Application of Color Featuring and Deep Learning in Maize Plant Detection
Abstract
1. Introduction
2. Methodologies
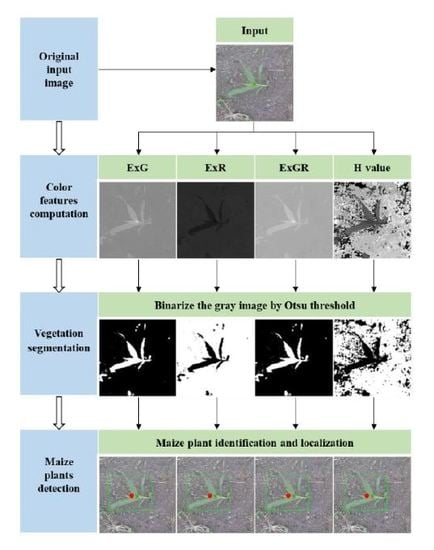
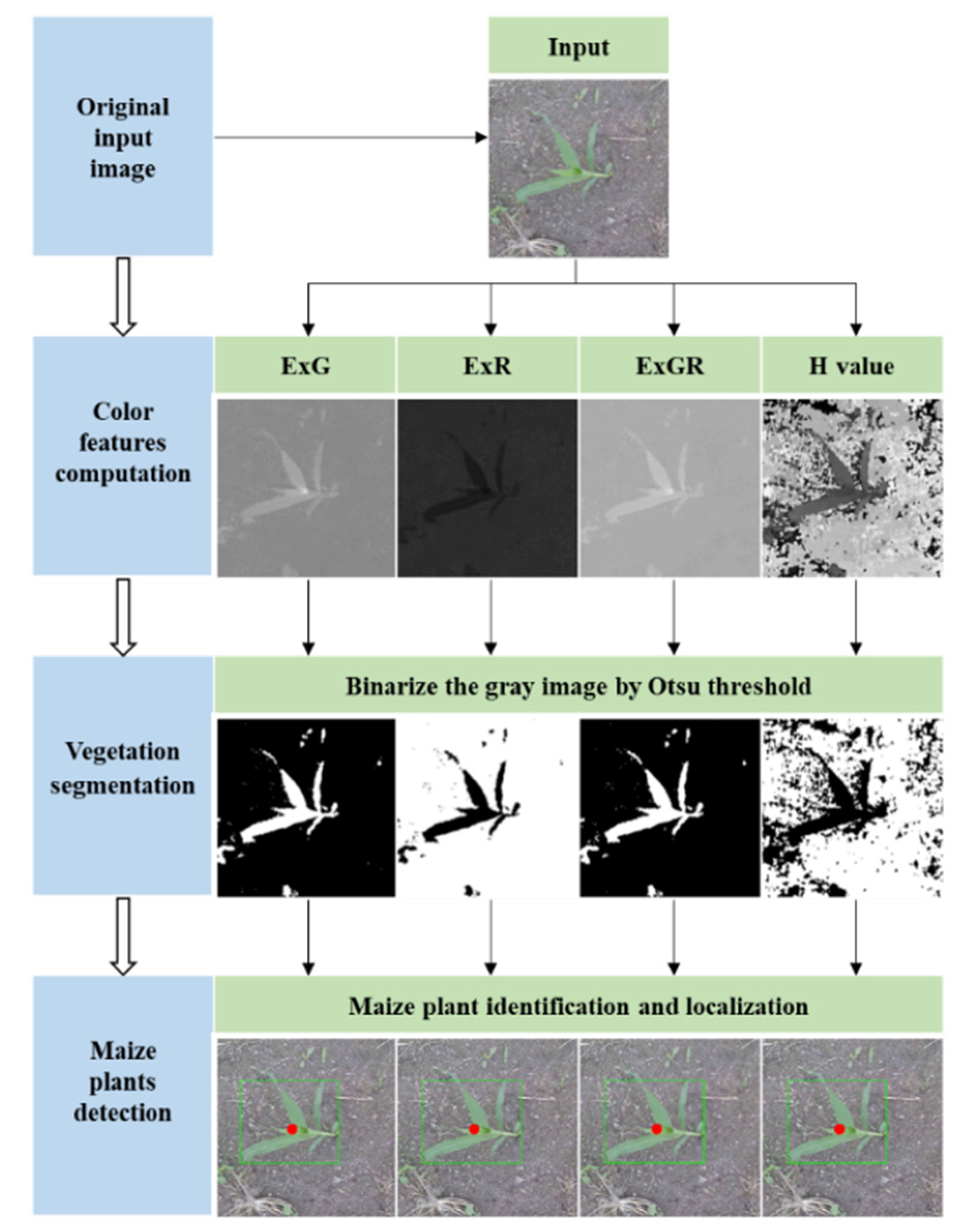
2.1. Color Index–Based Methods
2.1.1. Color Index Computation
2.1.2. Otsu Thresholding
2.1.3. Maize Plant Discrimination
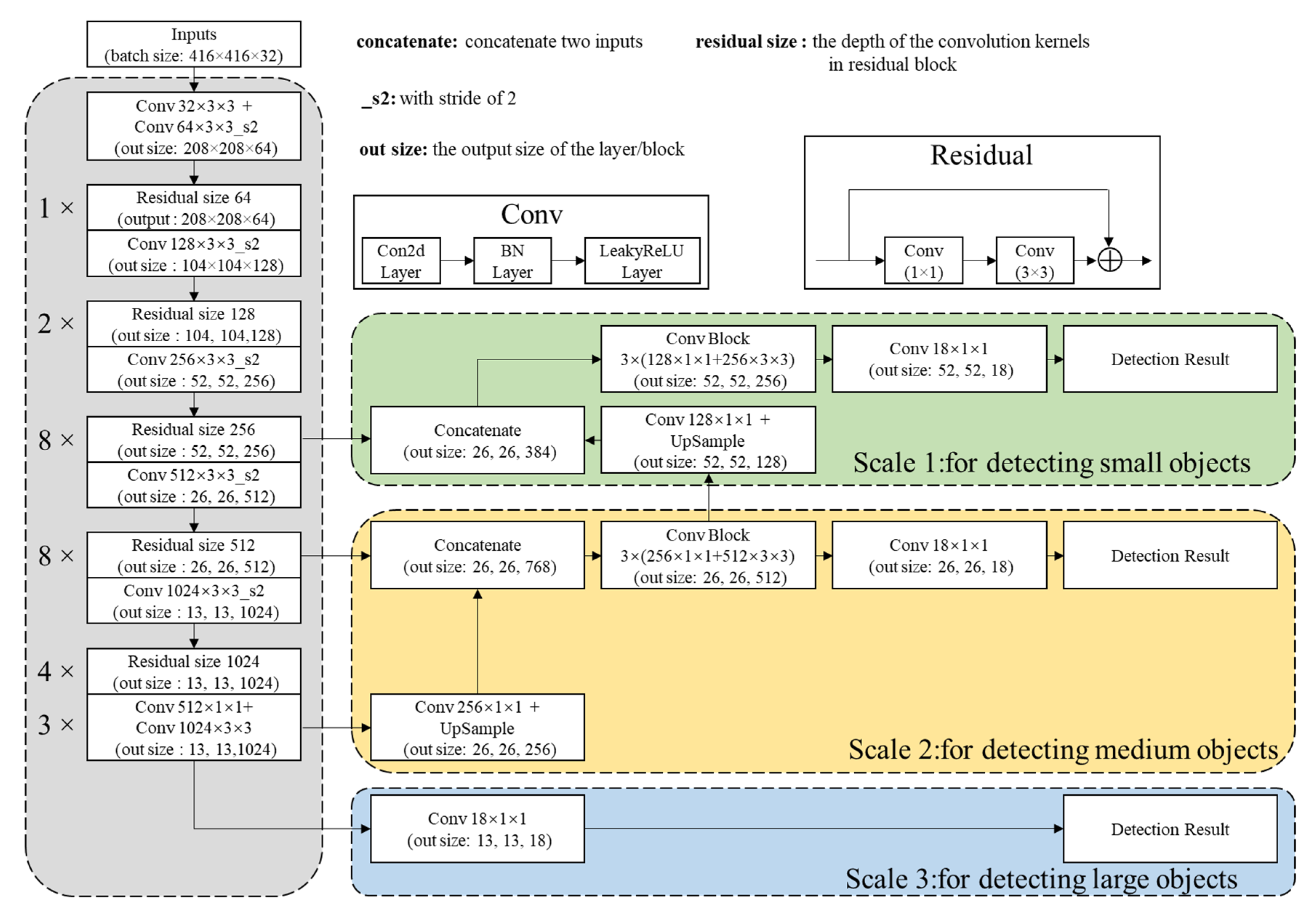
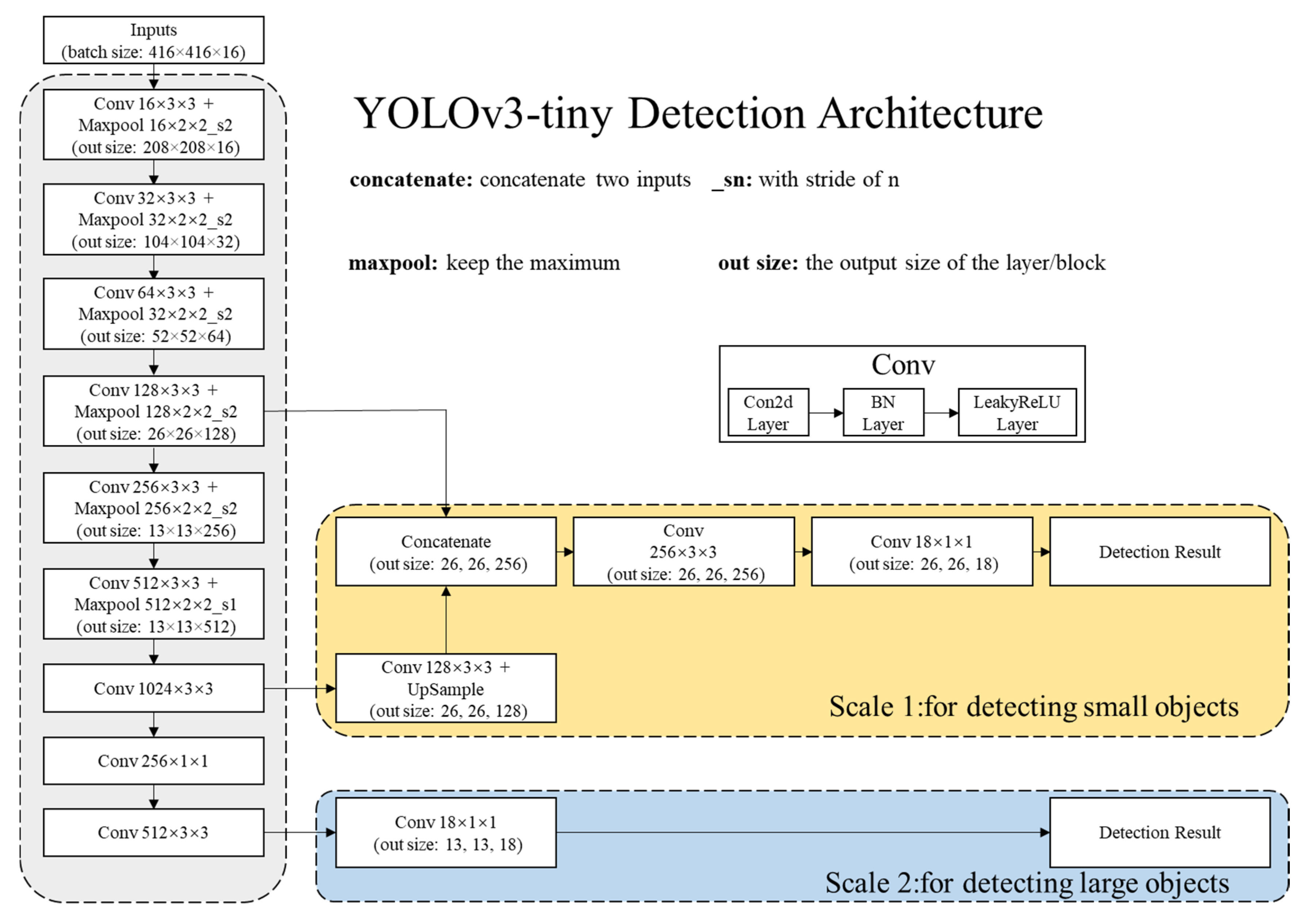
2.2. DL Methods
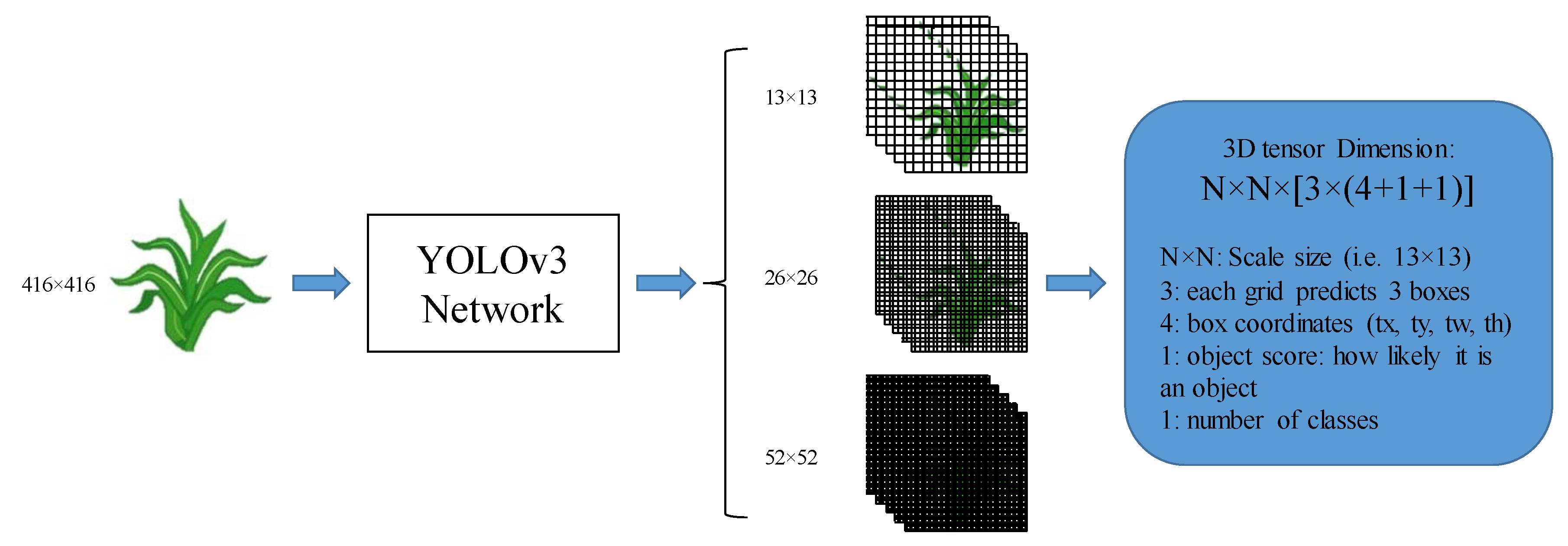
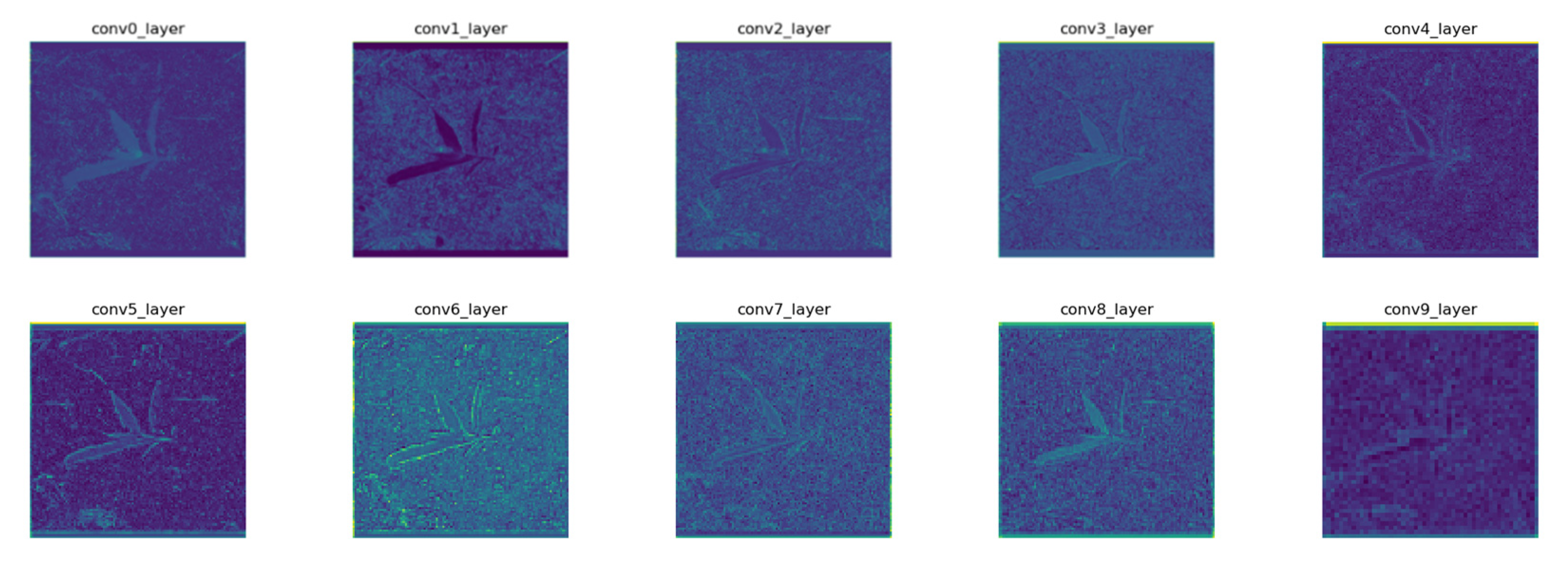
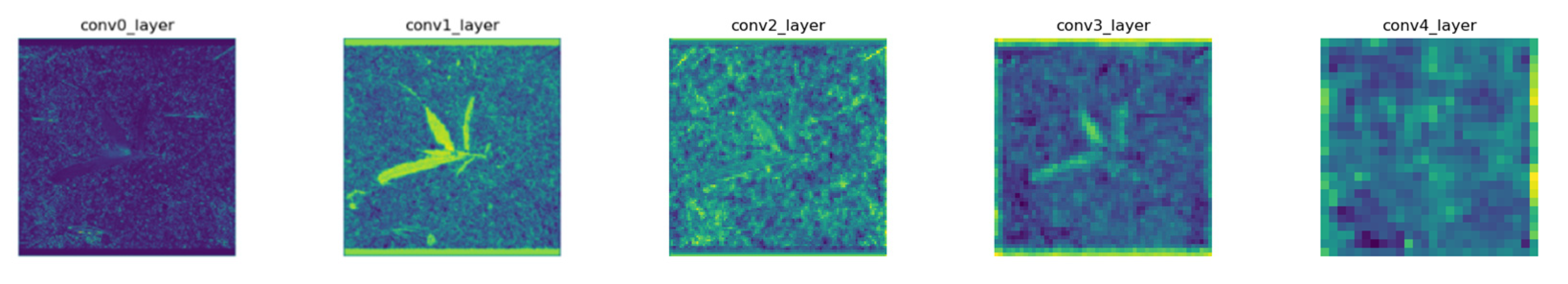
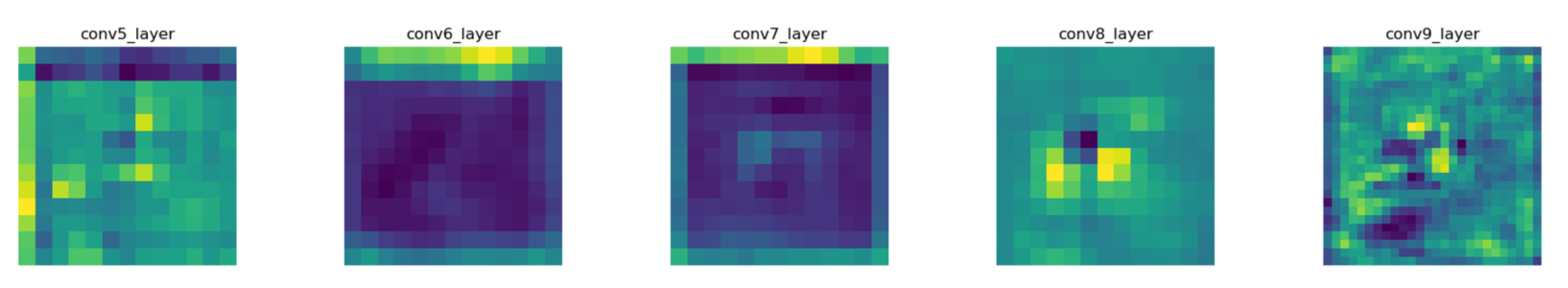
2.2.1. Network Architectures
2.2.2. Detection Principle
2.2.3. Loss Function Definitions
- S2 denotes the number of grid cells in feature maps;
- B denotes the number of prediction bounding boxes of each grid cell;
- ;
- ;
- ;
- ;
- predictioni,j = ();
- ;
- .
2.2.4. Network Training
2.3. Performance Evaluation
3. Experiments and Results
3.1. Image Data Collection
3.2. Image Data Augmentation
3.2.1. Data Augmentation: Image Brightness
3.2.2. Data Augmentation: Image Definition
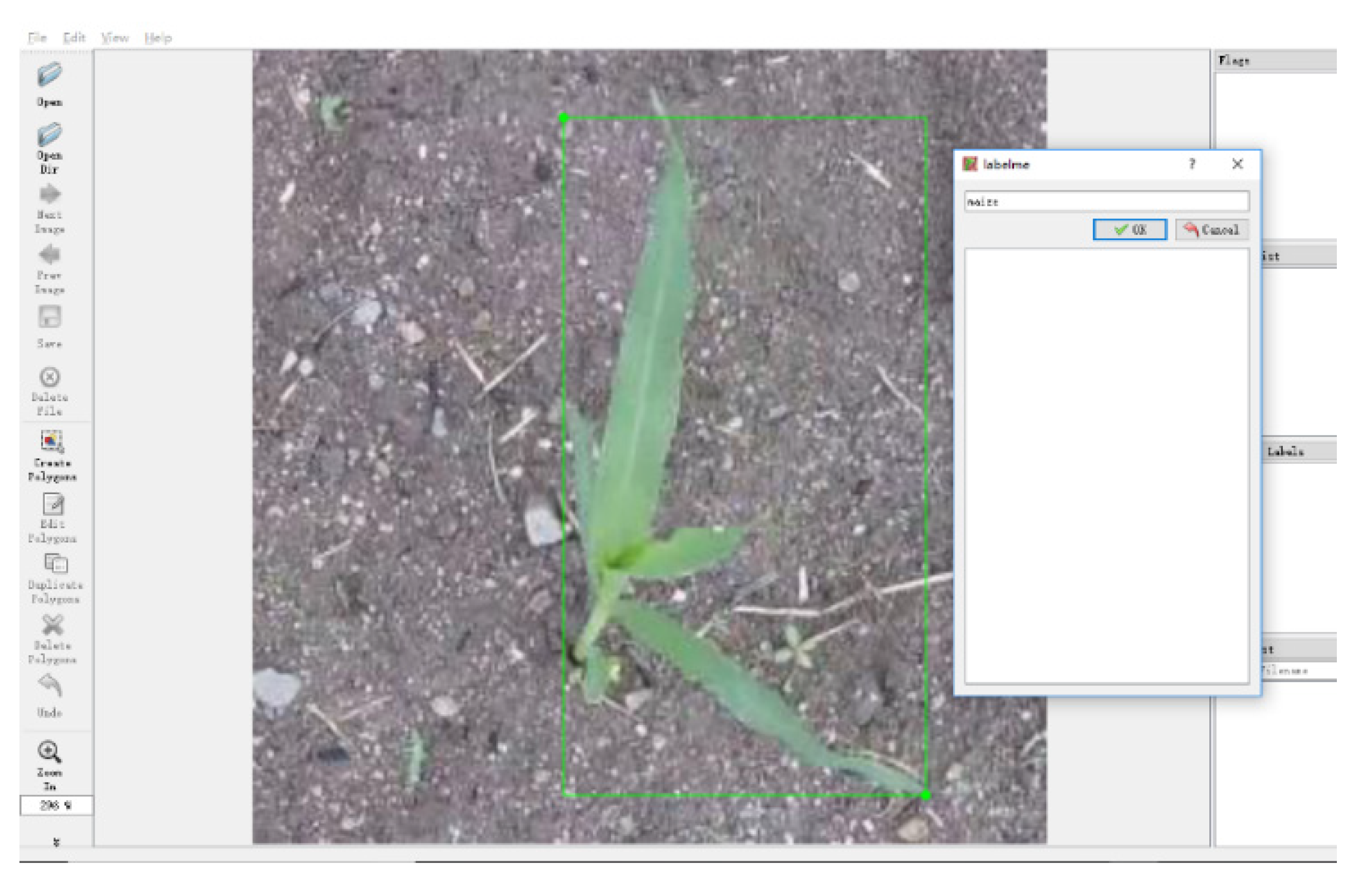
3.2.3. Image Data Annotation
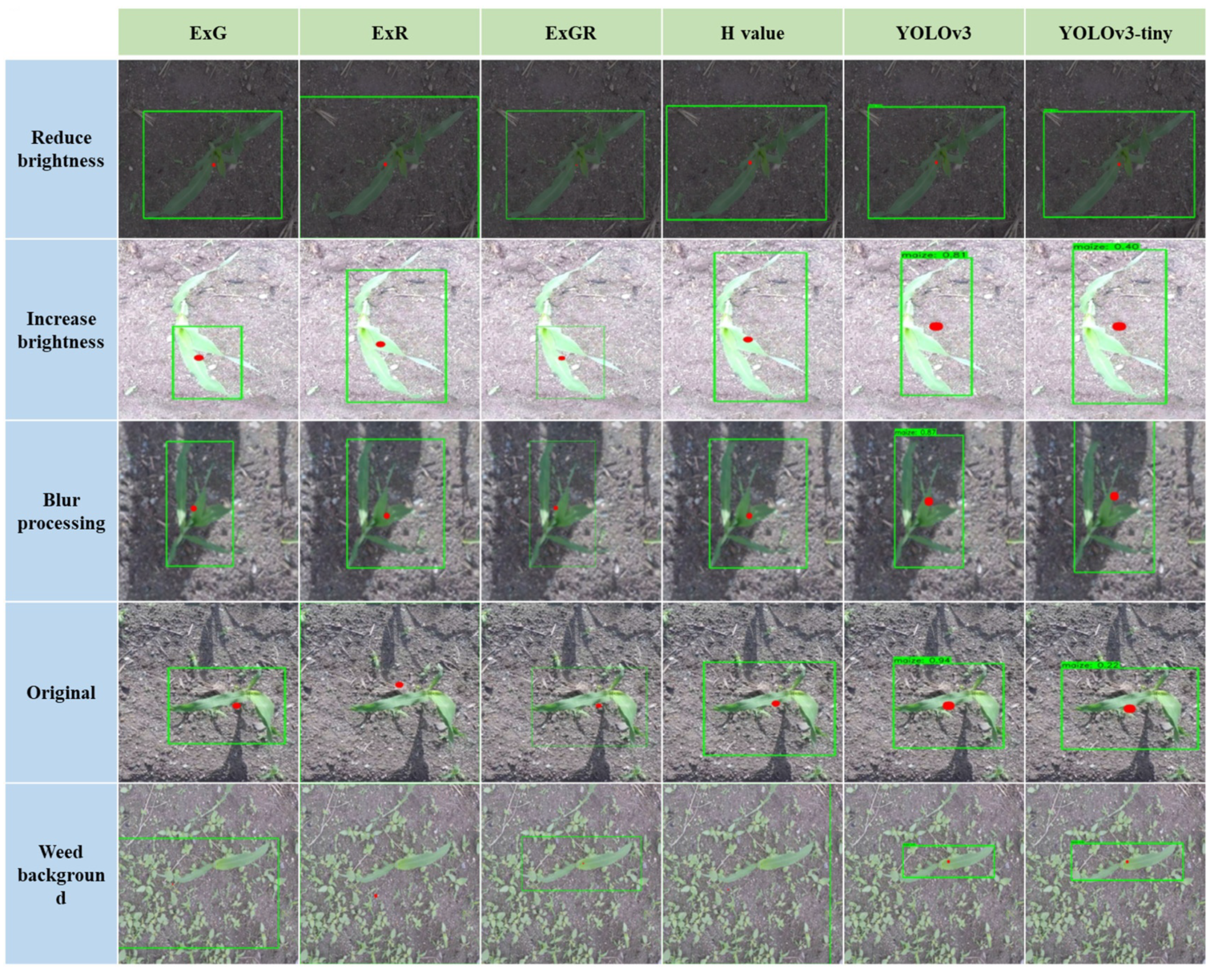
3.3. Results and Discussion
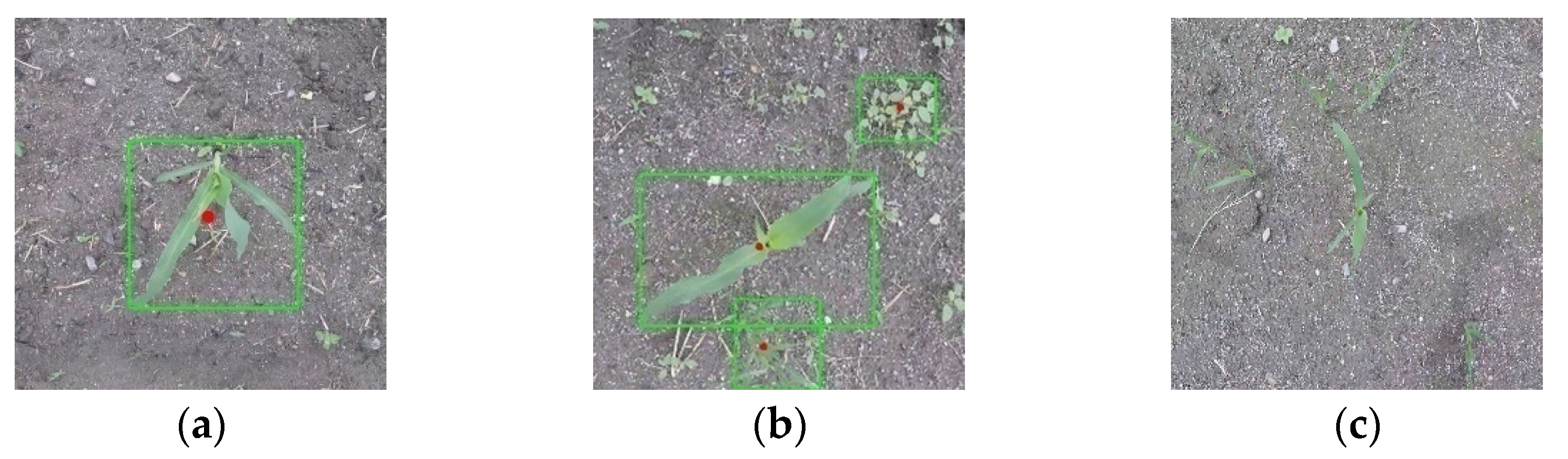
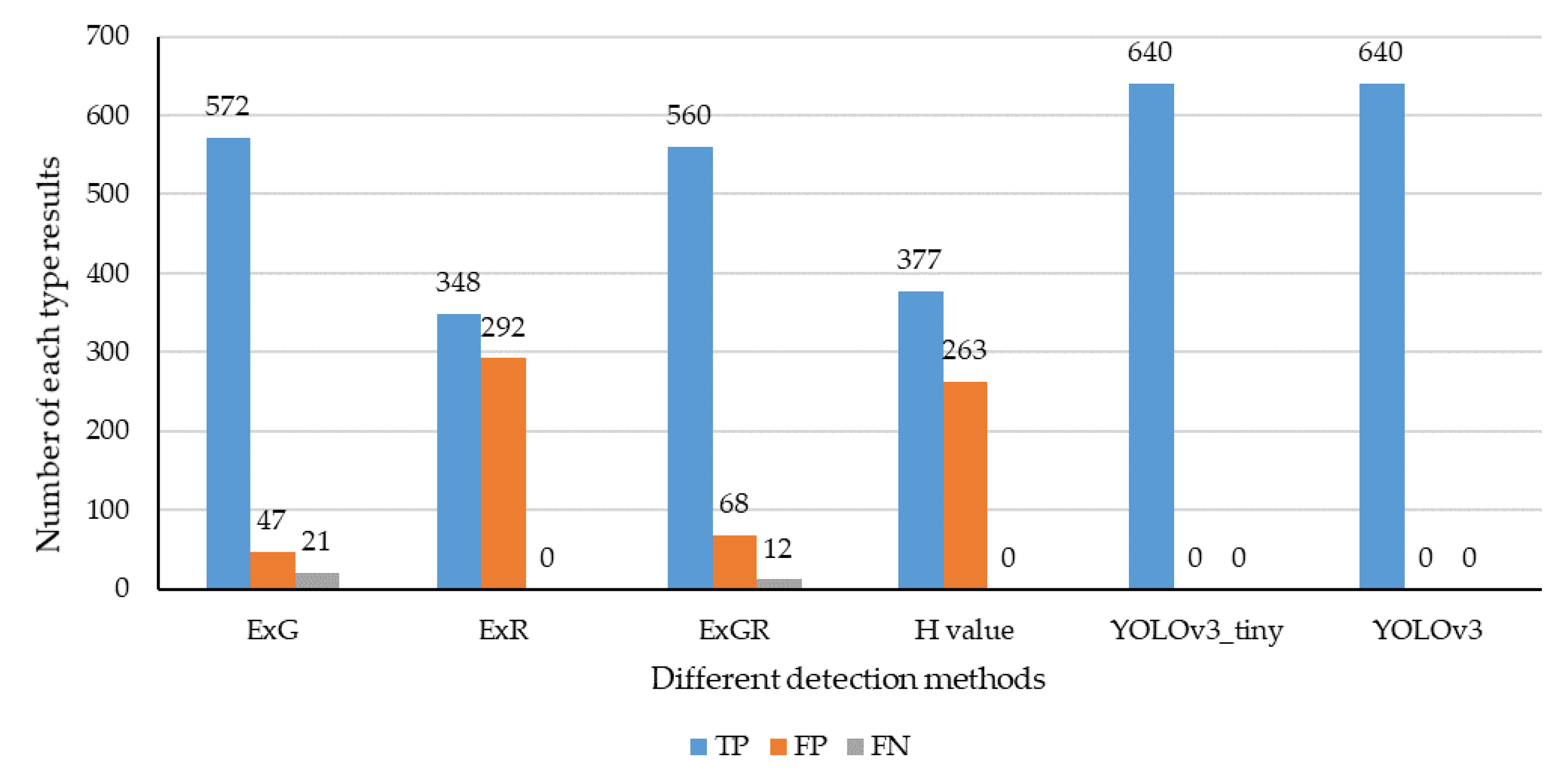
3.3.1. Identification Accuracy
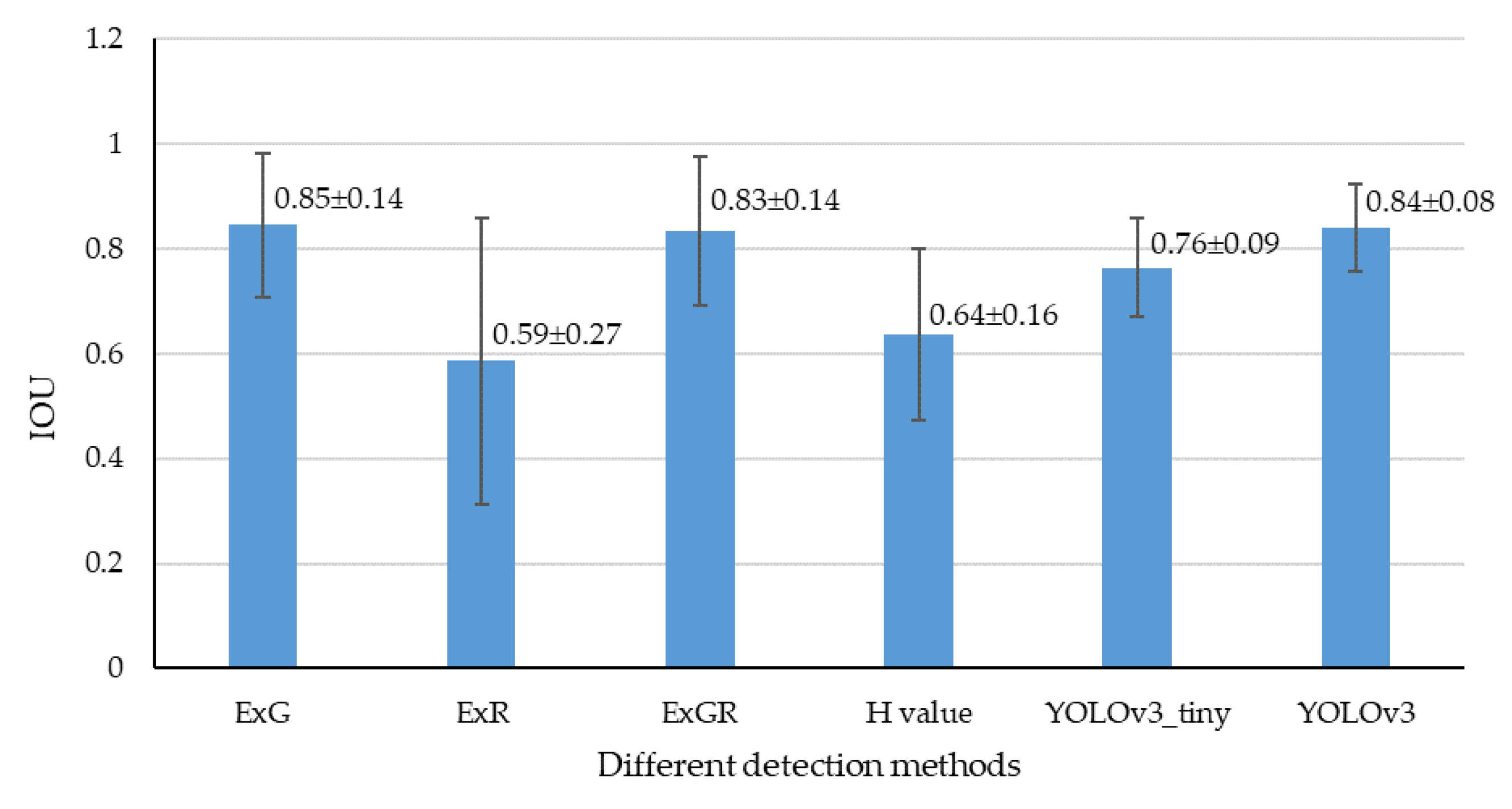
3.3.2. Localization Accuracy
3.3.3. Robustness Analysis
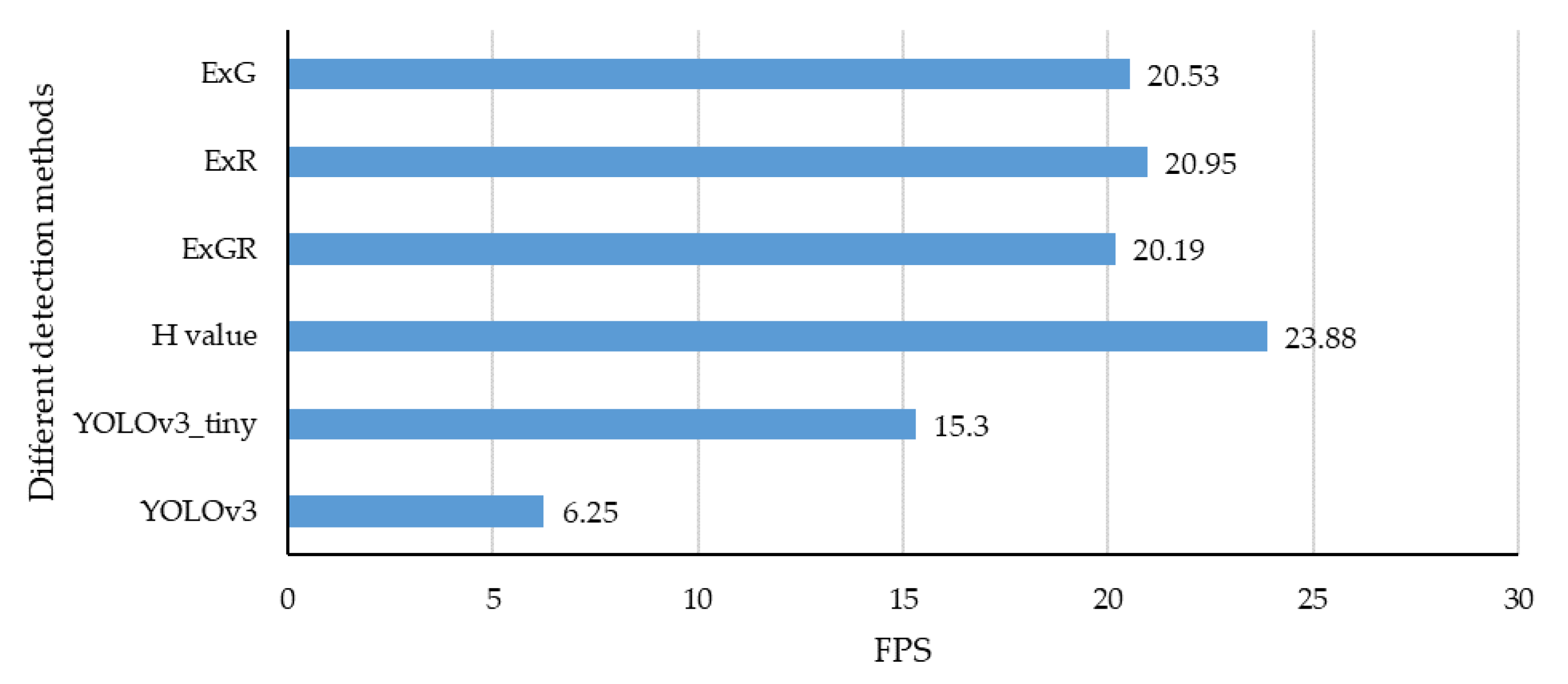
3.3.4. Detection Speed
3.3.5. Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pierce, F.J.; Nowak, P. Aspects of precision agriculture. Adv. Agron. 1999, 67, 1–85. [Google Scholar]
- Pahlmann, I.; Böttcher, U.; Kage, H. Evaluation of small site–specific N fertilization trials using uniformly shaped response curves. Eur. J. Agron. 2016, 76, 87–94. [Google Scholar] [CrossRef]
- Robert, P.C. Precision agriculture: A challenge for crop nutrition management. Plant Soil 2002, 247, 143–149. [Google Scholar] [CrossRef]
- Robertson, M.J.; Llewellyn, R.S.; Mandel, R.; Lawes, R.; Bramley, R.G.V.; Swift, L.; Metz, N.; O’Callaghan, C. Adoption of variable rate fertiliser application in the Australian grains industry: Status, issues and prospects. Precis. Agric. 2012, 13, 181–199. [Google Scholar] [CrossRef]
- Basso, B.; Cammarano, D.; Fiorentino, D.; Ritchie, J.T. Wheat yield response to spatially variable nitrogen fertilizer in Mediterranean environment. Eur. J. Agron. 2013, 51, 65–70. [Google Scholar] [CrossRef]
- Basso, B.; Dumont, B.; Cammarano, D.; Pezzuolo, A.; Marinello, F.; Sartori, L. Environmental and economic benefits of variable rate nitrogen fertilization in a nitrate vulnerable zone. Sci. Total Environ. 2016, 545, 227–235. [Google Scholar] [CrossRef] [PubMed]
- López-Granados, F. Weed detection for site–specifc weed management: Mapping and real–time approaches. Weed Res. 2011, 51, 1–11. [Google Scholar] [CrossRef]
- Peteinatos, G.G.; Weis, M.; Andújar, D.; Rueda Ayala, V.; Gerhards, R. Potential use of ground–based sensor technologies for weed detection. Pest Manag. Sci. 2014, 70, 190–199. [Google Scholar] [CrossRef]
- Dammer, K.-H.; Intress, J.; Beuche, H.; Selbeck, J.; Dworak, V. Discrimination of Ambrosia artemisiifolia and Artemisia vulgaris by hyperspectral image analysis during the growing season. Weed Res. 2013, 53, 146–156. [Google Scholar] [CrossRef]
- Wang, A.C.; Zhang, W.; Wei, X.H. A review on weed detection using ground–based machine vision and image processing techniques. Comput. Electron. Agric. 2019, 158, 226–240. [Google Scholar] [CrossRef]
- Bakhshipour, A.; Jafari, A. Evaluation of support vector machine and artificial neural networks in weed detection using shape features. Comput. Electron. Agric. 2018, 145, 153–160. [Google Scholar] [CrossRef]
- García-Santillán, I.; Guerrero, J.M.; Montalvo, M.; Pajares, G. Curved and straight crop row detection by accumulation of green pixels from images in maize fields. Precis. Agric. 2018, 19, 18–41. [Google Scholar] [CrossRef]
- Hamuda, E.; Glavin, M.; Jones, E. A survey of image processing techniques for plant extraction and segmentation in the field. Comput. Electron. Agric. 2016, 125, 184–199. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Bargen, K.; Von Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. ASAE 1995, 38, 259–260. [Google Scholar] [CrossRef]
- Marcial-Pablo, M.D.; Gonzalez-Sanchez, A.; Jimenez-Jimenez, S.I.; Ontiveros-Capurata, R.E.; Ojeda-Bustamante, W. Estimation of vegetation fraction using RGB and multispectral images from UAV. Int. J. Remote Sens. 2019, 40, 420–438. [Google Scholar] [CrossRef]
- Meyer, G.E.; Hindman, T.W.; Lakshmi, K. Machine vision detection parameters for plant species identification. In Precision Agriculture and Biological Quality, Proceedings of SPIE; Meyer, G.E., DeShazer, J.A., Eds.; International Society for Optics and Photonics, Univ. of Nebraska/Lincoln: Lincoln, NE, USA, 1999. [Google Scholar]
- Meyer, G.E.; Camargo Neto, J.; Jones, D.D.; Hindman, T.W. Intensified fuzzy clusters for classifying plant, soil, and residue regions of interest from color images. Comput. Electron. Agric. 2004, 42, 161–180. [Google Scholar] [CrossRef]
- Hunt, E.R.; Cavigelli, M.; Daughtry, C.S.T.; McMurtrey, J.E.; Walthall, C.L. Evaluation of digital photography from model aircraft for remote sensing of crop biomass and nitrogen status. Precis. Agric. 2005, 6, 359–378. [Google Scholar] [CrossRef]
- Burgos-Artizzu, X.P.; Ribeiro, A.; Guijarro, M.; Pajares, G. Real–time image processing for crop/weed discrimination in maize fields. Comput. Electron. Agric. 2011, 75, 337–346. [Google Scholar] [CrossRef]
- Hassanein, M.; Lari, Z.; El-Sheimy, N.; Hassanein, M.; Lari, Z.; El-Sheimy, N. A new vegetation segmentation approach for cropped fields based on threshold detection from hue histograms. Sensors 2018, 18, 1253. [Google Scholar] [CrossRef]
- Sabzi, S.; Abbaspour-Gilandeh, Y.; García-Mateos, G. A fast and accurate expert system for weed identifcation in potato crops using metaheuristic algorithms. Comput. Ind. 2018, 98, 80–89. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray–level histograms. IEEE Trans. Syst. Man. Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Neto, J.C.; Meyer, G.E.; Jones, D.D.; Samal, A.K. Plant species identification using elliptic fourier leaf shape analysis. Comput. Electron. Agric. 2006, 50, 121–134. [Google Scholar] [CrossRef]
- Tang, Z.; Su, Y.; Er, M.J.; Qi, F.; Zhang, L.; Zhou, J. A local binary pattern based texture descriptors for classification of tea leaves. Neurocomputing 2015, 168, 1011–1023. [Google Scholar] [CrossRef]
- Larese, M.G.; Namías, R.; Craviotto, R.M.; Arango, M.R.; Gallo, C.; Granitto, P.M. Automatic classification of legumes using leaf vein image features. Pattern Recognit. 2014, 47, 158–168. [Google Scholar] [CrossRef]
- Guo, W.; Rage, U.K.; Ninomiya, S. Illumination invariant segmentation of vegetation for time series wheat images based on decision tree model. Comput. Electron. Agric. 2013, 96, 58–66. [Google Scholar] [CrossRef]
- Riegler-Nurscher, P.; Prankl, J.; Bauer, T.; Strauss, P.; Prankl, H. A machine learning approach for pixel wise classification of residue and vegetation cover under field conditions. Biosyst. Eng. 2018, 169, 188–198. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, G.; Gómez-Gil, J.; Navas-Gracia, L.M. Testing different color spaces based on hue for the environmentally adaptive segmentation algorithm (EASA). Comput. Electron. Agric. 2009, 68, 88–96. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, J.; Wang, Q. Mean–shift–based color segmentation of images containing green vegetation. Comput. Electron. Agric. 2009, 65, 93–98. [Google Scholar] [CrossRef]
- Zheng, L.; Shi, D.; Zhang, J. Segmentation of green vegetation of crop canopy images based on mean shift and Fisher linear discriminant. Pattern Recognit. Lett. 2010, 31, 920–925. [Google Scholar] [CrossRef]
- Guerrero, J.M.; Pajares, G.; Montalvo, M.; Romeo, J.; Guijarro, M. Support Vector Machines for crop/weeds identification in maize fields. Expert Syst. 2012, 39, 11149–11155. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Milioto, A.; Lottes, P.; Stachniss, C. Real–time Semantic Segmentation of Crop and Weed for Precision Agriculture Robots Leveraging Background Knowledge in CNNs. arXiv 2017, arXiv:1709.06764v1. [Google Scholar]
- Ruicheng, Q.; Ce, Y.; Ali, M.; Man, Z.; Brian, J.S.; Cory, D.H. Detection of Fusarium Head Blight in Wheat Using a Deep Neural Network and Color Imaging. Remote Sens. 2019, 11, 2658. [Google Scholar]
- Tian, Y.N.; Yang, G.D.; Wang, Z.; Wang, H.; Li, E.; Liang, Z.Z. Apple detection during different growth stages in orchards using the improved YOLO–V3 model. Comput. Electron. Agric. 2019, 157, 417–426. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Shaun, M.S.; Schumann, A.W.; Yu, J.L.; Nathan, S.B. Vegetation detection and discrimination within vegetable plasticulture row–middles using a convolutional neural network. Precis. Agric. 2019, 20, 1107–1135. [Google Scholar]
- Song, X.; Zhang, G.; Liu, F.; Li, D.; Zhao, Y.; Yang, J. Modeling spatio–temporal distribution of soil moisture by deep learning–based cellular automata model. J. Arid Land. 2016, 8, 734–748. [Google Scholar] [CrossRef]
- Lee, S.H.; Chan, C.S.; Mayo, S.J.; Remagnino, P. How deep learning extracts and learns leaf features for plant classification. Pattern Recognit. 2017, 71, 1–13. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Y.Q.; Gong, C.Y.; Chen, Y.Y.; Yu, H.H. Applications of Deep Learning for Dense Scenes Analysis in Agriculture: A Review. Sensors 2020, 20, 1520. [Google Scholar] [CrossRef]
- Chen, S.W.; Shivakumar, S.S.; Dcunha, S.; Das, J.; Okon, E.; Qu, C.; Kumar, V. Counting apples and oranges with deep learning: A data–driven approach. IEEE Rob. Autom. Lett. 2017, 2, 781–788. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Koirala, A.; Walsh, K.B.; Wang, Z.; McCarthy, C. Deep learning for real–time fruit detection and orchard fruit load estimation: Benchmarking of ‘MangoYOLO’. Precis. Agric. 2019, 20, 1107–1135. [Google Scholar] [CrossRef]
- Liu, Z.C.; Wang, S. Brokent Corn Detection Based on an Adjusted YOLO with Focal Loss. IEEE Access 2019, 7, 68281–68289. [Google Scholar] [CrossRef]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A database and web–based tool for image annotation. Int. J. Comput. Vis. 2008, 77, 157–173. [Google Scholar] [CrossRef]
- Meyer, G.E.; Camargo-Neto, J. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Hyeonwoo, N.; Seunghoon, H.; Bohyung, H. Learning Deconvolution Network for Semantic Segmentation. arXiv 2015, arXiv:1505.04366v1. [Google Scholar]
- Song, H.; Huizi, M.; William, J.D. Deep Compression: Compressing Deep Neural Networks with Pruning, Trained Quantization and Huffman Coding. arXiv 2016, arXiv:1510.00149v5. [Google Scholar]




| Parameter Types | Values |
|---|---|
| Video | 4K30, 1440p80, 1080p120, video stabilization |
| Connection | USB Type–C/micro–HDMI |
| Display | 2”/LCD touch display |
| Memory | Slot type: microSD/Max. slot capacity: 128 GB |
| Wireless | Bluetooth low energy/Wi–Fi: 802.11 b/g/n, 2.4 and 5 GHz |
| Location | GPS |
| Waterproof | 10 m (33 ft) |
| Battery | 1220 mAh (Removable), 4.40 V |
| Dataset Names | Operation Types | Number of Images |
|---|---|---|
| Training dataset | Original | 500 |
| Training dataset | Reduced brightness | 500 |
| Training dataset | Increased brightness | 500 |
| Training dataset | Blur processing | 500 |
| Validation dataset | Original | 160 |
| Validation dataset | Reduced brightness | 160 |
| Validation dataset | Increased brightness | 160 |
| Validation dataset | Blur processing | 160 |
| Test dataset | Original | 160 |
| Test dataset | Reduced brightness | 160 |
| Test dataset | Increased brightness | 160 |
| Test dataset | Blur processing | 160 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Sun, H.; Li, M.; Iida, M. Application of Color Featuring and Deep Learning in Maize Plant Detection. Remote Sens. 2020, 12, 2229. https://doi.org/10.3390/rs12142229
Liu H, Sun H, Li M, Iida M. Application of Color Featuring and Deep Learning in Maize Plant Detection. Remote Sensing. 2020; 12(14):2229. https://doi.org/10.3390/rs12142229
Chicago/Turabian StyleLiu, Haojie, Hong Sun, Minzan Li, and Michihisa Iida. 2020. "Application of Color Featuring and Deep Learning in Maize Plant Detection" Remote Sensing 12, no. 14: 2229. https://doi.org/10.3390/rs12142229
APA StyleLiu, H., Sun, H., Li, M., & Iida, M. (2020). Application of Color Featuring and Deep Learning in Maize Plant Detection. Remote Sensing, 12(14), 2229. https://doi.org/10.3390/rs12142229