, 1338. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, J.L.; Zhang, L. Trends in automatic individual tree crown detection and delineation—Evolution of LiDAR data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Donoghue, D.N.M.; Watt, P.J.; Cox, N.J.; Wilson, J. Remote sensing of species mixtures in conifer plantations using LiDAR height and intensity data. Remote Sens. Environ. 2007, 110, 509–522. [Google Scholar] [CrossRef]
- Rockwood, L.D.; Rudie, W.A.; Ralph, A.S.; Zhu, Y.J.; Winandy, E.J. Energy product options for Eucalyptus species grown as short rotation woody crops. Int. J. Mol. Sci. 2008, 9, 1361. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization (FAO) of the United Nations. Global Forest Resources Assessment 2005—Main Report; FAO Forestry Paper; FAO: Rome, Italy, 2005. [Google Scholar]
- McMahon, D.E.; Jackson, R.B. Management intensification maintains wood production over multiple harvests in tropical Eucalyptus plantations. Ecol. Appl. 2019, 29, e01879. [Google Scholar] [CrossRef]
- Crecente-Campo, F.; Tomé, M.; Soares, P.; Diéguez-Aranda, U. A generalized nonlinear mixed-effects height–diameter model for Eucalyptus globulus L. in northwestern Spain. For. Ecol. Manag. 2010, 259, 943–952. [Google Scholar] [CrossRef]
- Gonçalves-Seco, L.; González-Ferreiro, E.; Diéguez-Aranda, U.; Fraga-Bugallo, B.; Crecente, R.; Miranda, D. Assessing the attributes of high-density Eucalyptus globulus stands using airborne laser scanner data. Int. J. Remote Sens. 2011, 32, 9821–9841. [Google Scholar] [CrossRef]
- Guerra-Hernández, J.; Cosenza, D.N.; Rodriguez, L.C.E.; Silva, M.; Tomé, M.; Díaz-Varela, R.A.; González-Ferreiro, E. Comparison of ALS- and UAV (SfM)-derived high-density point clouds for individual tree detection in Eucalyptus plantations. Int. J. Remote Sens. 2018, 39, 5211–5235. [Google Scholar] [CrossRef]
- Oliveira, L.T.d.; Carvalho, L.M.T.d.; Ferreira, M.Z.; Oliveira, T.C.d.A.; Batista, V.T.F.P. Influência da idade na contagem de árvores de Eucalyptussp. com dados de Lidar (Portuguese). Cerne 2014, 20, 557–565. [Google Scholar] [CrossRef]
- Ferraz, A.; Bretar, F.; Jacquemoud, S.; Gonçalves, G.; Pereira, L.; Tomé, M.; Soares, P. 3-D mapping of a multi-layered Mediterranean forest using ALS data. Remote Sens. Environ. 2012, 121, 210–223. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Watson, C.S. Evaluating tree detection and segmentation routines on very high resolution UAV LiDAR data. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7619–7628. [Google Scholar] [CrossRef]
- Torresan, C.; Berton, A.; Carotenuto, F.; Chiavetta, U.; Miglietta, F.; Zaldei, A.; Gioli, B. Development and Performance Assessment of a Low-Cost UAV Laser Scanner System (LasUAV). Remote Sens. 2018, 10, 1094. [Google Scholar] [CrossRef]
- Zhou, J.; Proisy, C.; Descombes, X.; Le Maire, G.; Nouvellon, Y.; Stape, J.-L.; Viennois, G.; Zerubia, J.; Couteron, P. Mapping local density of young Eucalyptus plantations by individual tree detection in high spatial resolution satellite images. For. Ecol. Manag. 2013, 301, 129–141. [Google Scholar] [CrossRef]
- Amon, P.; Riegl, U.; Rieger, P.; Pfennigbauer, M. UAV-based laser scanning to meet special challenges in LiDAR surveying. In Proceedings of the Geomatics Indaba 2015—Stream 2, Ekurhuleni, South Africa, 11–13 August 2015; pp. 138–147. [Google Scholar]
- Liu, Q.; Li, S.; Li, Z.; Fu, L.; Hu, K. Review on the Applications of UAV-Based LiDAR and Photogrammetry in Forestry. Sci. Silvae Sin. 2017, 53, 134–148. [Google Scholar] [CrossRef]
- Petrie, G. Current developments in airborne laser scanners suitable for use on lightweight UAVs: Progress is being made! GeoInformatics 2013, 16, 16–22. [Google Scholar]
- Pilarska, M.; Ostrowski, W.; Bakuła, K.; Górski, K.; Kurczyński, Z. The potential of light laser scanners developed for unmanned aerial vehicles—The review and accuracy. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 42, 87–95. [Google Scholar] [CrossRef]
- Salach, A.; Bakuła, K.; Pilarska, M.; Ostrowski, W.; Górski, K.; Kurczyński, Z. Accuracy assessment of point clouds from LiDAR and dense image matching acquired using the UAV platform for DTM creation. ISPRS Int. J. GeoInf. 2018, 7, 342. [Google Scholar] [CrossRef]
- Barnes, C.; Balzter, H.; Barrett, K.; Eddy, J.; Milner, S.; Suárez, C.J. Individual tree crown delineation from airborne laser scanning for diseased larch forest Ssands. Remote Sens. 2017, 9, 231. [Google Scholar] [CrossRef]
- Xunta de Galicia; Instituto Gallego de Promoción Económica (IGAPE). Oportunidades Industria 4.0 en Galicia—Diagnóstico Sectorial: Madera/Forestal (Spanish); 2017. Available online: http://www.igape.es/es/ser-mas-competitivo/galiciaindustria4-0/estudos-e-informes/item/download/58_60810179c81085089705d6dc680b42e7 (accessed on 15 October 2019).
- Chas Amil, M.L. Forest fires in Galicia (Spain): Threats and challenges for the future. J. For. Econ. 2007, 13, 1–5. [Google Scholar] [CrossRef]
- Jiménez, E.; Vega, J.A.; Fernández-Alonso, J.M.; Vega-Nieva, D.; Ortiz, L.; López-Serrano, P.M.; López-Sánchez, C.A. Estimation of aboveground forest biomass in Galicia (NW Spain) by the combined use of LiDAR, LANDSAT ETM+ and National Forest Inventory data. IFor. Biogeosci. For. 2017, 10, 590–596. [Google Scholar] [CrossRef]
- Gobierno de España; Ministerio de Agricultura Pesca y Alimentación. Avance de Estadística Forestal 2017 (Spanish); Centro de Publicaciones del Ministerio de Agricultura, Pesca y Alimentación: Madrid, Spain, 2017. [Google Scholar]
- Xunta de Galicia; Consellería do Medio Rural. 1a Revisión del Plan Forestal de Galicia. Documento de diagnóstico del monte y el sector forestal gallego (Spanish); 2018. Available online: https://mediorural.xunta.gal/sites/default/files/temas/forestal/plan-forestal/1_REVISION_PLAN_FORESTAL_CAST.pdf (accessed on 22 October 2019).
- Picos, J. Producción de eucalipto en Galicia. Guía de supervivencia. In Proceedings of the Jornadas CIDEU (Spanish), Huelva, Spain, 21–23 October 2009. [Google Scholar]
- Corbelle Rico, E.J.; Tubío Sánchez, J.M. Productivismo y abandono: Dos caras de la transición forestal en Galicia (España), 1966–2009 (Spanish). Bosque (Valdivia) 2018, 39, 457–467. [Google Scholar] [CrossRef]
- Álvarez-Díaz, M.; González-Gómez, M.; Otero-Giráldez, M.S. Main determinants of export-oriented bleached Eucalyptus kraft pulp (BEKP) demand from the north-western regions of Spain. For. Policy Econ. 2018, 96, 112–119. [Google Scholar] [CrossRef]
- Rapidlasso GmbH. LASTools. Available online: https://rapidlasso.com//lastools (accessed on 27 June 2019).
- Pacific Northwest Research Station—USDA Forest Service FUSION/LDV LIDAR Analysis and Visualization Software. Available online: http://forsys.sefs.uw.edu/FUSION/fusion_overview.html (accessed on 4 November 2019).
- QGIS—A Free and Open Source Geographic Information System. Available online: https://qgis.org (accessed on 18 February 2019).
- McGaughey, R.J. FUSION/LDV: Software for LIDAR Data Analysis and Visualization. 2018. Available online: http://forsys.sefs.uw.edu/Software/FUSION/FUSION_manual.pdf (accessed on 23 October 2019).
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geosci. Model. Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef]
- Huang, H.; Gong, P.; Cheng, X.; Clinton, N.; Li, Z. Improving measurement of forest structural parameters by co-registering of high resolution aerial imagery and low density LiDAR data. Sensors 2009, 9, 1541. [Google Scholar] [CrossRef]


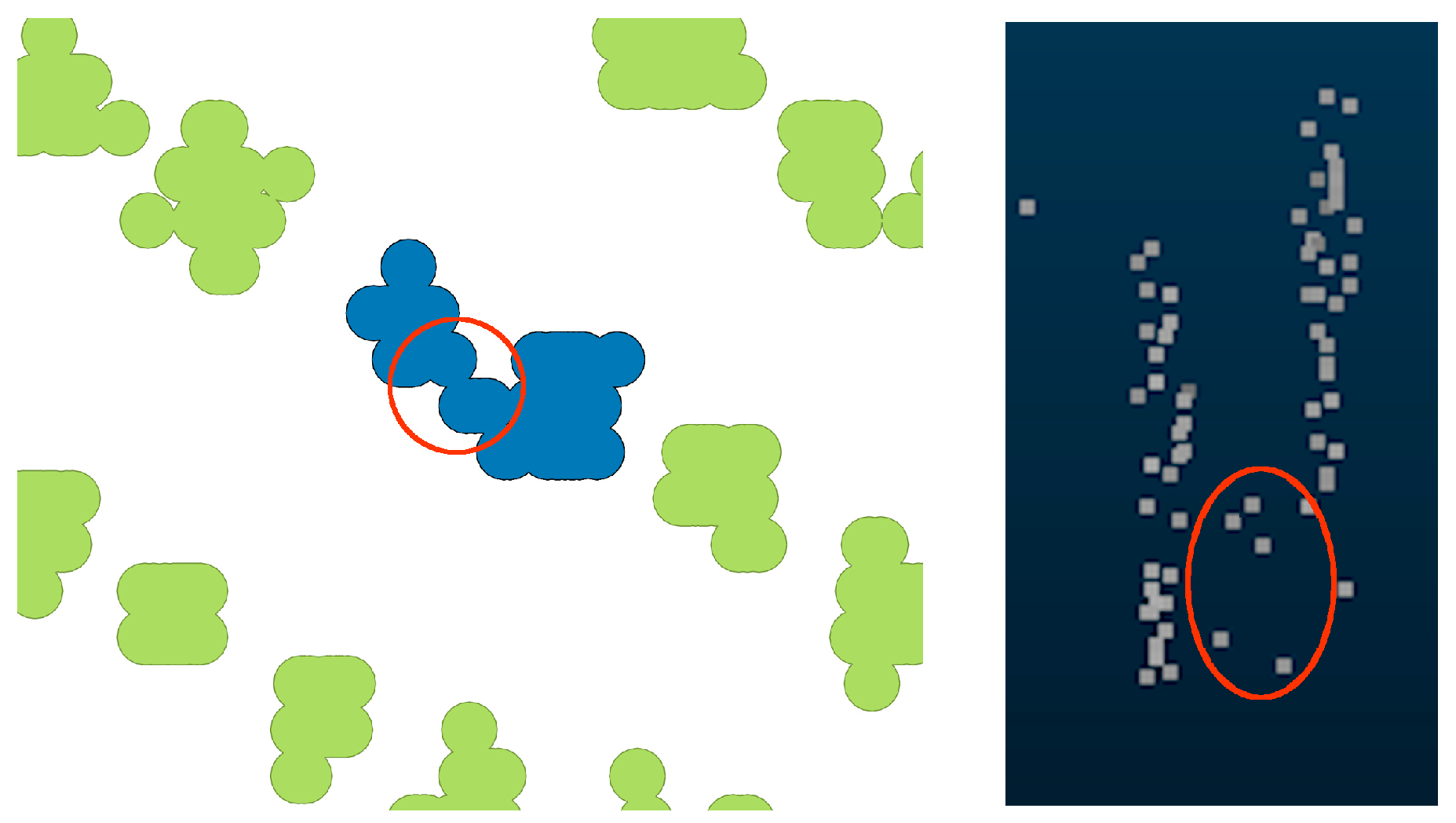

| Ref. | Plantation Characteristics | Data Acquisition System | Point Density (pulses/m2) | ITD Approach | Quality of Results 1 |
|---|---|---|---|---|---|
| [11] | Age 4–7 years Spacing: 3.0 × 3.0 m 3.0 × 3.3 m | Airplane | 5 | CHM–LM | DR: 65%–92% |
| [9] | Spacing: 2 × 3 m Density: 1667 trees/ha | Airplane – Riegl LMS-Q680I | 5 | Iterative LM algorithm on images from different spectral bands/Adding the use of aerial images | RMSE: 6.8%/5.2% |
| [41] | Height: 7.0–29.2 m | Airplane – Optech ALTM 2033 | 4 | CHM–LM | RMSE: 733 trees/ha Bias: 234 trees/ha |
| [42] | Age: 7 years Spacing: 3.70 × 2.50 m Density: 1081 trees/ha | Airplane – Leica ALS80-HP | 43.3 | CHM–LM | DR: 96.7% OE: 1.3% CE: 13.7% |
| [43] | Age: 3 years Spacing: 4 × 3 m Age: 5/7 years Spacing: 5 × 2.4 m | Airplane –Optech ALTM 3100 | 0.5–5.0 | CHM–LM | DR: 98.8% in 3-year stands; 99.2% in 5-year stands; 98.6% in 7-year stands |
| [44] | Age: 1–13 years | Airplane – LiteMapper 5600 | 9.5 | Adaptive mean shift algorithm directly on the point cloud | CE: 9.2% |
| [45] | Age: 4 years Spacing 3 × 3 m | UAV – IBEO Lux | 61–163 | Voxel space detection and delineation (VDD)/LM based on seeded k-means clustering (CDPD) | DR: VDD 99%/CDPD 100.6% OE: VDD 6.4%/CDPD 4.6% CE: VDD 7.3%/CDPD 5.5% |
| Layer | Threshold Value (m) | |
|---|---|---|
| West Plot | East Plot | |
| Ground–Shrub | 0.25 | 0.25 |
| Shrub–Stem | 2.00 | 3.00 |
| Stem–Canopy | 7.00 | 6.00 |
| Method | Number of Trees | Average Tree Density (trees/ha) | ||
|---|---|---|---|---|
| West Plot | East Plot | West Plot | East Plot | |
| CHM – 0.5 m pixel size (Fusion) | 1046 | 3626 | 30 | 145 |
| CHM – 2.0 m pixel size (Fusion) | 607 | 2378 | 17 | 95 |
| CHM – 0.5 m pixel size (QGIS) | 6036 | 16,593 | 177 | 633 |
| CHM – 2.0 m pixel size (QGIS) | 1341 | 3334 | 74 | 120 |
| Method 1 | 6264 | 18,892 | 184 | 755 |
| Method 2 | 3940 | 13,699 | 115 | 547 |
| Method | Number of Trees | DR (%) |
|---|---|---|
| CHM – 0.5 m pixel size (Fusion) | 52 | 21.5 |
| CHM – 2.0 m pixel size (Fusion) | 37 | 15.3 |
| CHM – 0.5 m pixel size (QGIS) | 257 | 106.2 |
| CHM – 2.0 m pixel size (QGIS) | 54 | 22.3 |
| Method 1 | 251 | 103.7 |
| Method 2 | 275 | 113.6 |
| Manually identified | 242 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picos, J.; Bastos, G.; Míguez, D.; Alonso, L.; Armesto, J. Individual Tree Detection in a Eucalyptus Plantation Using Unmanned Aerial Vehicle (UAV)-LiDAR. Remote Sens. 2020, 12, 885. https://doi.org/10.3390/rs12050885
Picos J, Bastos G, Míguez D, Alonso L, Armesto J. Individual Tree Detection in a Eucalyptus Plantation Using Unmanned Aerial Vehicle (UAV)-LiDAR. Remote Sensing. 2020; 12(5):885. https://doi.org/10.3390/rs12050885
Chicago/Turabian StylePicos, Juan, Guillermo Bastos, Daniel Míguez, Laura Alonso, and Julia Armesto. 2020. "Individual Tree Detection in a Eucalyptus Plantation Using Unmanned Aerial Vehicle (UAV)-LiDAR" Remote Sensing 12, no. 5: 885. https://doi.org/10.3390/rs12050885
APA StylePicos, J., Bastos, G., Míguez, D., Alonso, L., & Armesto, J. (2020). Individual Tree Detection in a Eucalyptus Plantation Using Unmanned Aerial Vehicle (UAV)-LiDAR. Remote Sensing, 12(5), 885. https://doi.org/10.3390/rs12050885