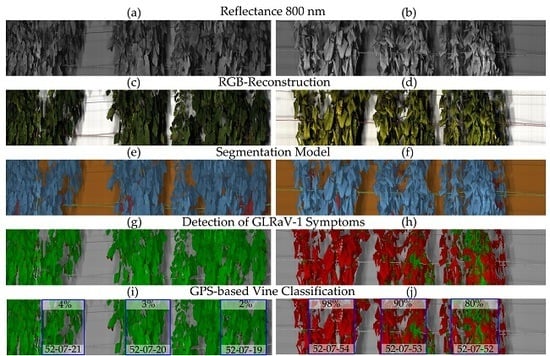
1. Introduction
Grapevine is considered to be one of the most important hosts for plant viruses with around 70 different virus and virus-like agents documented [
1]. Among these, viruses associated with grapevine leafroll disease (GLD) are the most widespread and of high economic importance worldwide [
2]. Currently, six distinct grapevine leafroll associated viruses (GLRaVs)—numbered GLRaV-1, -2, -3, -4, -7, and -13—are known. GLRaVs belong to the family
Closteroviridae and are separated into three genera: (i)
Ampelovirus (GLRaV-1, -3, -4, -13), (ii)
Closterovirus (GLRaV-2), and (iii)
Velarivirus (GLRaV-7). Within the genus
Ampelovirus, two subgroups were formed with GLRaV- 1, -3 and -13 assigned to subgroup I and GLRaV-4 assigned to subgroup II [
3]. GLRaVs are mostly limited to phloem-associated cells and are unevenly distributed in plants [
4]. Since grapevines are vegetatively propagated to maintain clonal integrity, these viruses are typically introduced into vineyards through contaminated planting material [
5] with subsequent local dispersal of some GLRaVs by insect vectors like mealybug and soft scale species [
6].
GLRaVs can occur as single or mixed infections of different combinations, and as a consequence thereof, symptom manifestation and severity are highly variable [
7]. In general, GLRaV-1, -3, and most strains of GLRaV-2 induce strong GLD symptoms, whereas GLRaV-4 and -7 elicit only mild or no symptoms [
8]. So far, little is known about the newly assigned GLRaV-13 concerning pathogenicity and symptomatology [
9]. Symptom development typically begins around veraison in mature leaves at the bottom sections of canes and then expands gradually upwards along the shoots as the growing season progresses. In red cultivars, colored spots develop, which enlarge and coalesce over time, so that most of the interveinal leaf surface exhibits a red or reddish-purple discoloration with a narrow strip of leaf tissue around the primary and secondary veins remaining green. In late autumn, leaf margins begin to roll downward giving the disease its common name [
4]. Symptoms of white-berried cultivars, which are expressed as mild yellowing or chlorotic mottling, are often subtle, thus hindering visual disease assessment. Moreover, some white
Vitis vinifera cultivars, most rootstocks and wild
Vitis species may be completely symptomless, making symptom-based disease diagnosis not sufficiently reliable [
1].
To date, no curative in-field treatment for GLD is known. Consequently, only prophylactic measures can be applied to reduce the spread of the disease. Besides planting certified virus-free vines, identification of infected plants and subsequent uprooting is one of the most common approaches [
10]. In this context, several diagnostic methods have been developed for robust and reliable disease detection, with serological and molecular analyses being the most relevant [
11,
12]. Serological techniques like enzyme-linked immunosorbent assay (ELISA) are used to detect viral proteins allowing for high-throughput screening [
11]. Alternatively, molecular methods based on reverse transcription followed by polymerase chain reaction (RT-PCR) are applied for the detection of viral genomic material, providing high sensitivity [
12]. Irrespective of the technique used, unknown viruses or new strains of known viruses can remain undetected leading to false-negative results [
11,
12].
In recent years, spectral sensors have proven to be a promising tool for disease diagnosis being independent from genetic and phenotypic information about pathogens [
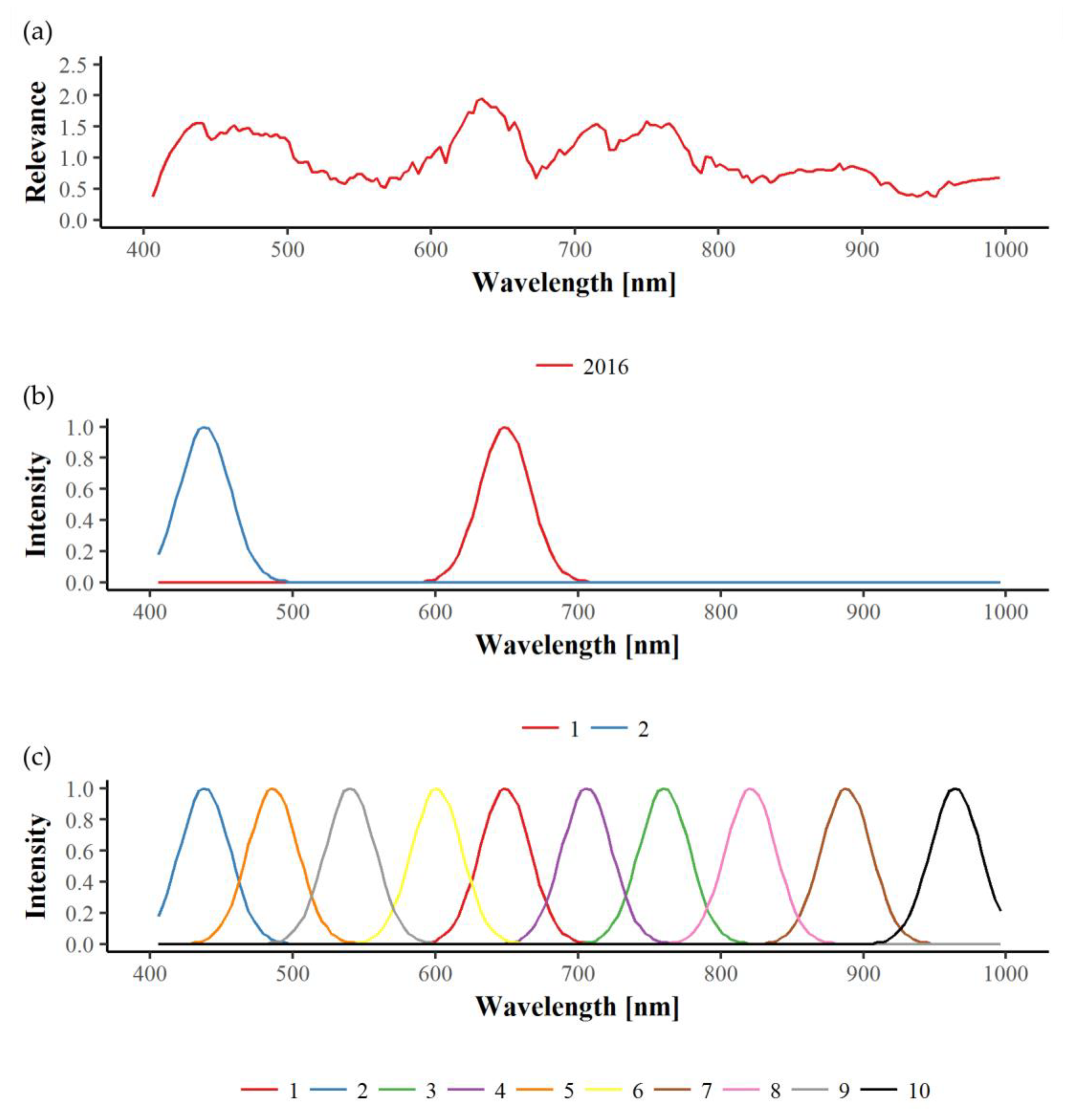
13]. These sensors capture reflectance characteristics of plants in a large part of the electromagnetic spectrum, typically in the visible (VIS; 400–700 nm), near-infrared (NIR; 700–1000 nm), and short-wave infrared range (SWIR; 1000–2500 nm) [
14]. For analysis either the whole spectral range (hyperspectral) or selected bands only (multispectral) can be used [
15]. Leaf spectral patterns vary in response to biochemical and biophysical alterations caused by abiotic and biotic stresses such as virus infections [
16]. Since spectral sensors allow the objective and non-invasive assessment of plant traits, they are well suited to follow dynamic processes like symptom development.
Sensor technology has already been implemented to differentiate spectral reflectance patterns of virus-infected and non-infected plants. In field tests, multispectral and hyperspectral approaches for the detection of tulip breaking virus (TBV) in tulips as well as potato virus Y (PVY) in potato and tomato plants have successfully been performed [
17,
18,
19]. Several studies have been conducted using greenhouse plants for the detection of tomato spotted wilt virus (TSWV) [
20], potato yellow vein virus (PYVV) [
21], cucumber green mottle mosaic virus (CGMMV) [
22], or tomato yellow leaf curl virus (TYLCV) [
23]. In the work of Afonso et al. [
24], two citrus cultivars infected with citrus tristeza virus (CTV)—a closterovirus closely related to GLRaV-2—were examined prior to symptom expression showing good classification rates. For the detection of GLD symptoms, Naidu et al. [
25] used a portable spectroradiometer (350–2500 nm) to collect hyperspectral reflectance data of detached healthy, asymptomatic, and symptomatic leaves, thereby reaching classification accuracies of up to 81%. In a comparable work, Sinha et al. [
26] investigated GLD symptoms during two years under field conditions. Pagay et al. [
27] also used a portable spectroradiometer to detect GLD in asymptomatic leaves directly in the vineyard. Similar studies were performed by Hou et al. [
28] and MacDonald et al. [
29], both following an airborne remote sensing approach to obtain multispectral and hyperspectral images, respectively. All of these works have been examining red-berried cultivars infected with GLRaV-3—except for Hou et al. [
28] who did not further specify the GLRaV investigated.
This study focuses on the detection of GLRaV-1 and -3 in both white- and red-berried Vitis cultivars using hyperspectral imaging in the range of 400–2500 nm. For this purpose, plants were recorded under controlled laboratory conditions as well as directly in the field during three consecutive years to (i) develop disease detection models for the discrimination of symptomatic, asymptomatic, and healthy plants, (ii) identify relevant wavelengths for the simulation of a multispectral approach, and (iii) differentiate GLRaV-1 and -3 symptoms.
4. Discussion
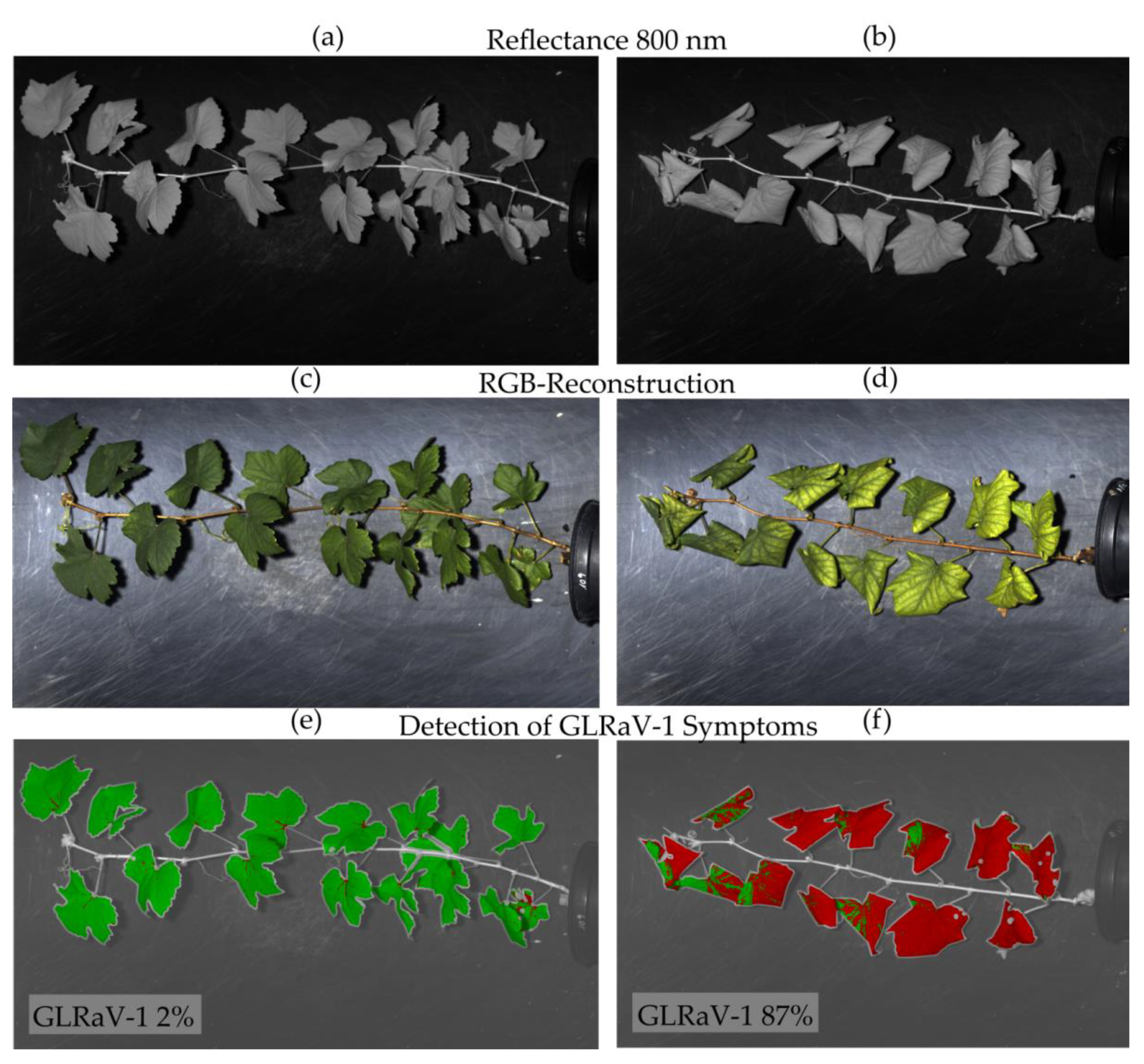
In the present study, hyperspectral imaging is used for the detection of GLRaV-1 and -3 in different white and red grapevine cultivars. Since symptom development is highly dependent on environmental factors, scion-rootstock combination, and the cultivar itself [
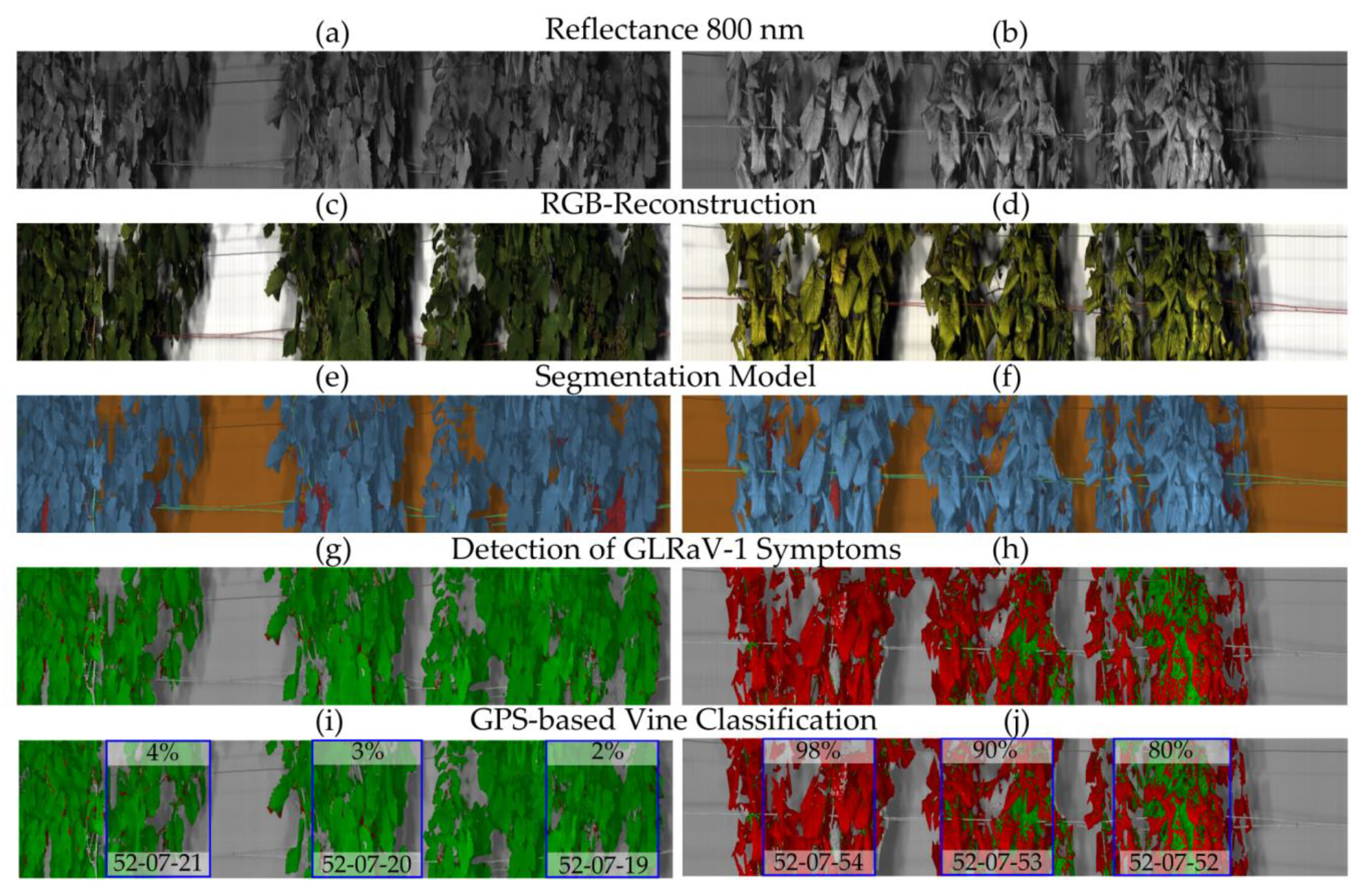
4], as a first step, ungrafted greenhouse plants were used to evaluate the potential of hyperspectral imaging for GLD detection. Thereby, symptomatic vines could be successfully distinguished from healthy vines with TPRs of 82–100% and corresponding low FPRs of 0–14%. Although growing vines under greenhouse conditions allows environmental factors to be closely controlled, phenotypes rarely agree with those in the field as grapevines are typically large perennial plants. Therefore, hyperspectral sensors were also applied in the field over three consecutive years. In order to reduce disruptive factors the Phenoliner phenotyping platform was used for standardized data acquisition [
35] and vineyards in close proximity to each other were selected to minimize environmental effects. While disease detection models for greenhouse plants were developed separately per cultivar due to the small set of plant-virus combinations, in models for field-grown grapevines more than 150
Vitis vinifera cultivars and interspecific crossings were combined. Since spectral reflectance is known to differ by cultivar as recently demonstrated by Gutiérrez et al. [
45], we wanted to develop robust detection models that should not be influenced by varietal effects. In red cultivars, GLRaV-3 symptomatic vines can be differentiated from healthy vines, reaching CAs of 84–96%; similar results were obtained for GLRaV-1. Symptom detection is also successful for white cultivars, which had not been investigated in previous studies, with CAs between 83% and 98% for GLRaV-3 and 76–93% for GLRaV-1. Hence, in-field disease detection is possible, although in most cases slightly better for red than white cultivars. This is not surprising as the contrast between reddish discoloration of symptomatic leaves and green healthy leaves in red-berried cultivars is more pronounced than that of yellowish discolorations in white cultivars. These effects were also observed by Albetis et al. [
46] using airborne multispectral imaging for the detection of
Flavescence dorée—a grapevine disease exhibiting similar symptoms as GLD. They obtained better results for red than white cultivars which was due to higher confusion between symptomatic and healthy pixels in white cultivars.
The different symptom expression of GLD can be explained by an increase of anthocyanin concentration in infected red cultivars [
47], which does not occur in white-berried grapevines due to several mutations in the flavonoid biosynthetic pathway genes [
48]. Therefore, detection models were developed separately for red and white cultivars. As expected, symptoms induced by GLRaV-1 and -3 in red and white cultivars, respectively, can be clearly distinguished in VNIR but also in SWIR. Interestingly, Al-Saddik et al. [
49] combined leaves of different white and red cultivars that showed typical
Flavescence dorée symptoms in a binary classification approach (healthy vs. symptomatic). Thereby, joined symptomatic leaves reached only slightly lower classification accuracies than single cultivars. Additional studies are required to determine whether this concept could also be successful for GLD detection.
In this context, model robustness should also be evaluated in further studies by applying them to unknown plant material that was not considered in model development. Thereby, effects of different locations (e.g., environment, soil) can be analyzed and disease detection could be tested under commercial conditions with only one cultivar per vineyard. Furthermore, it would be interesting to compare the models developed in this study with more than 150 cultivars with models developed specifically for one vineyard or one cultivar in order to further evaluate the potential of one universal GLD detection model.
Although GLD symptoms can be clearly distinguished between white and red cultivars, visual differentiation between the divergent GLRaVs is impossible [
2]. However, Berdugo et al. [
50] showed the potential of spectral imaging for the discrimination of viral diseases in cucumber plants. They were able to differentiate cucumber mosaic virus (CMV) and cucumber green mottle mosaic virus (CGMMV) up to accuracies of 100% using VNIR. Therefore, we first developed detection models separately for GLRaV-1 and -3 symptoms and, in a next step, developed models to discriminate plants infected with the two viruses. In both white and red cultivars, differentiation is possible with CAs of 76–100%. Since GLRaV-1 and -3 are closely related and can be distinguished using hyperspectral imaging, spectral data of infections caused by their different genetic variant groups [
51,
52] should also be evaluated in order to analyze the disease in more detail. Furthermore, spectral data of additional GLRaVs, e.g., GLRaV-2 with most of its strains producing strong GLD symptoms [
53], or mixed infections, which are common for GLRaVs [
7], could be included in future work in order to follow a broader approach.
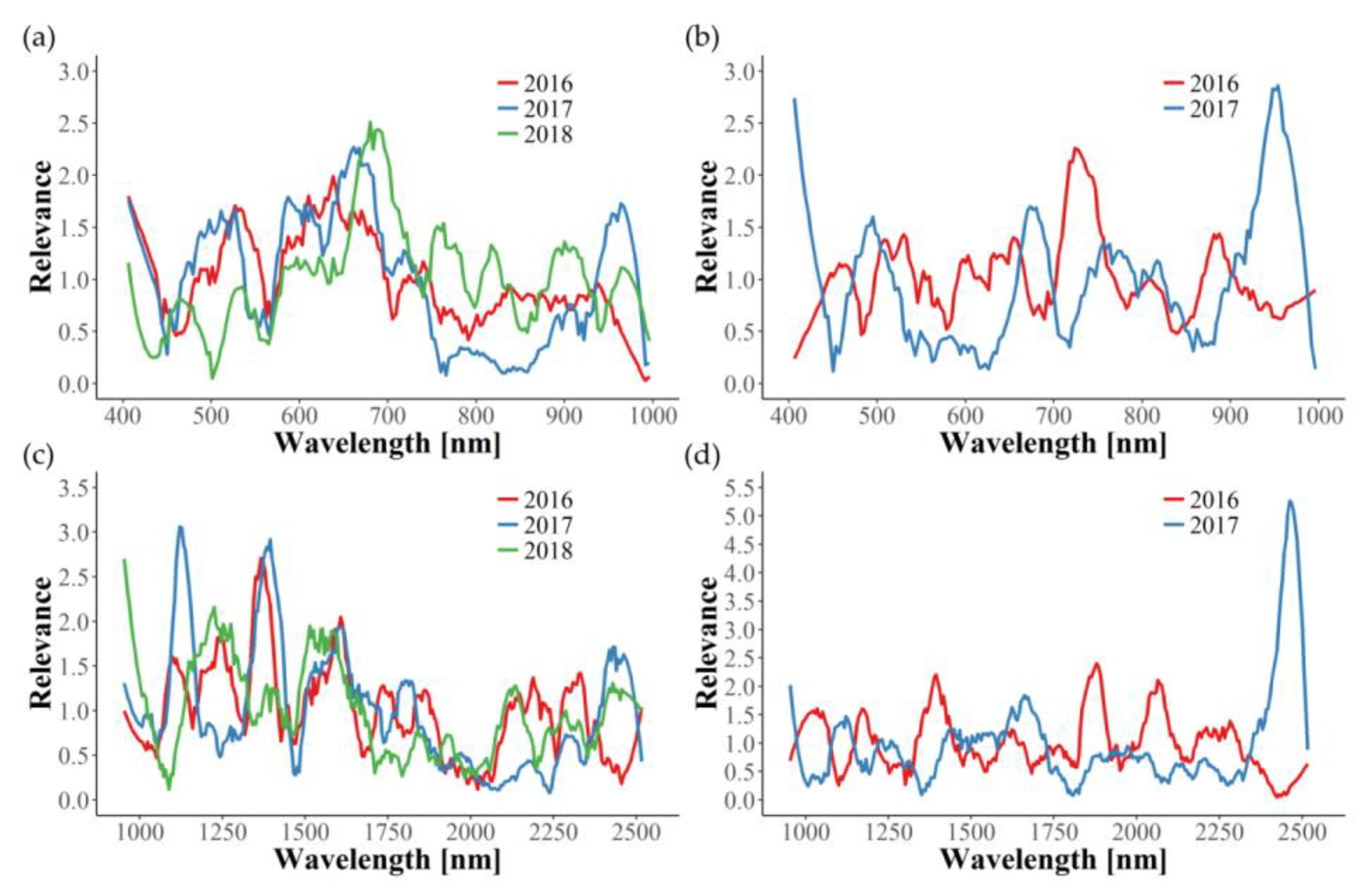
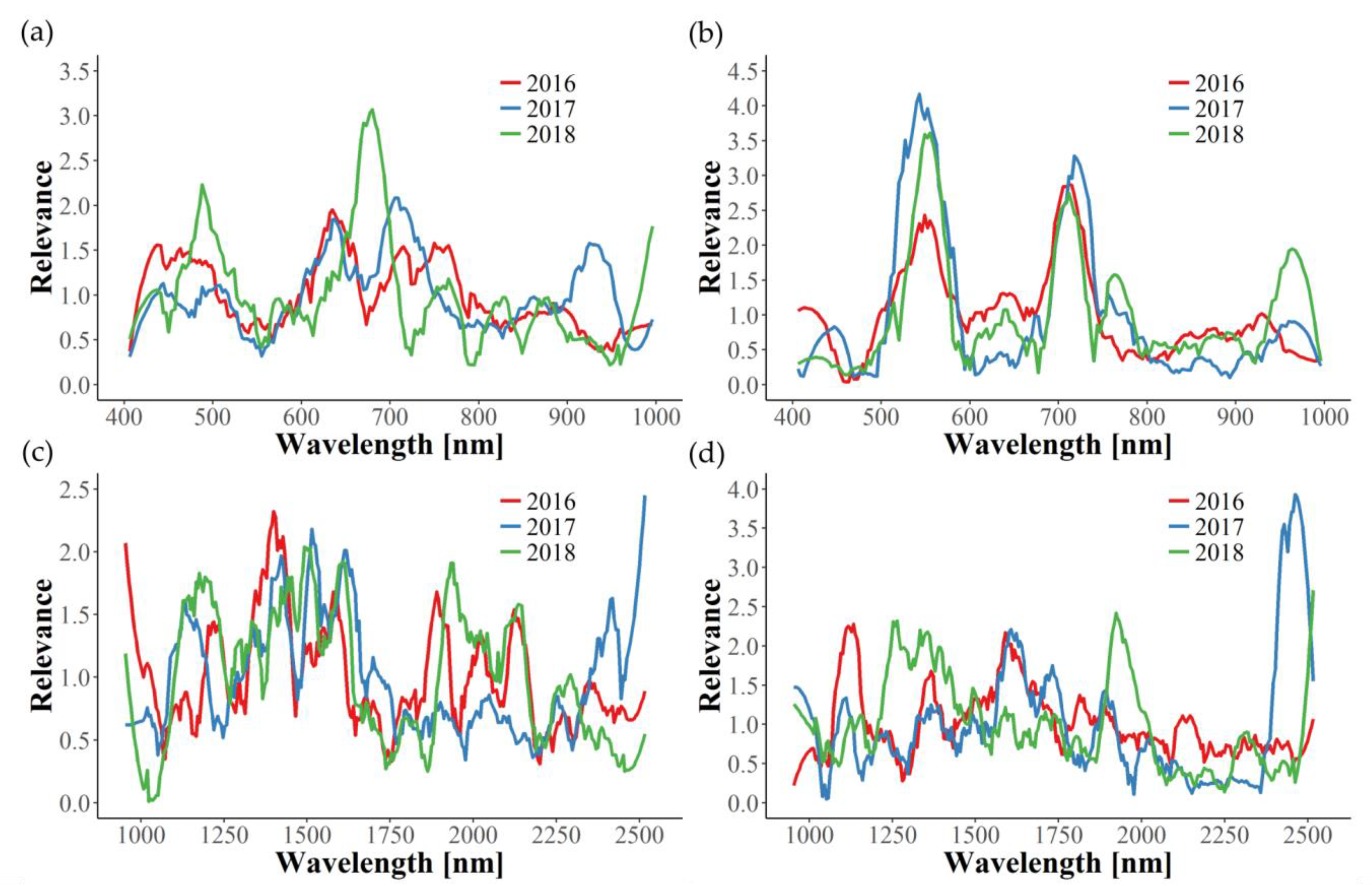
In this study, relevance profiles were calculated for the in-field disease detection, providing information about the importance of wavelengths for the differentiation tasks. In most cases, relevance profiles of the three years examined are not consistent, since many wavelengths seem to be of significance in only one or two years. Apparently, year-dependent environmental conditions interfere with the effects caused by GLD infection, thus hindering the development of one uniform disease detection model. The only exception in this work is the identification of GLRaV-1 in red cultivars using the VNIR wavelength range. Here, similar graphs can be observed for the three years, with wavelengths around 550 and 710 nm being the most important. Naidu et al. [
25] also observed changes in these spectral regions for GLRaV-3 symptomatic Cabernet Sauvignon and Merlot plants. In our work, the relevance of 550 and 710 nm could also be seen for GLRaV-3 infected red cultivars in 2016, but not in 2017. Therefore, further experimental years would be helpful in determining important spectral regions of GLRaV-3 infection. Gitelson et al. [
54] found that reflectance in these two spectral bands (550 ± 15 and 700 ± 7.5 nm) is well suited to estimate leaves’ anthocyanin content non-destructively and, subsequently, introduced the anthocyanin reflectance index (ARI). Thus, our findings can be clearly correlated to the increase of anthocyanin concentration in red cultivars upon GLD symptom development [
47].
Through the identification of application specific wavelengths data dimensionality can be reduced and, as a consequence thereof, computational analysis and processing becomes more efficient. Using optimal spectral bands is a common approach that has successfully been implemented for the detection of three sugar beet diseases [
55], anthracnose on strawberries [
56], yellow rust on winter wheat [
57], and powdery mildew as well as
Flavescence dorée on grapevines [
58,
59]. Moshou et al. [
57] and Yeh et al. [
56] demonstrated that the number of selected spectral bands significantly influences classification, which is in accordance with our results—classification improves as more wavelengths are considered and is comparable to using the whole spectral range. This was also reported by Wang et al. [
20] who identified eight wavelengths that achieved the same plant-level accuracies as the entire spectrum for the detection of tomato spotted wilt virus. Although most relevance profiles in our study vary between the three years, classification within one year is always successful when using the identified spectral bands. Al-Saddik et al. [
60] developed spectral disease indices (SDIs) for the detection of
Flavescence dorée, but their best wavelengths selected were different from one case to another and, therefore, no universal SDI was found to be applicable. Moreover, Sinha et al. [
26] tried to transfer relevant wavelengths from one year to another for identification of GLRaV-3 in Cabernet Sauvignon vines, but results were very divergent and transfer was only partially satisfying.
However, symptom detection is only the first step in disease management. There appears to be a delay of at least one growing season between virus inoculation and symptom manifestation [
61], and given the fact that some cultivars, rootstocks and most wild
Vitis species do not exhibit symptoms at all [
4], identification of symptomless carriers is crucial to reduce pathogen reservoirs in vineyards. GLRaVs are known to affect primary and secondary metabolism in asymptomatic plants leading to alterations in the composition of leaf biochemicals and subsequently causing changes in reflectance spectra [
62,
63]. The detection of asymptomatic plants was successful under greenhouse conditions even though only few plants could be used for modeling. However, in-field identification was challenging in both VNIR and SWIR for GLRaV-1 and -3, respectively. Naidu et al. [
25] also used leaves from field-grown vines to detect GLRaV-3 before developing symptoms and gained significantly lower classification results (max. 75%) in comparison to symptomatic leaves. A possible explanation is that GLRaVs are unevenly distributed within grapevines and virus titer differs in the course of a growing season [
64,
65,
66]. In contrast to field-grown grapevines, greenhouse plants consisted of only one shoot, which could lead to higher virus concentrations and better distribution, resulting in satisfying CAs. In general, detection of asymptomatic vines would not only be helpful for winegrowers in commercial vineyards but could also be a useful tool for nurseries as it might improve their ability to provide virus-free plant material to growers. Although the presented system is applicable to mother vineyards, further efforts are required to optimize classification accuracies for better in-field detection of infected, but symptomless, mother plants. Moreover, field nurseries could be screened, but here too, further improvements and a different experimental setup would be necessary as the plants are rather small and densely planted. However, young grafted plants could be analyzed after removing them from callusing boxes since most of them developed several leaves or even shoots by then [
67]. Spectra of these plants could be assessed in a similar approach as was presented in this study for greenhouse plants that showed promising results with regard to asymptomatic disease detection.