Modelling Site Index in Forest Stands Using Airborne Hyperspectral Imagery and Bi-Temporal Laser Scanner Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Selection of Field Plots
2.3. Field Measurements
2.3.1. Plot Positioning
2.3.2. Tree Measurements
2.3.3. Registration of Field Reference Site Index
2.4. Airborne Laser Scanner Data
Calculation of ALS Variables
2.5. Hyperspectral Data
2.5.1. Preprocessing (Atmospheric Correction and Normalization)
2.5.2. Selection of Pixels
2.6. Datasets and Analyses
2.6.1. Screening Variable Importance Using Partial Least Squares Regression
2.6.2. Modelling H40 Site Index Using Bi-Temporal ALS Data
2.6.3. Modelling H40 Site Index Using Hyperspectral Data and Combined Data
3. Results
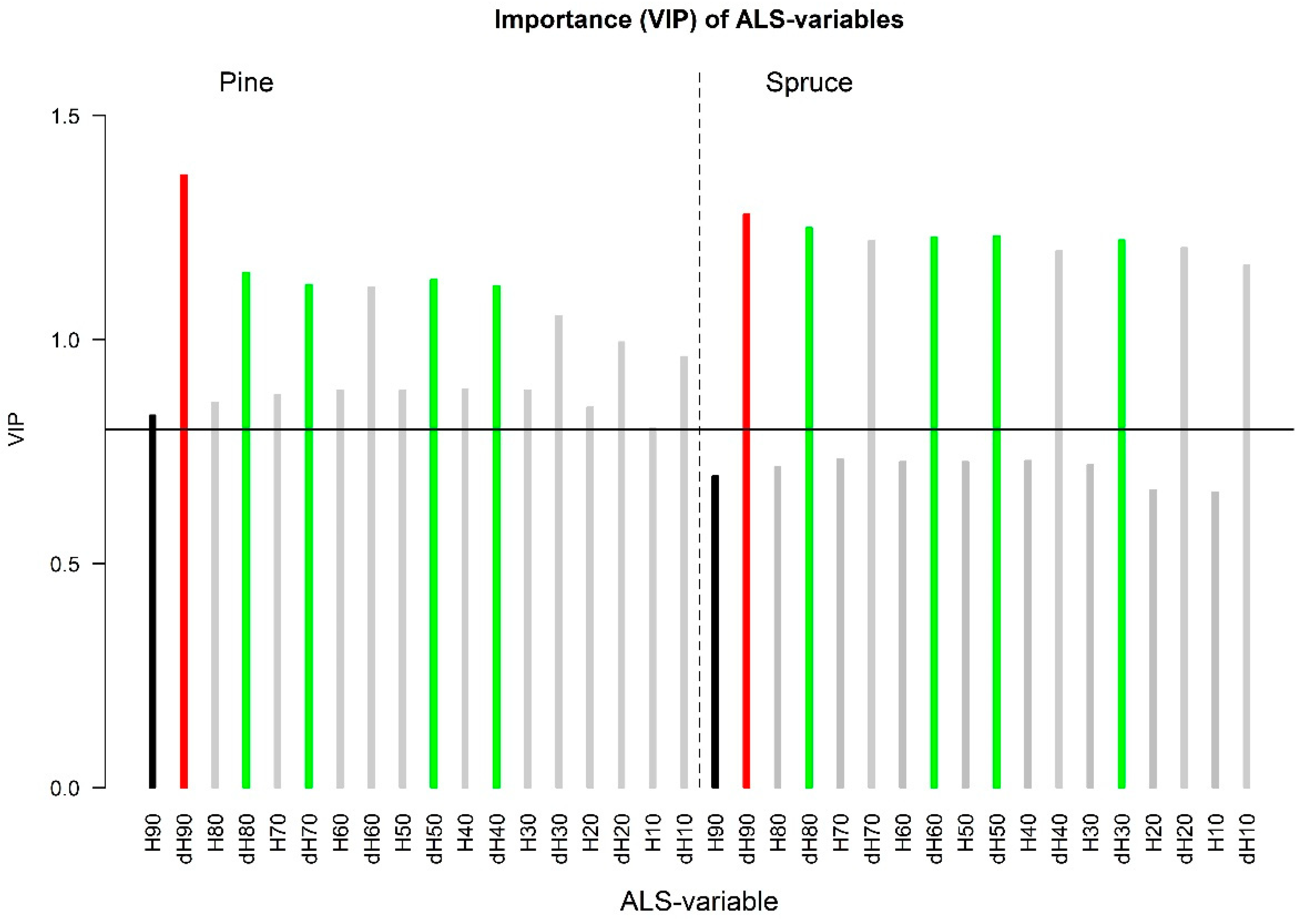
3.1. Screening of Variable Importance
3.1.1. ALS Data
3.1.2. Hyperspectral Data
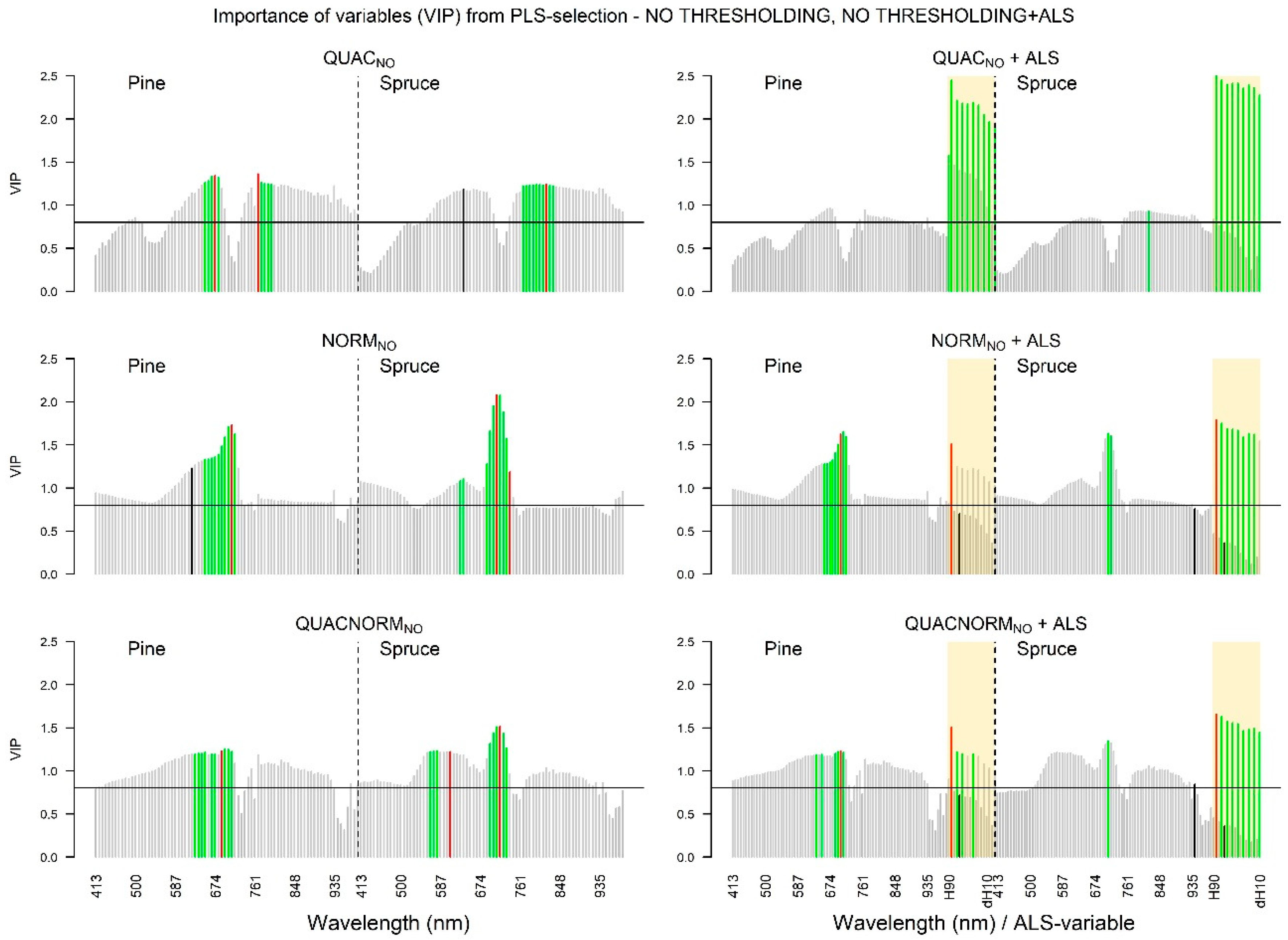
3.1.3. VIP Values: NDVI Selection Threshold
3.1.4. VIP Values: NO Selection Threshold
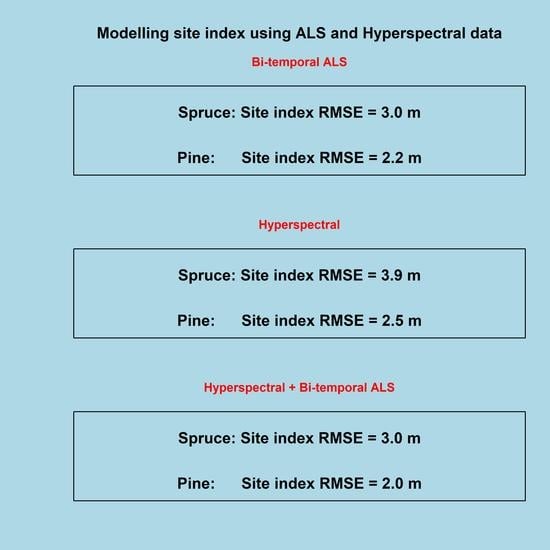
3.1.5. The Relative Effect of Introducing ALS Data
3.2. Modelling
3.2.1. ALS Data
3.2.2. Hyperspectral Data: NDVI Selection Threshold
3.2.3. Hyperspectral Data: NO Selection Threshold
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Skovsgaard, J.P.; Vanclay, J.K. Forest site productivity: A review of spatial and temporal variability in natural site conditions. For. Int. J. For. Res. 2013, 86, 305–315. [Google Scholar] [CrossRef]
- Pretzsch, H.; Grote, R.; Reineking, B.; Rötzer, T.; Seifert, S. Models for forest ecosystem management: A european perspective. Ann. Bot. 2008, 101, 1065–1087. [Google Scholar] [CrossRef] [PubMed]
- Rivas, J.J.C.; González, J.G.Á.; Aguirre, O.; Hernández, F.J. The effect of competition on individual tree basal area growth in mature stands of pinus cooperi blanco in durango (mexico). Eur. J. For. Res. 2005, 124, 133–142. [Google Scholar] [CrossRef]
- Bollandsås, O.M.; Næsset, E. Weibull models for single-tree increment of norway spruce, scots pine, birch and other broadleaves in norway. Scand. J. For. Res. 2009, 24, 54–66. [Google Scholar] [CrossRef]
- Hasenauer, H.; Monserud, R.A. Biased predictions for tree height increment models developed from smoothed ‘data’. Ecol. Model. 1997, 98, 13–22. [Google Scholar] [CrossRef]
- Hasenauer, H.; Monserud, R.A.; Gregoire, T.G. Using simultaneous regression techniques with individual-tree growth models. For. Sci. 1998, 44, 87–95. [Google Scholar]
- Sharma, R.P.; Brunner, A. Modeling individual tree height growth of norway spruce and scots pine from national forest inventory data in norway. Scand. J. For. Res. 2017, 32, 501–514. [Google Scholar] [CrossRef]
- Sharma, R.P.; Brunner, A.; Eid, T.; Øyen, B.-H. Modelling dominant height growth from national forest inventory individual tree data with short time series and large age errors. For. Ecol. Manag. 2011, 262, 2162–2175. [Google Scholar] [CrossRef]
- Pienaar, L.V.; Rheney, J.W. Modeling stand level growth and yield response to silvicultural treatments. For. Sci. 1995, 41, 629–638. [Google Scholar]
- Boisvenue, C.; Running, S.W. Impacts of climate change on natural forest productivity—Evidence since the middle of the 20th century. Glob. Chang. Biol. 2006, 12, 862–882. [Google Scholar] [CrossRef]
- Sharma, R.P.; Brunner, A.; Eid, T. Site index prediction from site and climate variables for norway spruce and scots pine in norway. Scand. J. For. Res. 2012, 27, 619–636. [Google Scholar] [CrossRef]
- Skovsgaard, J.P.; Vanclay, J.K. Forest site productivity: A review of the evolution of dendrometric concepts for even-aged stands. For. Int. J. For. Res. 2008, 81, 13–31. [Google Scholar] [CrossRef]
- Tveite, B. Bonitetskurver for Gran; Norsk Institutt for Skogforskning: Ås, Norway, 1977; pp. 1–84. [Google Scholar]
- Tveite, B.; Braastad, H. Bonitering for gran, furu og bjørk. Nor. Skogbr. 1981, 27, 17–22. [Google Scholar]
- Eid, T. Use of uncertain inventory data in forestry scenario models and consequential incorrect harvest decisions. Silva Fenn. 2000, 34, 89–100. [Google Scholar] [CrossRef]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Næsset, E. Area-based inventory in norway—From innovation to an operational reality. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 215–240. [Google Scholar]
- Gatziolis, D.; Fried, J.S.; Monleon, V.S. Challenges to estimating tree height via lidar in closed-canopy forests: A parable from western oregon. For. Sci. 2010, 56, 139–155. [Google Scholar]
- Packalén, P.; Mehtätalo, L.; Maltamo, M. Als-based estimation of plot volume and site index in a eucalyptus plantation with a nonlinear mixed-effect model that accounts for the clone effect. Ann. For. Sci. 2011, 68, 1085. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, X. Site quality assessment of a pinus radiata plantation in victoria, australia, using lidar technology. South. For. J. For. Sci. 2012, 74, 217–227. [Google Scholar] [CrossRef]
- Tompalski, P.; Coops, N.C.; White, J.C.; Wulder, M.A.; Pickell, P.D. Estimating forest site productivity using airborne laser scanning data and landsat time series. Can. J. Remote Sens. 2015, 41, 232–245. [Google Scholar] [CrossRef]
- Kandare, K.; Ørka, H.O.; Dalponte, M.; Næsset, E.; Gobakken, T. Individual tree crown approach for predicting site index in boreal forests using airborne laser scanning and hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2017, 60, 72–82. [Google Scholar] [CrossRef]
- Holopainen, M.; Vastaranta, M.; Yu, X.; Haapanen, R.; Hyyppä, J.; Kaartinen, H.; Viitala, R.; Hyyppä, H. Site type estimation using airborne laser scanning and stand register data. Photogramm. J. Finl. 2010, 22, 16–32. [Google Scholar]
- Vehmas, M.; Eerikäinen, K.; Peuhkurinen, J.; Packalén, P.; Maltamo, M. Airborne laser scanning for the site type identification of mature boreal forest stands. Remote Sens. 2011, 3, 100–116. [Google Scholar] [CrossRef]
- Næsset, E. Practical large-scale forest stand inventory using a small-footprint airborne scanning laser. Scand. J. For. Res. 2004, 19, 164–179. [Google Scholar] [CrossRef]
- Eid, T. Standwise Control of Forest Management Planning Data in Cutting Class III–V; Meddelelser fra Skogforskningen: Ås, Norway, 1992; pp. 1–78. [Google Scholar]
- Eid, T. Kontroll av Skogbruksplandata fra “Understøttet Fototakst”; Aktuelt fra Skogforsk: Ås, Norway, 1996; pp. 1–21. [Google Scholar]
- Eid, T.; Nersten, S. Sammenligning av skogbruksplandata og kontrolldata. In Problemer Omkring Registreringer og Planlegging for en Skogeiendom i Birkenes Kommune; Eid, T., Ed.; Skogforsk: Ås, Norway, 1996; Volume 47, pp. 1–34. [Google Scholar]
- St-Onge, B.; Vepakomma, U. Assessing forest gap dynamics and growth using multi-temporal laser-scanner data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2004, 140, 173–178. [Google Scholar]
- Bollandsås, O.M.; Gregoire, T.G.; Næsset, E.; Øyen, B.-H. Detection of biomass change in a norwegian mountain forest area using small footprint airborne laser scanner data. Stat. Methods Appl. 2013, 22, 113–129. [Google Scholar] [CrossRef]
- Ene, L.T.; Næsset, E.; Gobakken, T.; Bollandsås, O.M.; Mauya, E.W.; Zahabu, E. Large-scale estimation of change in aboveground biomass in miombo woodlands using airborne laser scanning and national forest inventory data. Remote Sens. Environ. 2017, 188, 106–117. [Google Scholar] [CrossRef]
- Næsset, E.; Bollandsås, O.M.; Gobakken, T.; Gregoire, T.G.; Ståhl, G. Model-assisted estimation of change in forest biomass over an 11year period in a sample survey supported by airborne lidar: A case study with post-stratification to provide “activity data”. Remote Sens. Environ. 2013, 128, 299–314. [Google Scholar] [CrossRef]
- Strîmbu, V.F.; Ene, L.T.; Gobakken, T.; Gregoire, T.G.; Astrup, R.; Næsset, E. Post-stratified change estimation for large-area forest biomass using repeated als strip sampling. Can. J. For. Res. 2017, 47, 839–847. [Google Scholar] [CrossRef]
- Hyyppä, J.; Yu, X.; Rönnholm, P.; Kaartinen, H.; Hyyppä, H. Factors affecting laser-derived object-oriented forest height growth estimation. Photogramm. J. Finl. 2003, 18, 16–31. [Google Scholar]
- Næsset, E.; Gobakken, T. Estimating forest growth using canopy metrics derived from airborne laser scanner data. Remote Sens. Environ. 2005, 96, 453–465. [Google Scholar] [CrossRef]
- Hollaus, M.; Eysn, L.; Maier, B.; Pfeifer, N. Site index assessment based on multi-temporal als data. In Proceedings of the Silvilaser 2015, La Grande Motte, France, 28–30 September 2015. [Google Scholar]
- Kvaalen, H.; Solberg, S.; May, J. Aldersuavhengig Bonitering Med Laserscanning av Enkelttrær; Rapport fra NIBIO: Ås, Norway, 2015; pp. 1–31. [Google Scholar]
- Gobakken, T.; Næsset, E. Assessing effects of laser point density, ground sampling intensity, and field sample plot size on biophysical stand properties derived from airborne laser scanner data. Can. J. For. Res. 2008, 38, 1095–1109. [Google Scholar] [CrossRef]
- Noordermeer, L.; Bollandsås, O.M.; Gobakken, T.; Næsset, E. Direct and indirect site index determination for norway spruce and scots pine using bitemporal airborne laser scanner data. For. Ecol. Manag. 2018, 428, 104–114. [Google Scholar] [CrossRef]
- Baltsavias, E.; Gruen, A.; Eisenbeiss, H.; Zhang, L.; Waser, L.T. High-quality image matching and automated generation of 3d tree models. Int. J. Remote Sens. 2008, 29, 1243–1259. [Google Scholar] [CrossRef]
- Véga, C.; St-Onge, B. Mapping site index and age by linking a time series of canopy height models with growth curves. For. Ecol. Manag. 2009, 257, 951–959. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the southern alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and lidar data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Ørka, H.O.; Dalponte, M.; Gobakken, T.; Næsset, E.; Ene, L.T. Characterizing forest species composition using multiple remote sensing data sources and inventory approaches. Scand. J. For. Res. 2013, 28, 677–688. [Google Scholar] [CrossRef]
- Smith, M.; Martin, M.E.; Plourde, L.; Ollinger, S.V. Analysis of hyperspectral data for estimation of temperate forest canopy nitrogen concentration: Comparison between an airborne (aviris) and a spaceborne (hyperion) sensor. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1332–1337. [Google Scholar] [CrossRef]
- Smith, M.-L.; Ollinger, S.V.; Martin, M.E.; Aber, J.D.; Hallett, R.A.; Goodale, C.L. Direct estimation of aboveground forest productivity through hyperspectral remote sensing of canopy nitrogen. Ecol. Appl. 2002, 12, 1286–1302. [Google Scholar] [CrossRef]
- Eid, T.; Tuhus, E. Models for individual tree mortality in norway. For. Ecol. Manag. 2001, 154, 69–84. [Google Scholar] [CrossRef]
- Næsset, E. Determination of mean tree height of forest stands by digital photogrammetry. Scand. J. For. Res. 2002, 17, 446–459. [Google Scholar] [CrossRef]
- Anon. Terrascan User’s Guide; Terrasolid Ltd.: Jyvaskyla, Finland, 2016. [Google Scholar]
- Axelsson, P. Dem generation from laser scanner data using adaptive tin models. Int. Arch. Photogramm. Remote Sens. 2000, 33, 110–117. [Google Scholar]
- Cao, F.; Yang, Z.; Ren, J.; Jiang, M.; Ling, W.-K. Does Normalization Methods Play a Role for Hyperspectral Image Classification? 2017. Available online: https://arxiv.org/abs/1710.02939 (accessed on 29 April 2019).
- Bernstein, L.S.; Adler-Golden, S.M.; Sundberg, R.L.; Levine, R.Y.; Perkins, T.C.; Berk, A.; Ratkowski, A.J.; Felde, G.; Hoke, M.L. A new method for atmospheric correction and aerosol optical property retrieval for vis-swir multi- and hyperspectral imaging sensors: Quac (quick atmospheric correction). In Proceedings of the 2005 IEEE International Geoscience and Remote Sensing Symposium, IGARSS ’05, Seoul, Korea, 29 July 2005; pp. 3549–3552. [Google Scholar]
- Rani, N.; Mandla, V.R.; Singh, T. Evaluation of atmospheric corrections on hyperspectral data with special reference to mineral mapping. Geosci. Front. 2017, 8, 797–808. [Google Scholar] [CrossRef]
- Yu, B.; Ostland, M.; Gong, P.; Pu, R. Penalized discriminant analysis of in situ hyperspectral data for conifer species recognition. IEEE Trans. Geosci. Remote Sens. 1999, 37, 2569–2577. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Ene, L.T.; Gobakken, T.; Næsset, E. Tree crown delineation and tree species classification in boreal forests using hyperspectral and als data. Remote Sens. Environ. 2014, 140, 306–317. [Google Scholar] [CrossRef]
- Martens, H.; Martens, M. Multivariate Analysis of Quality: An Introduction; John Wiley & Sons: Hoboken, NJ, USA, 2000. [Google Scholar]
- Wold, S.; Sjöström, M.; Eriksson, L. Pls-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Wold, S. Pls for multivariate linear modeling. In Chemometric Methods in Molecular Design; Waterbeemd, H.V.D., Ed.; VCH Verlagsgesellschaft mbH: Weinheim, Germany, 1994; pp. 195–218. [Google Scholar]
- McRoberts, R.E.; Næsset, E.; Gobakken, T.; Bollandsås, O.M. Indirect and direct estimation of forest biomass change using forest inventory and airborne laser scanning data. Remote Sens. Environ. 2015, 164, 36–42. [Google Scholar] [CrossRef]
- Anon. Sas® 9.4 Statements: Reference; SAS Institute Inc.: Cary, NC, USA, 2016. [Google Scholar]
- Montgomery, D.; Peck, E.; Vining, G. Introduction to Linear Regression Analysis; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Stephens, M.A. Edf statistics for goodness of fit and some comparisons. J. Am. Stat. Assoc. 1974, 69, 730–737. [Google Scholar] [CrossRef]
- Breusch, T.S.; Pagan, A.R. A simple test for heteroscedasticity and random coefficient variation. Econometrica 1979, 47, 1287–1294. [Google Scholar] [CrossRef]
- Bolton, D.K.; Coops, N.C.; Wulder, M.A. Measuring forest structure along productivity gradients in the canadian boreal with small-footprint lidar. Environ. Monit. Assess. 2013, 185, 6617–6634. [Google Scholar] [CrossRef]
- Fremstad, E. Vegetasjonstyper i Norge; Norsk Institutt for Naturforskning: Trondheim, Norway, 1997; pp. 1–279. [Google Scholar]
- Rautiainen, M.; Lukeš, P.; Homolová, L.; Hovi, A.; Pisek, J.; Mõttus, M. Spectral properties of coniferous forests: A review of in situ and laboratory measurements. Remote Sens. 2018, 10, 207. [Google Scholar] [CrossRef]
- Reichmuth, A.; Henning, L.; Pinnel, N.; Bachmann, M.; Rogge, D. Early detection of vitality changes of multi-temporal norway spruce laboratory needle measurements—The ring-barking experiment. Remote Sens. 2018, 10, 57. [Google Scholar] [CrossRef]
- Song, C.; Woodcock, C.E.; Seto, K.C.; Lenney, M.P.; Macomber, S.A. Classification and change detection using landsat tm data: When and how to correct atmospheric effects? Remote Sens. Environ. 2001, 75, 230–244. [Google Scholar] [CrossRef]
- Kim, S.-H.; Shin, J.-I.; Yoo, H.-R.; Lee, K.-S. Effect of Atmospheric Correction for the Land Cover Classification Using Hyperspectral Data. Proceedings of Asian Association on Remote Sensing—27th Asian Conference on Remote Sensing, ACRS 2006, Ulaanbaatar, Mongolia, 9–13 October 2006. [Google Scholar]
- Hoffbeck, J.P.; Landgrebe, D.A. Effect of radiance-to-reflectance transformation and atmosphere removal on maximum likelihood classification accuracy of high-dimensional remote sensing data. In Surface and Atmospheric Remote Sensing: Technologies, Data Analysis and Interpretation, Proceedings of the Geoscience and Remote Sensing Symposium (IGARSS ’94), Pasadena, CA, USA, 8–12 August 1994; IEEE: New York, NY, USA, 1994; Volume 4, pp. 2538–2540. [Google Scholar]
- Siebke, K.; Ball, M.C. Non-destructive measurement of chlorophyll b:A ratios and identification of photosynthetic pathways in grasses by reflectance spectroscopy. Funct. Plant Biol. 2009, 36, 857–866. [Google Scholar] [CrossRef]
- Agapiou, A.; Hadjimitsis, D.G.; Alexakis, D.D. Evaluation of broadband and narrowband vegetation indices for the identification of archaeological crop marks. Remote Sens. 2012, 4, 3892. [Google Scholar] [CrossRef]

| Variable | 1999 (n = 65 *) | 2010 (n = 81) | 2013 (n = 81) |
|---|---|---|---|
| SprucePROP | 0.45 (0.0–1.0) | 0.51 (0.0–1.0) | |
| PinePROP | 0.43 (0.0–1.0) | 0.40 (0.0–1.0) | |
| DecidPROP | 0.11 (0.0–0.5) | 0.09 (0.0–0.4) | |
| N (ha−1) | 1624 (350–4600) | 1686 (350–5700) | |
| HDOM (m) | 18.1 (8.3–28.8) | 20.4 (13.6–30.6) | 20.0 (11.8–30.4) |
| Stand age (yrs) | 85 (24–209) | ||
| H40 site index (m) | 14.5 (6.2–25.8) |
| Parameter | 1999 | 2011 |
|---|---|---|
| Dates | 8–9 June | 9 September |
| Instrument | Optech ALTM 1210 | Leica ALS70 |
| Average flying altitude (m above ground level) | 700 | 1500 |
| Flight speed (ms−1) | 71 | 70 |
| Pulse repetition frequency (kHz) | 10 | 181 |
| Scan frequency (Hz) | 21.0 | 37.7 |
| Half scan angle (degrees) | 14.0 | 20 |
| Pulse density on the ground (m−2) | 1.2 | 2.4 |
| Model | Dominant Species | Explanatory Variables | AD Test | BP Test | R2 | RMSE (m) | VIF |
|---|---|---|---|---|---|---|---|
| ALS | Spruce | P901999 dP90 | ns | ns | 0.70 | 3.04 | 1.7 |
| Pine | P901999 dP90 | ns | ns | 0.69 | 2.21 | 3.0 | |
| QUACNDVI | Spruce | 638 nm, 819 nm | ns | ns | 0.48 | 4.01 | 2.1 |
| Pine | 674 nm, 768 nm | ns | ns | 0.59 | 2.49 | 2.3 | |
| NORMNDVI | Spruce | 638 nm, 718 nm | ns | ns | 0.54 | 3.78 | 2.6 |
| Pine | 645 nm, 703 nm | ns | ns | 0.60 | 2.43 | 7.9 | |
| QUACNORMNDVI | Spruce | 710 nm | ns | ns | 0.47 | 3.93 | 1 |
| Pine | 689 nm | ns | ns | 0.56 | 2.48 | 1 | |
| QUACNDVI + ALS | Spruce | - | - | - | |||
| Pine | - | - | - | ||||
| NORMNDVI + ALS | Spruce | dP90, 710 nm | ns | ns | 0.63 | 3.36 | 2.4 |
| Pine | dP90, 710 nm | ns | * | 0.71 | 2.14 | 1.9 | |
| QUACNORMNDVI + ALS | Spruce | dP90, 943 nm | ns | ns | 0.75 | 2.80 | 1 |
| Pine | P701999, dP90, 652 nm | ns | ns | 0.77 | 1.88 | 4.6 | |
| QUACNO | Spruce | 638 nm, 819 nm | ns | ns | 0.47 | 4.04 | 2.0 |
| Pine | 674 nm, 768 nm | ns | ns | 0.56 | 2.59 | 1.9 | |
| NORMNO | Spruce | 710 nm, 739 nm | ns | ns | 0.52 | 3.80 | 1.2 |
| Pine | 623 nm, 710 nm | ns | ns | 0.60 | 2.43 | 1.3 | |
| QUACNORMNO | Spruce | 609 nm, 718 nm | ns | ns | 0.51 | 3.86 | 1.7 |
| Pine | 689 nm | ns | ns | 0.53 | 2.57 | 1 | |
| QUACNO + ALS | Spruce | - | - | - | |||
| Pine | - | - | - | ||||
| NORMNO + ALS | Spruce | P701999 dP90, 943 nm | ns | ns | 0.75 | 2.88 | 1.8 |
| Pine | P701999 dP90, 703 nm | ns | ns | 0.78 | 1.89 | 4.0 | |
| QUACNORMNO + ALS | Spruce | P701999 dP90, 943 nm | ns | ns | 0.76 | 2.86 | 2.6 |
| Pine | P701999 dP90, 703 nm | ns | ns | 0.75 | 1.91 | 4.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bollandsås, O.M.; Ørka, H.O.; Dalponte, M.; Gobakken, T.; Næsset, E. Modelling Site Index in Forest Stands Using Airborne Hyperspectral Imagery and Bi-Temporal Laser Scanner Data. Remote Sens. 2019, 11, 1020. https://doi.org/10.3390/rs11091020
Bollandsås OM, Ørka HO, Dalponte M, Gobakken T, Næsset E. Modelling Site Index in Forest Stands Using Airborne Hyperspectral Imagery and Bi-Temporal Laser Scanner Data. Remote Sensing. 2019; 11(9):1020. https://doi.org/10.3390/rs11091020
Chicago/Turabian StyleBollandsås, Ole Martin, Hans Ole Ørka, Michele Dalponte, Terje Gobakken, and Erik Næsset. 2019. "Modelling Site Index in Forest Stands Using Airborne Hyperspectral Imagery and Bi-Temporal Laser Scanner Data" Remote Sensing 11, no. 9: 1020. https://doi.org/10.3390/rs11091020
APA StyleBollandsås, O. M., Ørka, H. O., Dalponte, M., Gobakken, T., & Næsset, E. (2019). Modelling Site Index in Forest Stands Using Airborne Hyperspectral Imagery and Bi-Temporal Laser Scanner Data. Remote Sensing, 11(9), 1020. https://doi.org/10.3390/rs11091020