Abstract
To minimize pesticide dosage and its adverse environmental impact, Unmanned Aerial Vehicle (UAV) spraying requires precise individual canopy information. Branches from neighboring trees may overlap, preventing image-based artificial intelligence analysis from correctly identifying individual trees. To solve this problem, this paper proposes a segmentation and evaluation method for mingled fruit tree canopies with irregular shapes. To extract the individual trees from mingled canopies, the study fitted the projection curve distribution of the interlacing tree with Gaussian Mixture Model (GMM) and solved the matter of segmentation by estimating the GMM parameters. For the intermingling degree assessment, the Gaussian parameters were used to quantify the characteristics of the mingled fruit trees and then as the input for Extreme Gradient Boosting (XGBoost) model training. The proposed method was tested on the aerial images of cherry and apple trees. Results of the experiments show that the proposed method can not only accurately identify individual trees, but also estimate the intermingledness of the interlacing canopies. The root mean squares (R) of the over-segmentation rate (Ro) and under-segmentation rate (Ru) for individual trees counting were less than 10%. Moreover, the Intersection over Union (IoU), used to evaluate the integrity of a single canopy area, was greater than 88%. An 84.3% Accuracy (ACC) with a standard deviation of 1.2% was achieved by the assessment model. This method will supply more accurate data of individual canopy for spray volume assessments or other precision-based applications in orchards.
1. Introduction
Aerial spraying by Unmanned Aerial Vehicles (UAV) has great application potential in China for conventional orchard farms, especially those with trees growing on hills [1]. The widespread UAV spraying in orchards calls for automatic extraction of individual fruit trees and its parameters such as canopy size and shape to estimate the pesticide spray volumes [2,3,4].
At present, remotely sensed images are widely adopted for automatic individual tree identification because of the visualized results and noncontact data collecting [5]. There is an extensive amount of research done on the remotely sensed data for tree extraction, such as high-resolution satellite images [6,7] and RADAR data [8,9]. With the recent Unmanned Aerial Vehicle (UAV) developments, UAV images are more easily obtained and require less data processing [10]. Therefore, UAV images have become more commonly used within remote sensing studies focused on tree extraction [11,12,13].
A single tree is the basic unit for orchard farming. Accurately identifying individual trees is a prerequisite for tree extraction studies. Distinguishing individual trees in the case of contiguous tree canopies, however, has always been a difficult task in horticultural computer vision. Commonly, the Hough transform is used to isolate individual trees from UAV images by applying a canopy shadow, generated from optical data and a circular matching template [14]. The Hough transform is a feature extraction technique used in image analysis, computer vision, and digital image processing. It is used to identify lines in images and positions of arbitrary shapes, such as circles or ellipses. This method, however, is not ideal for interlaced orchards. After factoring in the possibility of intermingling branches in aerial images, this process still relies on a circular template to segregate the citrus canopies [15] and the clove canopies [16]. Although trees do grow following general patterns, branches extend, cross, and overlap unsystematically, rendering the template matching inaccurate and unfit for image segmentation. Convolutional neural networks are applied to semantically segregate pomegranate trees in UAV scenes [17]. Convolutional neural networks CNN(s) are a class of deep neural networks most commonly used to analyze visual imagery, but manual canopy boundaries are needed as its training sample input. Contour curvature or corner methods used for cells or particles segmentation [18,19,20] also have difficulties separating the contiguous trees in images.
Additionally, degree assessment of intermingledness of canopies helps decide whether to spray continuously. Traditional data information extracted from image data include vegetation index [21,22], crown delineation [23,24], and tree quantity [25,26]. The intermingledness, or an index for the intermingledness, for contiguous canopies, however, has rarely been reported.
The objective of this study, therefore, is to propose a method that can separate interlacing tree canopies and evaluate the degree of such phenomena. Past studies showed that Gaussian Mixture Models (GMMs) can be used to analyze pixel distribution in images of agriculture scenes [5,27]. In this study, therefore, a GMM was chosen to characterize the projection histogram of contiguous tree canopies. GMM is a probabilistic model for representing normally distributed subpopulations within an overall population. Since GMM mainly utilizes the positional information of the trees contained in the projection histogram, it is robust to the shape of the fruit trees. GMM not only segregates the contiguous trees but also outputs Gaussian parameters to quantify the features of mingled canopies, providing training data for Extreme Gradient Boosting (XGBoost) to evaluate intermingledness. XGBoost is a decision-tree-based ensemble Machine Learning algorithm that uses a gradient boosting framework, which is recently proven to be very powerful in other research areas [28]. Its advantages over other machine learning methods are properly discussed as well in this study. The findings from this study will also contribute to crop feature acquisition based on machine vision in general.
2. Test Sites and Image Acquisition
This study was conducted on the commercial cherry orchard of Zhongnong Futong in Tongzhou, Beijing and the apple orchard of Orchard Research and Demonstration Park in Taian, Shandong. These two sites were selected because of their row-planting pattern for the orchards, which is the norm in this industry. The cherry trees are spaced 4 m per plant with a 5-m path between rows and the apple trees 3 m by 4 m, respectively. The crown diameters ranged from about 2 to 4 m. The spatial relationship of the trees was categorized into (1) individuals, (2) slightly interlacing, and (3) greatly interlacing, as shown in Figure 1.

Figure 1.
The illustrations of images under different lighting conditions and interlacing situations: (a) cherry trees, sunny day; (b) cherry trees, cloudy day; (c) apple trees, sunny day; (d) apple trees, cloudy day.
The images of cherry and apple trees were taken in June 2017 and October 2018, using DJI Phantom 3 UAV (Shenzhen Dajiang Innovation Technology Co., Ltd, China) (Figure 2). This drone is equipped with 3-axis gimbal stabilization and a Sony EXMOR 1/2.3-inch CMOS digital camera, outputting RGB orthophotos of 3000 × 4000 pixels. The UAV filmed horizontally at a constant speed (5 m/s) and a height range of 20–60 m.

Figure 2.
Image acquisition: (a) a-1: cherry orchard at Zhongnong Futong; a-2: apple orchard in Taian; a-3: photographing device; a-4: data collection team. (b) the Unmanned Aerial Vehicle (UAV) flight route.
3. Methodology
The proposed method can be summarized by a three-part processing framework: interlacing canopy extraction, image segmentation, and evaluation. The first step was the preliminary extraction of interlacing regions. The background was removed by applying the Excess Green Index (EXG) [29] and Otsu’s method [30]; both of them are the common methods for image preprocessing purposes. The target regions were extracted using the length threshold of the circumscribed rectangle of the connected area. The aim of the second step was to separate interlacing canopies. A GMM was built by fitting the projection histogram curve in the horizontal direction of the extracted interlacing regions. We then manually determined the initial parameters and applied the Expectation Maximization (EM) algorithm [31] to find the parameters of this GMM. Once the model parameters were obtained, the intersection of the Gaussian curve was computed as the segmentation thresholds. The third stage was to assess intermingling degrees. The images were labeled to create a training set, and the canopy feature parameters extracted from the Gaussian components were put into the XGBoost training model. An overall flowchart is shown in Figure 3.
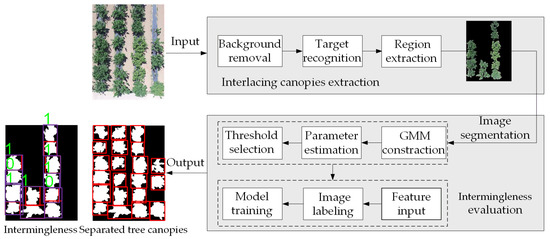
Figure 3.
Overall flowchart of the proposed method. The input image is the original image, and the output image is the result of segmentation and evaluation of the canopy image: the extracted trees are enclosed within the red boxes for better illustration; the Extreme Gradient Boosting (XGBoost) classification model output will be 1 for the greatly intermingling canopies or 0 for the slightly intermingling canopies.
3.1. Extraction of Interlacing Canopy Region
The extraction of interlacing canopies consists of two steps: removing the background of the images and excluding the already identified individual trees to get the interlacing canopies regions.
3.1.1. Background Removal
The background in the images was removed based on color differences, as the plants were distinctively green. The ExG index and Otsu’s method were applied because they were well tested in image segmentation of different agricultural scenarios [32,33,34]. Figure 4b shows the resulting binary images after applying ExG and Otsu. The ExG index is defined as follows:
where r, g, and b are the color components; R, G, and B are the normalized RGB values ranging from 0 to 1.
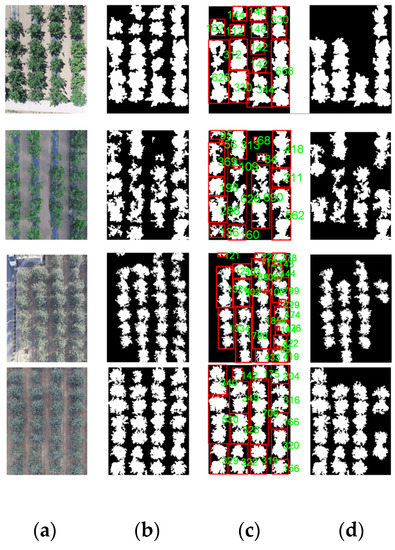
Figure 4.
Extraction process of interlacing canopy region: (a) original image; (b) background removal; (c) intermingling canopy identification; (d) intermingling canopy extraction.
3.1.2. Interlacing Canopy Region Extraction
After the background removal, the target regions were identified to obtain interlacing canopies. Methods such as contour features, bump detection, and contour stripping [35,36] could not be used to identify interlacing tree canopies because the trees do not have regular shape. Figure 4 shows that intermingled trees would occupy more area on their respective rows than single trees. The target canopies were thus picked out based on the length of the circumscribed rectangle of the maximum connected area. Contiguous canopies were estimated according to Equation (2):
where p is the pixel value within the connected area; l is the length of the connected area; l is the mean value; and r is the number of connected areas; i = 1,2,3…r.
3.2. Analysis of Projection Curve of Individual Tree
A GMM is a universal model to estimate the unknown probability density function and is generally used to estimate probability density distribution of the target in images [37]. When analyzing a tree’s distribution characteristics in binary images, the projection curve of individual trees was similar to the Gaussian distribution (Figure 5). Further tests were necessary to confirm whether the projection histogram of individual trees follows the Gaussian distribution. With goodness-of-fit tests [38], this theory was verified as shown.
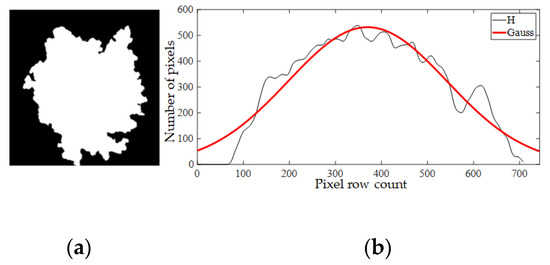
Figure 5.
(a) An individual tree; (b) Projection histogram in the horizontal direction. x-label represents pixel row count of an image from top to bottom. y-label represents number of pixels on each row.
The verification process:
(1) For the binary image bw(i, j) in the horizontal direction, projection histogram can be defined as H = (H1, H2, H3,…, Hi), Hi the projection value of the image on the i row:
where i is the row number of the image, j is the column number, bw(i, j) is the pixel value of the binary image at the (i, j), h is the height of the image, w is the width of the images.
The original image was cropped into individual trees and converted into a binary image for the test. A total of 370 samples were obtained after the projection. We assumed that each sample obeys H~N(µ,σ2). Probability density distribution of H:
where µ and σ are the mean value and the standard deviation and H is projection histogram.
By calculating the likelihood function—Equation (5)—we got an estimate of the mean and variance by the Equation. (6).
where µ and σ are the mean value and the standard deviation; Hi is the projection value of the image on the i row; L(µ,σ2) is the likelihood function; w is the image width; h is the height of image; and , is the estimated mean and variance.
(2) The chi-square (χ2) goodness-of-fit test [39] with a significant level of 0.5 was used to test the population normality. Each sample was divided into [ai-1, ai), a total of k groups, χ2 is calculated as follows:
In this formula, , where fe is the actual frequency; fk is the theoretical frequency; and p(ai) can be calculated by Equation (4).
Since two estimators are obtained, the degree of freedom is:
χ20.05 can be acquired in χ2 the distribution table; test rules are
Figure 6 shows a hypothesis test of distribution for a total of 370 trees. According to the statistics, 89.6% of fruit trees projection histograms satisfied the assumption of Gaussian distribution. Five group examples of estimating parameters and statistics are presented in Table 1. Therefore, the projection histograms of fruit trees could be modeled as Gaussian probability density functions, which would provide the theoretical basis of image segmentation in the next step.
Figure 6.
Statistical test of 370 sampled distributions.

Table 1.
Examples of the goodness-of-fit test for some samples.
3.3. GMM for Image Segmentation
A Gaussian Mixture Model was used to characterize the projected histogram distribution of the interlacing fruit trees, where the kth Gaussian distribution represents the kth plant in the interlacing areas. The k value indicates the number of fruit trees in the range of the intermingling fruit trees, and τk, µk, and σk2 are weight, mean value, and variance, respectively in the kth region. The GMM is defined as:
where θk = (τk, µk, σk2, k ) is the unknown parameter to be determined, in which τk is the proportion of each distribution; µk is the mean value, σk2 is the variance; k is the number of Gaussian components; H is the projection histogram; and Hi is the projection value of the image on the i row.
Multiple local minimum points in the data made it difficult to determine the threshold value. To solve this problem, a relationship between the GMM and the intermingling trees was established to seek an estimate of the GMM parameters for the threshold value. Figure 7 shows the projection histogram of interlacing fruit trees with a multimodal distribution.
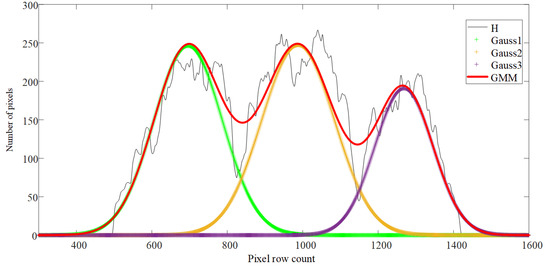
Figure 7.
Diagram of Gaussian Mixture Model (GMM) distribution. The red curve is the GMM; the black curve is the projection curve (H); and Gauss 1, Gauss 2, and Gauss 3 (the “+” curve) are the components of the GMM.
3.3.1. EM Algorithm for GMM Parameter Estimation
EM can effectively solve the model parameters. However, the EM algorithm is greatly affected by the initial parameters. In order to improve the accuracy of the parameter estimation, good initial parameters were obtained by analyzing the potential target range of each tree on the histogram. The steps were as follows.
(1) The scanline was established to scan the projection histogram curve by a line from bottom to top. The scan region was limited between the bottom of the projected histogram and the end position as shown in Figure 8. The end position was defined as line S = H + Hstd/2. The median H and standard deviation Hstd were obtained using the expression
where H is the projection histogram; Hi the projection value of the image on the i row; and h is the height of the image.
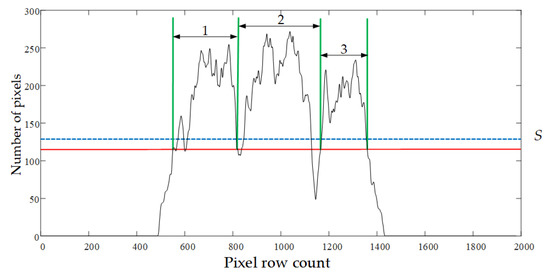
Figure 8.
Target areas scanned on the projection histogram. The end positions (blue line) and the final scan line position (red line).
(2) To identify the target region, the intersections of the scan line and the projection curve were recorded during the scanning process. The scanline stopped scanning when the number of intersections reached the maximum quantity. These intersections divided the projection curve into several intervals. The first and the second intersections were defined as the start and endpoint of the first target region, and the rest of the regions were defined by repeating this process. The average width of each interval was calculated by counting the pixels within each region. Regions that exceeded the average width become potential target regions, and the remaining intervals outside of each region were merged. If the newly merged interval had a width greater than the average value, it would be marked as a new target region. Otherwise, it would be merged into the nearest potential target region. Figure 8 shows an example of three target regions obtained this way (the domain marked 1, 2, and 3).
(3) Once the target regions were set, the parameters of each region were calculated. θ0 = (τk, µk, σk2, k) represented the initial parameter, where k is the number of target regions; µk, σk2 is the mean value and the variance of pixels within each region; and τk = 1/k. The final parameters θ were obtained using the EM iteration initial parameters. The input of EM algorithm is as follows: observation sample of Hi; implicit variable of z = 1,2,3,…, zi; joint distribution p(H, z| θ); conditional distribution p(z, H | θ). The E step of the EM algorithm is as follows: the parameter is θn after the nth iteration, and the conditional probability expectation of the joint distribution is:
The M step of the EM algorithm is as follows: maximizing the expected function Q, i.e., finding a θ(t + 1) meets:
These two steps were repeated until the algorithm converged. The Gaussian components in the Gaussian mixture model were obtained as shown in Figure 9.
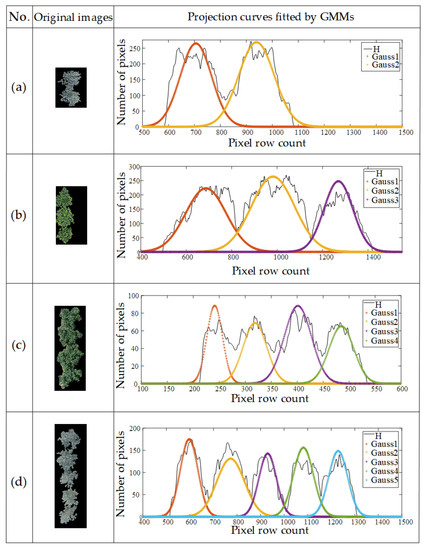
Figure 9.
Examples of parameter solving: (a) two touching cherry trees; (b) three touching apple trees; (c) four touching cherry trees; (d) five touching apple trees.
3.3.2. Threshold Value Determination
The final step for intermingling canopy segmentation was selecting a reasonable threshold to decompose the GMM. In this step, the T1 threshold computation method was introduced [40,41] to determine the segmentation point. As shown in Figure 10, the expressions of two adjacent Gaussian distributions are f1(µ1, σ12) and f2(µ2, σ22,). The following equations were used for solving the threshold (T):
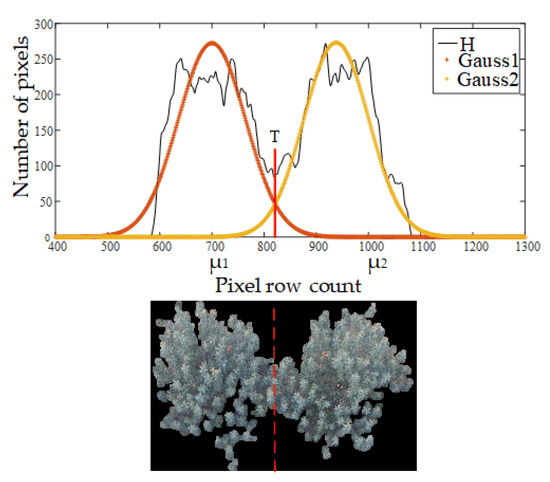
Figure 10.
An example of a threshold for image segmentation.
Figure 10 shows an example of the threshold (marked as a red solid line) after solving Equation (13) and the separation of two intermingling trees after applying the results (marked as a red dotted line).
3.4. Assessment of Intermingle Degrees
The projection histogram of the intermingled fruit trees was fitted to a GMM in this study. This means the degree of intermingledness of the contiguous trees can be estimated by the Gaussian component parameters in a GMM model. In this section, the XGBoost algorithm as a machine learning system for evaluation models will be discussed.
3.4.1. XGBoost Algorithm
Boosting is a technology that integrates multiple weak classifiers into one robust classifier. As an efficient implementation of Boosting algorithm, XGBoost algorithm not only solves the problem of parallel training of the model but also optimizes the parameters according to the second derivative, which improves the training efficiency [42]. At the same time, XGBoost adds regularization items to the loss function, which improves the generalization performance of the training model. The algorithm flow is as follows:
Assuming the model has k decision trees to predict the output, the can be estimated according to Equation (14)
where is the predicted value, xi is the input variable, and xi ∈ Rm. fk(xi) = ωq(xi), q: Rm → T, ω ∈ RT represents the prediction function to k decision trees. q(xi) is the structure of each tree that maps xi to the corresponding leaf index. T is the number of leaves in the tree, and ω is the leaf weight.
To learn the set of functions used in the model, the following regularized objective function should be minimized:
where Ω( fk) is a regular term, which helps to prevent over-fitting and to simplify models produced, γ is a complex parameter, λ is a fixed coefficient, and l is the loss function.
This algorithm was provided by the creators of the algorithm.
3.4.2. Parameters Input
As shown in Figure 11, the Gaussian parameters σ and f(µ) were used to characterize the shape features (width, height) of each component in the interlacing area. , f(T) represents overlapping degrees of the contiguous trees. In addition, x is the crown area of individual trees, which came from counting pixels in the images with isolated canopies. The photographed height was introduced as the scale parameter of the image as shown in Equation (16):
where Heightmax and Heightmin are the highest and the lowest photographed heights, respectively.
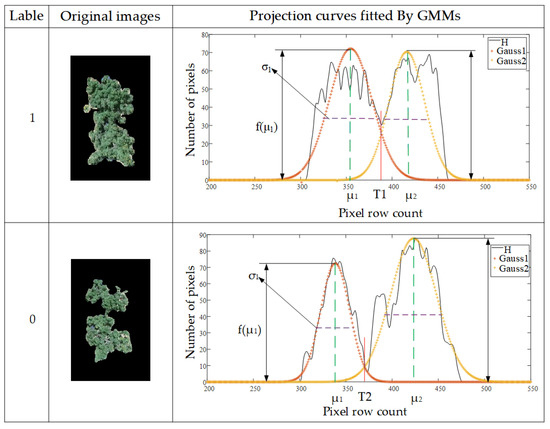
Figure 11.
Examples of labeling and the characterizing parameters.
In summary, (σ, f(µ), , f(T), x, hm) were selected as feature inputs, and the resized images were selected as training data for XGBoost model. Figure 11 shows examples of characterization parameters, and Table 2 shows partial sample feature inputs and its corresponding label.

Table 2.
Examples of the feature parameters and their labels.
3.4.3. Creation of Training Sets and Setting Parameters
In this study, 2173 images were labeled. Densely interlaced trees were labeled 1, and barely touching trees were labeled 0, as shown in Figure 11. To ensure the generalization ability of the XGBoost model, we applied the sklearn.grid_search package to search for optimal parameters. Some key parameters are displayed as follows:
{ Learning_rate of 0.002; N_estimators of 3000; Max_depth of 5; Gama of 0.2; Subsample of 0.8; Objective: binary: logistic }
3.5. Evaluation Indices
3.5.1. Evaluation Indices for Segmentation Performance
We evaluated and compared the segmentation results of the proposed method quantitatively. Specifically, we checked the number of trees and their pixels extracted from the images. Since there is no standard database or general evaluation methods in this specific topic of canopy isolation, we used four evaluation indices, namely Ro (over-segmentation rate), Ru (under-segmentation rate), R (root mean square) and the Intersection over Union (IoU), to evaluate the results of the canopy segmentation. Ro is the percentage of the extra trees extracted by the method, indicating the algorithm over-divided its targets. Ru is the ratio of the number of missing trees, indicating the algorithm under-divided its targets. R is the root mean square of the Ro and Ru and is the global metric of the accuracy of canopy counting. IoU (also known as the Jaccard index) compares the area of the segmented canopies to the actual area of the trees, which shows whether the canopy segmentation is complete. The actual quantity and area of the trees were manually counted using Adobe Photoshop as the ground truth. The calculation process is described as follows:
where Np is the over-counted trees and Nn is the unrecognized number of real trees in the segmentation result; N is the number of trees detected by the segmentation method; and Ni is the number of actual trees present in the images. S is the area of the trees produced by the segmentation algorithm, and Si is the real area of the trees calculated by manually tracing the borders using Adobe Photoshop. is the shared area by S and Si. is the total area by S and Si. An IoU value higher than the 0.70 generally indicates an excellent agreement between S and Si.
3.5.2. Evaluation Indices for Intermingling Degrees between Interlacing Canopies
A receiver operating characteristic curve (ROC), as well as area under the curve (AUC) and accuracy (ACC), respectively, were used for evaluation in the model training and test. An ROC curve [43] is the most commonly used way to visualize the performance of a binary classifier, and AUC [44] is a criterion used to quantitatively evaluate the training effect of the classification models. The ROC curve can be drawn by the (FPR, TPR) coordinates of each threshold of each model. The calculations of FPR and TPR are as follows:
where FP are the negative samples predicted to be positive by the model, N is the true number of the negative samples, TP is the positive samples predicted to be negative by the model, and P is the true number of positive samples.
AUC is calculated as shown in Equation (23).
ACC is a criterion used to judge the quality of classification in the final model test, which reflects the proportion of positives and negatives accurately identified by the model. It can be obtained by Equation (24).
where T is the number of samples classified correctly, and N and P are the values mentioned above.
4. Results and Discussion
4.1. Segmentation Result Analysis of Interlacing Canopies Using Three Competing Segmentation Methods
4.1.1. Competing Segmentation Methods
Watershed transform is a common solution for separating touching objects. Advanced instance segmentation methods such as convolutional neural networks (CNN) are also employed to isolate objects. In this study, our GMM method was compared with Watershed algorithms and CNN. To isolate canopies using the Watershed algorithm, both the traditional Watershed [45] algorithm and the improved Watershed algorithm were used to avoid over-segmentation. The improved Watershed steps are briefly described as follows: converting the original image into a binary image; calculating the distance transformation (Figure 12a); blurring the image to eliminate noises from the edges of the irregular canopy (Figure 12b); marking the local extremum of the canopy (Figure 12c); and performing the Watershed transformation (Figure 12d). Segmentation functions were then created by computing the local thresholds.
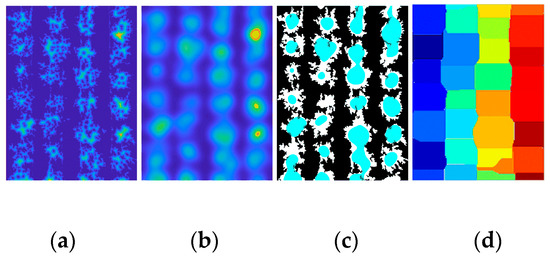
Figure 12.
The process of improved Watershed steps. (a) Distance transform; (b) image blur; (c) local extremum; and (d) color marker.
To separate interlacing canopies using CNN, the Mask Region Convolutional Neural Network ( Mask-RCNN) [46] was applied, which performed better than the Fast R-CNN [47] and Faster R-CNN [48], to generate segmentation masks for each instance of objects in the image. Mask R-CNN was implemented on Python 3.7, Keras, and TensorFlow. It is based on the Feature Pyramid Network (FPN) and the ResNet 101 backbone (Figure 13). Eight hundred seventy-five image training samples were created using Labelme. These datasets were stored in four different folders (mask, json, labemle_json, and pic). To train our dataset, the ResNet101 model was initialized using a pretrained parametric model on the MSCOCO dataset [49]. The training on the aviation tree dataset was set as follows: in the previous demo.py file, there were 81 classes. In our dataset, there were only two classes, tree canopy and background. The original data saved in the training session and Mask-RCNN folders, respectively, were changed. The accuracy was evaluated by a mean average precision with a threshold value of 0.5. All experiments were conducted on a graphics workstation with Ubuntu 18.04 operating system and NVIDIA RTX 2080TI graphics card installed.
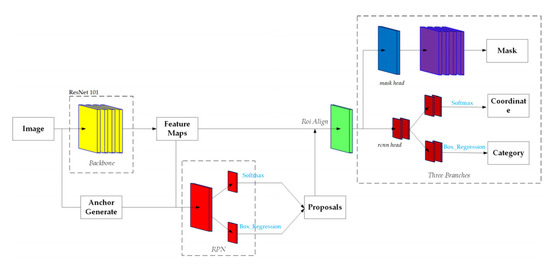
Figure 13.
The structure of Mask Region Convolutional Neural Network (Mask-RCNN).
4.1.2. Segmentation Result Using Four Methods
In order to evaluate the performance of the proposed method, 300 UAV images under different lighting conditions and different overlapping layouts were selected. Figure 14 shows an example of segmentation results. Figure 14a shows the original images of cherry trees taken on a sunny day and apple trees on a cloudy day. For convenience, detected canopies are mapped with red rectangles on the images (Figure 14b,c,e); the highlighted patches, or the masks, are the area of the individual canopies produced by Mask-RCNN (Figure 14d).
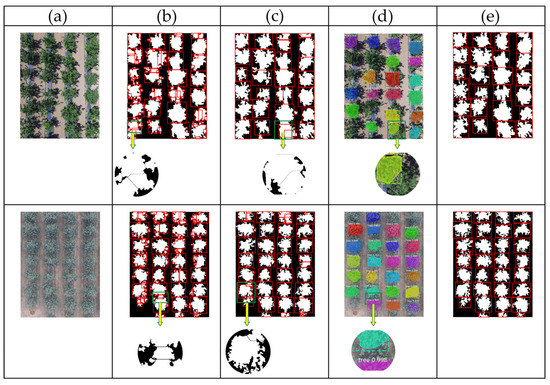
Figure 14.
Comparison of different segmentation results: (a) original images; (b) traditional Watershed algorithm; (c) improved Watershed algorithm; (d) Mask-RCNN algorithm; (e) our algorithm.
Figure 14b shows that the traditional watershed transform method does not adapt to irregular tree edges well. It over-separated the individual tree regions to many broken pieces, due to the variations of interlacing intensity of the trees. Figure 14c shows that the overall segmentation results using the improved Watershed algorithm are better than the traditional Watershed method in Figure 14b, since the exact local extremums are provided based on the fuzzy information. Its results are not satisfactory, however, as it failed to identify actual tree crown borders in the interlacing regions and it labeled some branches as extra tree units. According to Figure 14d, the generalization ability of Mask-RCNN was promising for isolating individual apple trees or cherry trees. Alas, the mask predictions generated by Mask-RCNN did not cover the entire area of the individual canopies because of the inconsistent shapes of the targets, not to mention that some trees were even completely missed. This experiment shows that our method has better separation results than the other algorithms and recognizes trees more accurately.
Table 3 shows the average segmentation results of 300 test images using different segmentation methods. The average Ro, Ru, R, and IoU values of the traditional Watershed methods were 80.8%, 84.3% 82.6%, and 64.3%, respectively. The quality of segmentation according to Ro, Ru, R, and IoU (47.3%, 16.5%, 35.4%, and 84.6%, respectively) improves significantly using the improved Watershed. The average Ro, Ru, and R of the Mask-RCNN algorithm were reduced by 67.6%, 6.6%, and 26.9%, respectively, compared to the traditional Watershed method. IoU had also improved 13.8%. However, the Ru metrics increased by 61.2%, and the IoU reduced by 6.5% relative to the improved Watershed analysis. The overall performance of the proposed method proves itself superior to the other three methods: the Watershed algorithms tended to over-separate and the Mask-RCNN performed poorly on the pixel prediction of tree canopies and the segmentation of the similar targets. Our method was more accurate, with the average Ro, Ru, R, and IoU values 3.1%, 5.3%, 4.3%, and 88.7%, respectively, proving itself a reliable input for the consequent evaluation of interlacedness.

Table 3.
Average results statistics for 300 apple images using different algorithms.
4.1.3. Segmentation Result in Different Scenarios
The orchard image samples used include several extreme cases, in which some trees may be heavily intermingled, and some barely have overlapping branches. The extremities were manually picked out and checked through the proposed method, as well as the other three formulas. This section compares the results in the more complex scenarios (Figure 15a), where canopy overlapping can be too hard to identify even for an expert, and in less complicated cases (Figure 15b).

Figure 15.
Segmentation results of canopies in different degrees of intermingledness: (a) image segmentation results of severely overlapping samples; (b) image segmentation results of lightly crossing samples. In each group, from left to right: original images, traditional Watershed, improved Watershed, Mask-RCNN, our algorithm results, and the ground truth.
Figure 15 shows the comparison between two types of overlapping scenarios using the traditional Watershed algorithm, the improved Watershed, Mask-RCNN, the proposed method, and the ground truth. The traditional Watershed for tree detection and tree crown separation failed in complex and less complex scenarios. The improved Watershed method performed better, but it would occasionally miss trees in the more complex scenarios. Mask-RCNN could not isolate individual plants when some overlapping canopies were covered by the same mask (Figure 15a). It missed some trees even in less complicated cases (Figure 15b). Our approach once again outperformed the other three, marking much more precise borders throughout patches with interweaving branches of various degrees. In this extreme scenario examination, the average Ro, Ru, R, and IoU values for the proposed method are 4.2%, 6.4%, 5.4%, and 87.9%, respectively, for the complex cases, and 1.7%, 3.3%, 2.6%, and 89.4%, respectively, for the less complex situations. Compared to the results in our previous test, the difference is negligible, demonstrating the robustness of our method. Overall, the proposed method of using GMM was feasible for identifying and isolating individual trees in continuous orchard rows and was most effective.
4.1.4. Segmentation Result of Multiple Interweaving Trees
The feasibility of the proposed method was then examined for handling multiple interweaving canopies in a row. The method was tested in orchards with different numbers of interweaving fruit trees, and the results were compared with the ground truth images. Figure 16 shows groups of the typical image segmentation results of the multiple interweaving trees in orchards (5 trees at most in our test areas). Specifically, Figure 16 includes two and five overlapping apple trees (Figure 16a,d) and three and four cherry trees (Figure 16b,c). The tested result indicates that the proposed method can effectively segment the interweaving regions. The image segmentation results of our methods are quite similar to the ground truth, visually speaking, indicating that the proposed method can effectively segment the interweaving regions. For quantitative verification, the test was performed on a total of 571 tree images containing different numbers of touching fruit trees. The final recognition results are shown in Figure 16.
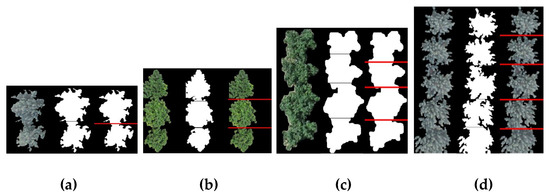
Figure 16.
Segmentation results of canopies based on the proposed method: (a) two interweaving apple trees; (b) three interweaving cherry trees; (c) four interweaving cherry trees; and (d) five interweaving apple trees. Each group, from left to right: original images, segmentation result, and the ground truth.
As shown in Figure 17, the proposed method was still able to accurately recognize the true boundaries of targets in the sample images, ranging from three touching trees to five. The corresponding levels of values of average Ro, Ru, R, and IoU were consistently above 87.78%, thanks to the fact that the accuracy of the solution of Gaussian mixture model does not have an obvious linear relationship with the number of Gaussian components. To be exact, the average of Ro, Ru, R, and IoU values were as high as 92.24%, 92.54%, 92.31%, and 89.56%, respectively. This shows our strategy is robust for image segmentation of interweaving areas with uncertain number of trees in complex or noncomplex conditions.
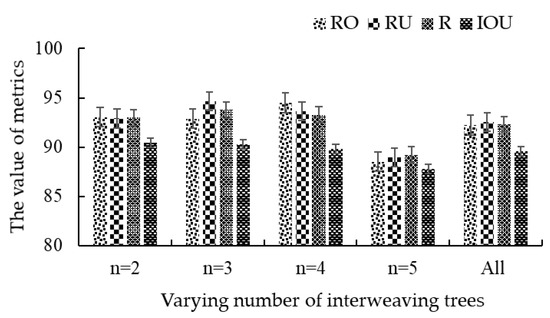
Figure 17.
Segmentation results of multiple interweaving trees based on the proposed method. n represents the number of fruit trees in the area.
4.2. Intermingledness Evaluation Result Analysis
4.2.1. Evaluation of Performance of Different Models
To test the learning ability of our model, the performance of XGBoost was compared with the other methods: Gradient Boosting Decision Tree (GBDT) + Random Forest (RF) model based on ensemble learning, XGBoost + Logistic regression (LR) model based on tree feature selection [50], and Deep Neural Network (DNN) model based on deep learning, which is a machine learning method based on artificial neural networks. Ensemble learning refers to an algorithm that combines base learners (such as decision trees and linear classifiers), the GBDT +RF and XGBoost+LR approaches are examples of such learning. Specifically, GBDT yields a predictive model in the form of an ensemble of decision trees. RF output the class for classification, regression and other tasks by constructing a multitude of decision trees at training time. LR is a linear model for capturing features. In order to use the GBDT + RF model, the results of outputs were obtained by weighted voting after training GBDT and RF independently. For the XGBoost + LR model, the trained XGBoost was sparsely encoded and input into LR for prediction results. The CNN model had ten layers in total, consisting of one input layer, one output layer, two dropout layers, and six fully connected layers. Table 4 shows the results of the 5-fold cross-validation on the test set using different models. The average ACC values of the XGBoost reached 84.3%, which was higher than the values of GDBT + RF and DNN (80.3% and 79.3%, respectively). While the average (AVG) ACC of the XGBoost algorithm was slightly lower than that of the XGBoost + LR (84.8%), the XGBoost model had the highest stability prediction, as its standard deviation (STD) of ACC was twice as high as XGBoost + LR (2.1%).

Table 4.
Comparison results of 5-fold cross-validation using four models.
Figure 18a shows the ROC curves and AUC results of the four evaluation models of their intermingledness Passessment. The ROC curve of XGBoost classifiers was close to the upper left corner and the AUC value was 93%. This means that the XGBoost had fewer errors. In comparison, the POC curve of XGBoost + LR and DNN classifier were lower than XGBoost’s, while GDBT + RF had lower AUC (91%). Figure 18b also shows the running time of the four evaluation models. XGBoost had the shortest running time (0.0104 s) of the four, while GDBT + RF had the longest (0.115 s). Overall, the XGBoost model outperformed the other three models, yielding the highest comprehensive performance on the test and validation sets.
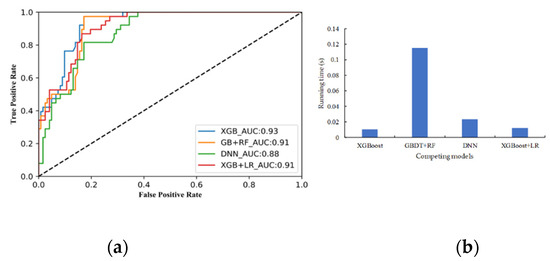
Figure 18.
(a) Receiver operating characteristic curve (ROC) and the area under the curve (AUC) of intermingledness evaluation model; (b) the running time of competing models.
4.2.2. Effect of Photographing Altitude on Model Accuracy
The pixel density per tree decreases as the photographing drone climbs in height, and the angles of shots taken may distort the pictures, affecting the kurtosis of the projection histograms. Additionally, the lowered resolution may have a smoothing effect on the images, blurring out the mixed pixels between overlapping trees. The accuracy of the models used to classify intermingledness may or may not decline. This study filmed images taken between 20 m and 60 m. To evaluate the possible effects of changing altitude, a new test set was generated with 240 images taken at different flight heights. Images were captured at approximately 20 m, 28 m, 36 m, 44 m, 52 m, and 60 m. Forty images, including 20 of the heavily interweaving patches and 20 of the less crossed areas, were selected at each flight height.
Figure 19 shows the accuracy of the model using photos taken at different flight heights. When the branches were not heavily crossed, the ACC value at 28 m was the highest. As the interweaving situation became more complex, however, the ACC declined. Pearson correlation coefficient (PCC) was introduced to measure the strength of the linear relationship between the model accuracy and the flight altitude [51]. Based on the data in Figure 19, the values of Pearson’s correlation were −0.257 and −0.326, respectively, under different intermingling conditions, and −0.260 for total test images. The results show that the photographing heights have no obvious effects on the model accuracy.
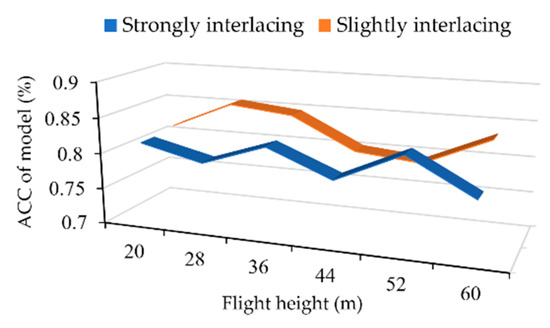
Figure 19.
The relationship between altitude of image taken and accuracy (ACC) of the model at different intermingling degrees.
5. Conclusions
The goal of this study was to correctly identify individual trees from contiguous canopies and evaluate the degree of intermingledness. We proposed a method utilizing a Gaussian Mixture Model and XGBoost. This method was able to pick out individual units by estimating the GMM parameters. The solved GMM parameters were then used to characterize the shape features of the trees for XGBoost input. Finally, supervised training was conducted based on the XGBoost classification algorithm to evaluate the severity of branch intermingling.
In the comparing experiments, the average Ro, Ru, R, and IoU values of the GMM algorithm segmentation were 3.1%, 5.3%, 4.3%, and 88.7%, respectively, indicating the proposed method could effectively segment the crossing canopies and produce the best results overall. The feasibility and validity of the method were also confirmed by testing with different interweaving conditions and multiple interweaving trees. As for the evaluation of intermingledness, our study showed that the average ACC and standard deviation of the XGBoost obtained on the test set was 84.3% and 1.2%, respectively, and performed well on the validation set (AUC of 93%). In addition, the proposed method was robust to handle images taken at varying heights, where the Pearson coefficient obtained was −0.260.
The above results are encouraging. UAV has great potential for horticultural tasks, which the study further attests. The proposed method is able to intake UAV images and output correctly identified plant information, paving the path for automating future precision-farming applications. Though the study is conducted based on apple trees and cherry trees, the diversity of the orchards shows that the algorithm is robust. As related technology advances, we will be able to further refine this methodology. If the input image has more than one kind of green plant, the preprocessing step in this method will reduce the accuracy of the results. Due to resource and time constraints, similar issues will be discussed in future work.
Author Contributions
Conceptualization, Z.C.; methodology, Z.C. and Y.C.; software, Y.C.; validation, Z.C. and Y.C.; formal analysis, Z.C.; investigation, Z.C. and Y.C.; resources, L.Q. and Y.W.; data curation, Z.C.; writing—Original draft preparation, Z.C.; writing—Review and editing, H.Z. and Y.W.; visualization, L.Q.; supervision, L.Q.; project administration, L.Q.; funding acquisition, L.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Plan of China (grant number 2017YFD0701400 and 2016YFD0200700).
Acknowledgments
The authors would like to thank the financial support provided by the National Key Research and Development Plan of China, and Sida Zheng for writing advice. Most of all, Zhenzhen Cheng wants to thank her partner Yifan Cheng for the constant encouragement and support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, Y.; Li, Y.; He, L.; Liu, F.; Cen, H.; Fang, H. Near ground platform development to simulate UAV aerial spraying and its spraying test under different conditions. Comput. Electron. Agric. 2018, 148, 8–18. [Google Scholar] [CrossRef]
- Lan, Y.; Thomson, S.J.; Huang, Y.; Hoffmann, W.C.; Zhang, H. Current status and future directions of precision aerial application for site-specific crop management in the USA. Comput. Electron. Agric. 2010, 74, 34–38. [Google Scholar] [CrossRef]
- Mogili, U.R.; Deepak, B.B.V.L. Review on Application of Drone Systems in Precision Agriculture. Proced. Comput. Sci. 2018, 133, 502–509. [Google Scholar] [CrossRef]
- Maes, W.H.; Steppe, K. Perspectives for Remote Sensing with Unmanned Aerial Vehicles in Precision Agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Cao, Z.-G.; Wu, X.; Bai, X.D.; Qin, Y.; Zhuo, W.; Xiao, Y.; Zhang, X.; Xue, H. Automatic image-based detection technology for two critical growth stages of maize: Emergence and three-leaf stage. Agric. For. Meteorol. 2013, 174–175, 65–84. [Google Scholar] [CrossRef]
- Gomes, M.F.; Maillard, P.; Deng, H. Individual tree crown detection in sub-meter satellite imagery using Marked Point Processes and a geometrical-optical model. Remote Sens. Environ. 2018, 211, 184–195. [Google Scholar] [CrossRef]
- Li, W.; Dong, R.; Fu, H.; Yu, L. Large-Scale Oil Palm Tree Detection from High-Resolution Satellite Images Using Two-Stage Convolutional Neural Networks. Remote Sens. 2019, 11, 11. [Google Scholar] [CrossRef]
- Liu, K.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Estimating forest structural attributes using UAV-LiDAR data in Ginkgo plantations. ISPRS J. Photogramm. Remote Sens. 2018, 146, 465–482. [Google Scholar] [CrossRef]
- Yin, D.; Wang, L. Individual mangrove tree measurement using UAV-based LiDAR data: Possibilities and challenges. Remote Sens. Environ. 2019, 223, 34–49. [Google Scholar] [CrossRef]
- Rasmussen, J.; Ntakos, G.; Nielsen, J.; Svensgaard, J.; Poulsen, R.N.; Christensen, S. Are vegetation indices derived from consumer-grade cameras mounted on UAVs sufficiently reliable for assessing experimental plots? Eur. J. Agron. 2016, 74, 75–92. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Surový, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- Nyamgeroh, B.B.; Groen, T.A.; Weir, M.J.C.; Dimov, P.; Zlatanov, T. Detection of forest canopy gaps from very high resolution aerial images. Ecol. Indic. 2018, 95, 629–636. [Google Scholar] [CrossRef]
- Durfee, N.; Ochoa, C.G. The Use of Low-Altitude UAV Imagery to Assess Western Juniper Density and Canopy Cover in Treated and Untreated Stands The Use of Low-Altitude UAV Imagery to Assess Western Juniper Density and Canopy Cover in Treated and Untreated Stands. Forests 2019, 10, 296. [Google Scholar] [CrossRef]
- Koc-San, D.; Selim, S.; Aslan, N.; San, B.T. Automatic citrus tree extraction from UAV images and digital surface models using circular Hough transform. Comput. Electron. Agric. 2018, 150, 289–301. [Google Scholar] [CrossRef]
- Recio, J.A.; Hermosilla, T.; Ruiz, L.A.; Palomar, J. Automated extraction of tree and plot-based parameters in citrus orchards from aerial images. Comput. Electron. Agric. 2013, 90, 24–34. [Google Scholar] [CrossRef]
- Roth, S.I.B.; Leiterer, R.; Volpi, M.; Celio, E.; Schaepman, M.E.; Joerg, P.C. Automated detection of individual clove trees for yield quantification in northeastern Madagascar based on multi-spectral satellite data. Remote Sens. Environ. 2019, 221, 144–156. [Google Scholar] [CrossRef]
- Niu, H.; Zhao, T.; Chen, Y. Tree Canopy Differentiation Using Instance-aware Semantic Segmentation. In Proceedings of the 2018 ASABE Annual International Meeting, Detroit, MI, USA, 29 July–1 August 2018; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2018; p. 1. [Google Scholar]
- Yao, Q.; Liu, Q.; Dietterich, T.G.; Todorovic, S.; Lin, J.; Diao, G.; Yang, B.; Tang, J. Segmentation of touching insects based on optical flow and NCuts. Biosyst. Eng. 2013, 114, 67–77. [Google Scholar] [CrossRef]
- Lin, P.; Chen, Y.M.; He, Y.; Hu, G.W. A novel matching algorithm for splitting touching rice kernels based on contour curvature analysis. Comput. Electron. Agric. 2014, 109, 124–133. [Google Scholar] [CrossRef]
- Liu, T.; Chen, W.; Wang, Y.; Wu, W.; Sun, C.; Ding, J.; Guo, W. Rice and wheat grain counting method and software development based on Android system. Comput. Electron. Agric. 2017, 141, 302–309. [Google Scholar] [CrossRef]
- Senthilnath, J.; Kandukuri, M.; Dokania, A.; Ramesh, K.N. Application of UAV imaging platform for vegetation analysis based on spectral-spatial methods. Comput. Electron. Agric. 2017, 140, 8–24. [Google Scholar] [CrossRef]
- Ishida, T.; Kurihara, J.; Viray, F.A.; Namuco, S.B.; Paringit, E.C.; Perez, G.J.; Takahashi, Y.; Marciano, J.J. A novel approach for vegetation classification using UAV-based hyperspectral imaging. Comput. Electron. Agric. 2018, 144, 80–85. [Google Scholar] [CrossRef]
- Ardila, J.P.; Bijker, W.; Tolpekin, V.A.; Stein, A. Multitemporal change detection of urban trees using localized region-based active contours in VHR images. Remote Sens. Environ. 2012, 124, 413–426. [Google Scholar] [CrossRef]
- Wagner, F.H.; Ferreira, M.P.; Sanchez, A.; Hirye, M.C.M.; Zortea, M.; Gloor, E.; Phillips, O.L.; de Souza Filho, C.R.; Shimabukuro, Y.E.; Aragão, L.E.O.C. Individual tree crown delineation in a highly diverse tropical forest using very high resolution satellite images. ISPRS J. Photogramm. Remote Sens. 2018, 145, 362–377. [Google Scholar] [CrossRef]
- Bazi, Y.; Malek, S.; Alajlan, N.; Alhichri, H. An Automatic Approach for Palm Tree Counting in UAV Images. In Proceedings of the 2014 IEEE Geoscience and Remote Sensing Symposium, Quebec, QC, Canada, 13–18 July 2014; pp. 537–540. [Google Scholar]
- Hassaan, O.; Nasir, A.K.; Roth, H.; Khan, M.F. Precision Forestry: Trees Counting in Urban Areas Using Visible Imagery based on an Unmanned Aerial Vehicle. IFAC-PapersOnLine 2016, 49, 16–21. [Google Scholar] [CrossRef]
- Li, L.; Mu, X.; Macfarlane, C.; Song, W.; Chen, J.; Yan, K.; Yan, G. A half-Gaussian fitting method for estimating fractional vegetation cover of corn crops using unmanned aerial vehicle images. Agric. For. Meteorol. 2018, 262, 379–390. [Google Scholar] [CrossRef]
- Cheng, Z.; Qi, L.; Wu, Y.; Cheng, Y.; Yang, Z.; Gao, C. Parameter Optimization on Swing Variable Sprayer of Orchard Based on RSM. Nongye Jixie Xuebao/Trans. Chin. Soc. Agric. Mach. 2017, 48. [Google Scholar] [CrossRef]
- Woebbecke, D.; Meyer, G.; Bargen, K.; Mortensen, D. Shape Features for Identifying Young Weeds Using Image Analysis. Trans. ASAE 1995, 38, 271–281. [Google Scholar] [CrossRef]
- Smith, P.; Reid, D.B.; Environment, C.; Palo, L.; Alto, P.; Smith, P.L. A Tlreshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum Likelihood from Incomplete Data via the EM Algorithm. J. R. Stat. Soc. Ser. B 1977, 39, 1–38. [Google Scholar]
- Zheng, L.; Shi, D.; Zhang, J. Segmentation of green vegetation of crop canopy images based on mean shift and Fisher linear discriminant. Pattern Recognit. Lett. 2010, 31, 920–925. [Google Scholar] [CrossRef]
- Guijarro, M.; Pajares, G.; Riomoros, I.; Herrera, P.J.; Burgos-Artizzu, X.P.; Ribeiro, A. Automatic segmentation of relevant textures in agricultural images. Comput. Electron. Agric. 2011, 75, 75–83. [Google Scholar] [CrossRef]
- Wang, A.; Zhang, W.; Wei, X. A review on weed detection using ground-based machine vision and image processing techniques. Comput. Electron. Agric. 2019, 158, 226–240. [Google Scholar] [CrossRef]
- Pastrana, J.C.; Rath, T. Novel image processing approach for solving the overlapping problem in agriculture. Biosyst. Eng. 2013, 115, 106–115. [Google Scholar] [CrossRef]
- Lu, M.; Xiong, Y.; Li, K.; Liu, L.; Yan, L.; Ding, Y.; Lin, X.; Yang, X.; Shen, M. An automatic splitting method for the adhesive piglets’ gray scale image based on the ellipse shape feature. Comput. Electron. Agric. 2016, 120, 53–62. [Google Scholar] [CrossRef]
- Coy, A.; Rankine, D.; Taylor, M.; Nielsen, D.C.; Cohen, J. Increasing the accuracy and automation of fractionalvegetation cover estimation from digital photographs. Remote Sens. 2016, 8, 474. [Google Scholar] [CrossRef]
- Ramachandran, K.M.; Tsokos, C.P. Chapter 6—Hypothesis Testing. In Mathematical Statistics with Applications in R, 2nd ed.; Ramachandran, K.M., Tsokos, C.P.B.T., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 311–369. ISBN 978-0-12-417113-8. [Google Scholar]
- Balakrishnan, N.; Voinov, V.; Nikulin, M.S. Chapter 2—Pearson’s Sum and Pearson-Fisher Test. In Chi-Squared Goodness of Fit Tests with Applications; Balakrishnan, N., Voinov, V., Nikulin, M.S.B.T., Eds.; Academic Press: Boston, MA, USA, 2013; pp. 11–26. ISBN 978-0-12-397194-4. [Google Scholar]
- Gonzalez, R.; Faisal, Z. Digital Image Processing, 2nd ed.; Pearson Education: London, UK, 2019; pp. 371–426. [Google Scholar]
- Liu, Y.; Mu, X.; Wang, H.; Yan, G. A novel method for extracting green fractional vegetation cover from digital images. J. Veg. Sci. 2012, 23, 406–418. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Hanley, J.A.; Mcneil, B. The Meaning and Use of the Area Under a Receiver Operating Characteristic (ROC) Curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef]
- Bradley, A. The Use of the Area Under the ROC Curve in the Evaluation of Machine Learning Algorithms. Pattern Recognit. 1996, 30, 1145–1159. [Google Scholar] [CrossRef]
- Meyer, F. Skeletons and watershed lines in digital spaces. In Proceedings of the 34th Annual International Technical Symposium on Optical and Optoelectronic Applied Science and Engineering, San Diego, CA, USA, 8–13 July 1990; SPIE: Bellingham, WA, USA, 1990; Volume 1350. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Honolulu, HI, USA, 22–25 July 2017; pp. 2980–2988. [Google Scholar]
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 39, 1137–1149. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C. Microsoft COCO: Common Objects in Context. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- He, X.; Pan, J.; Jin, O.; Xu, T.; Liu, B.; Xu, T.; Shi, Y.; Atallah, A.; Herbrich, R.; Bowers, S.; et al. Practical Lessons from Predicting Clicks on Ads at Facebook. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 1–9. [Google Scholar] [CrossRef]
- Mu, Y.; Liu, X.; Wang, L. A Pearson’s correlation coefficient based decision tree and its parallel implementation. Inf. Sci. 2017, 435, 40–58. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).