Estimating Forest Volume and Biomass and Their Changes Using Random Forests and Remotely Sensed Data
Abstract
1. Introduction
2. Materials and Methods
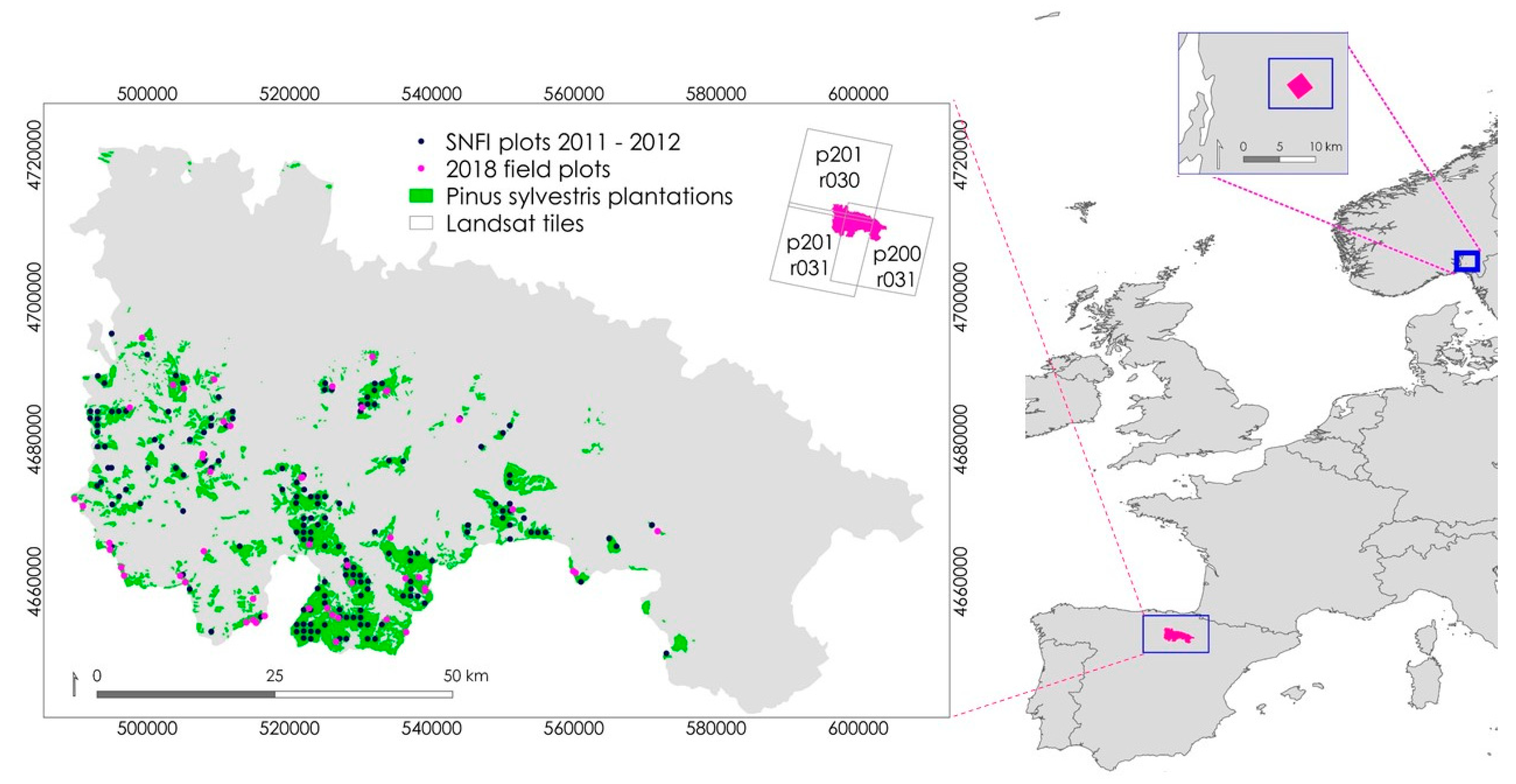
2.1. La Rioja, Spain
2.1.1. La Rioja Sampling Design
2.1.2. Remotely Sensed Data
2.2. Våler, Norway
2.3. Random Forests Prediction Models
2.4. Population Estimates and Inference
2.4.1. Expansion Estimator
2.4.2. Model Assisted Estimator
2.4.3. Model-Based Estimator
2.5. Population Change Estimation
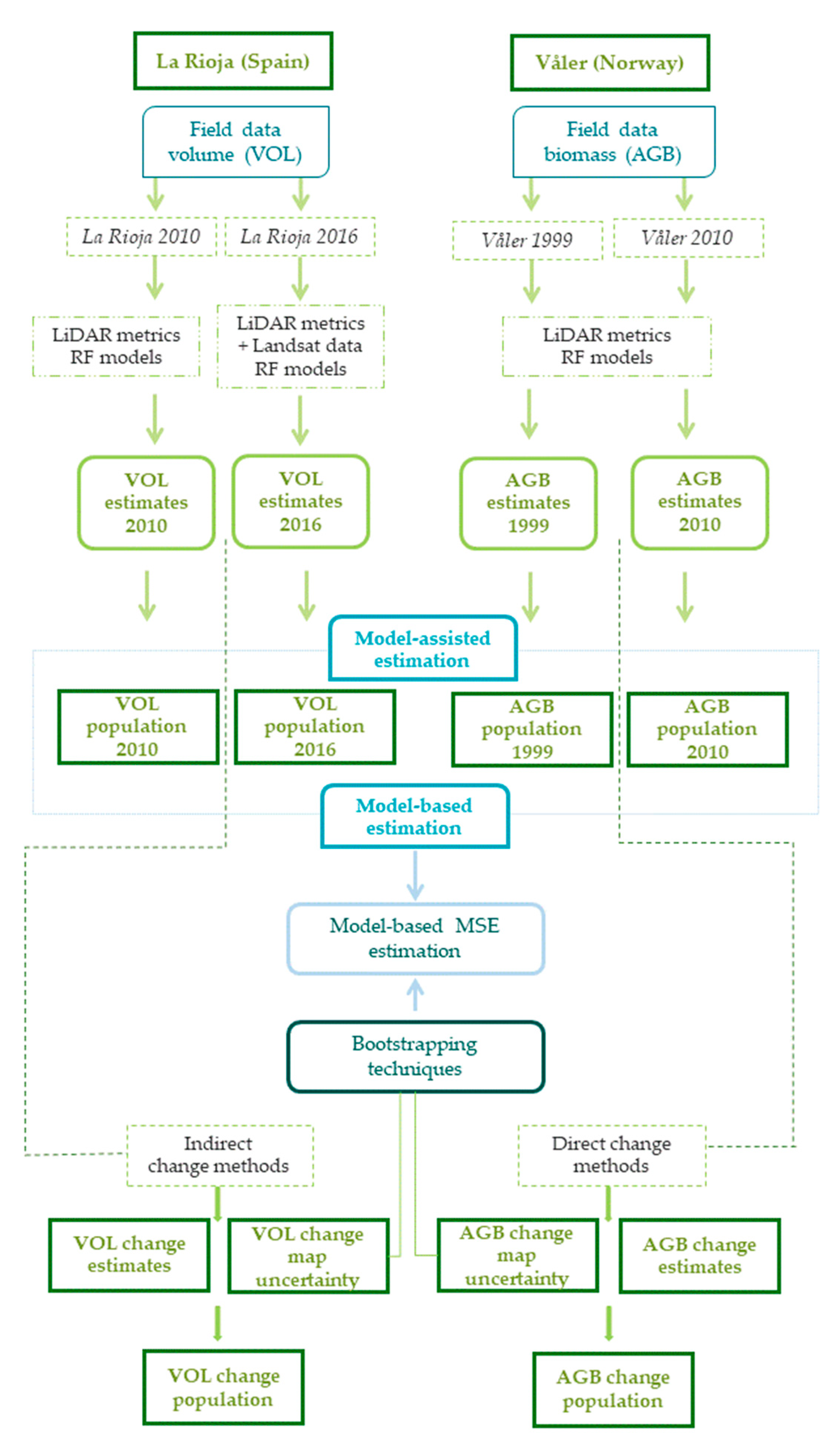
2.6. Overall Workflow
3. Results
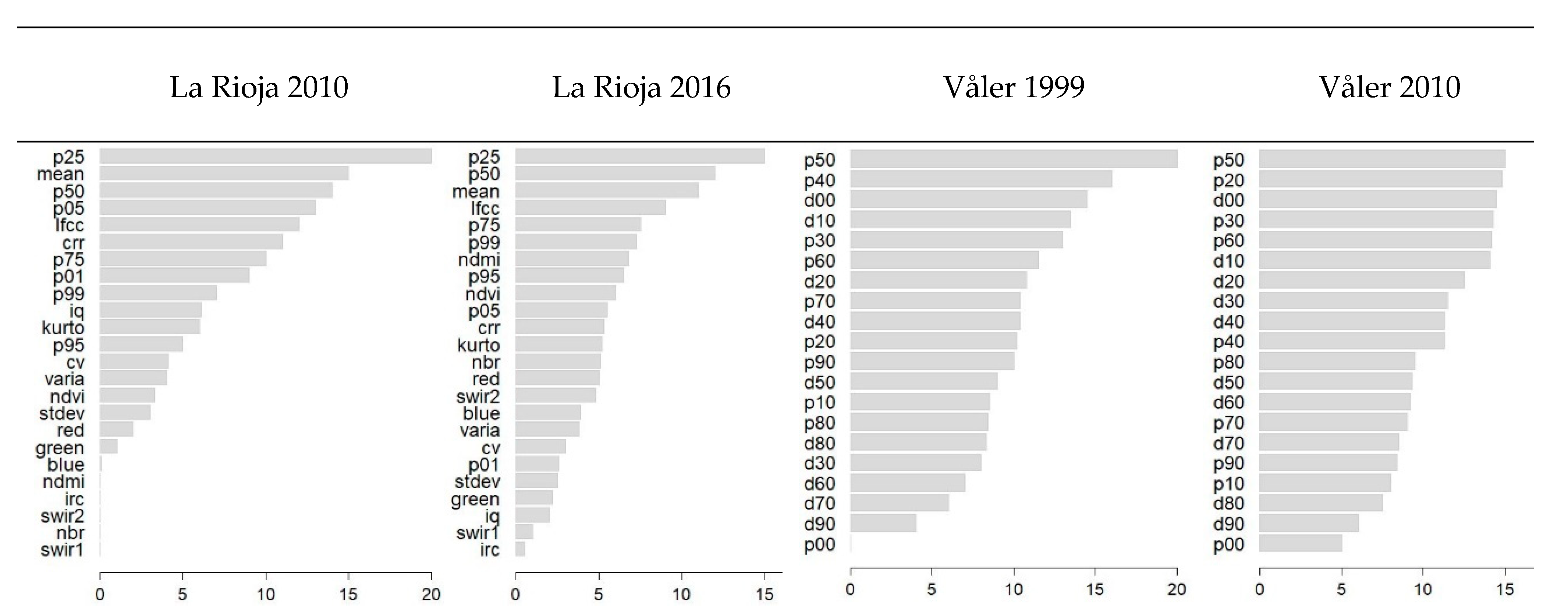
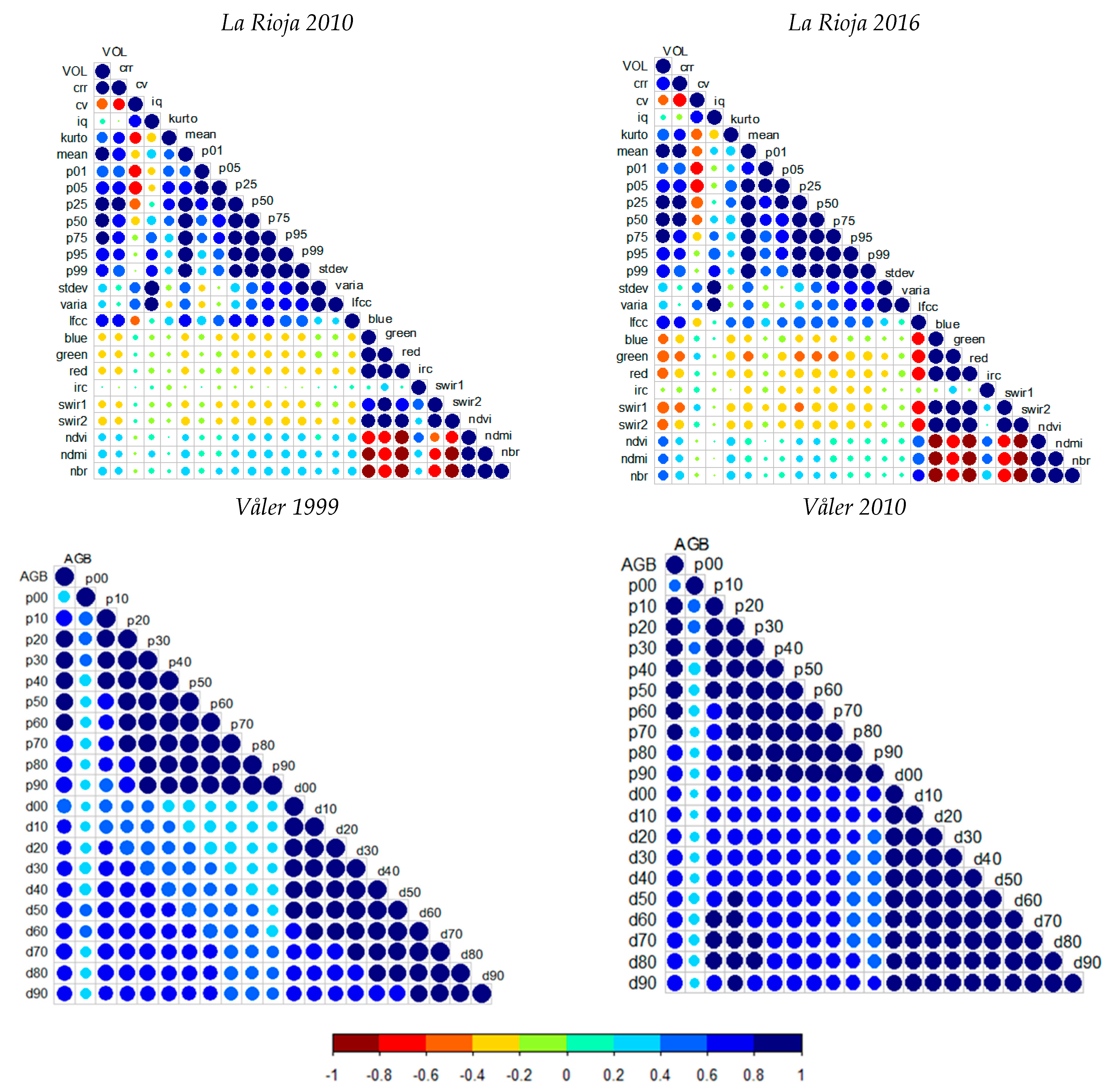
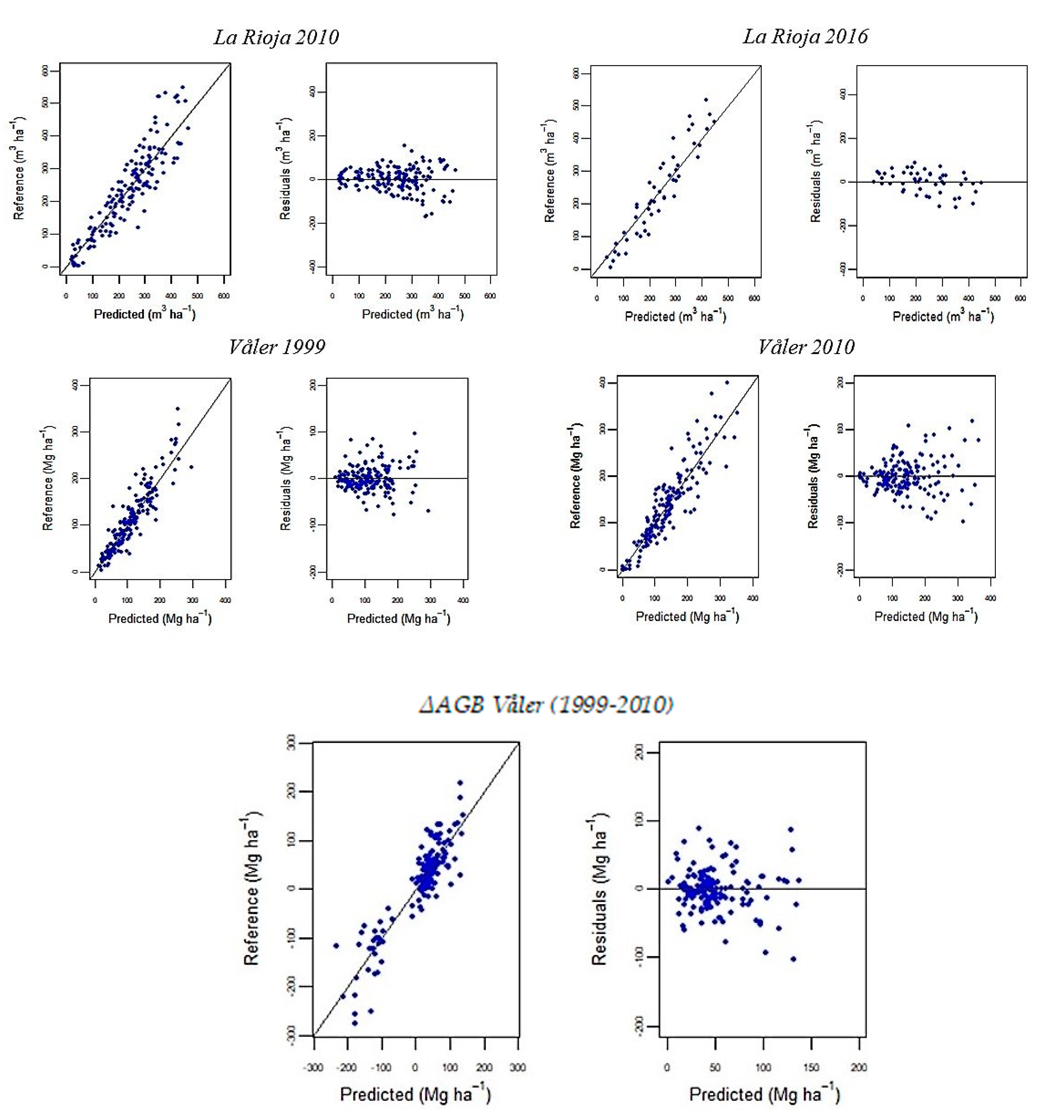
3.1. Random Forests Regression Models
3.2. Estimates of Population Parameters for Each Point in Time
3.3. Estimates of Population Parameters for Change
3.3.1. Estimates of Parameters
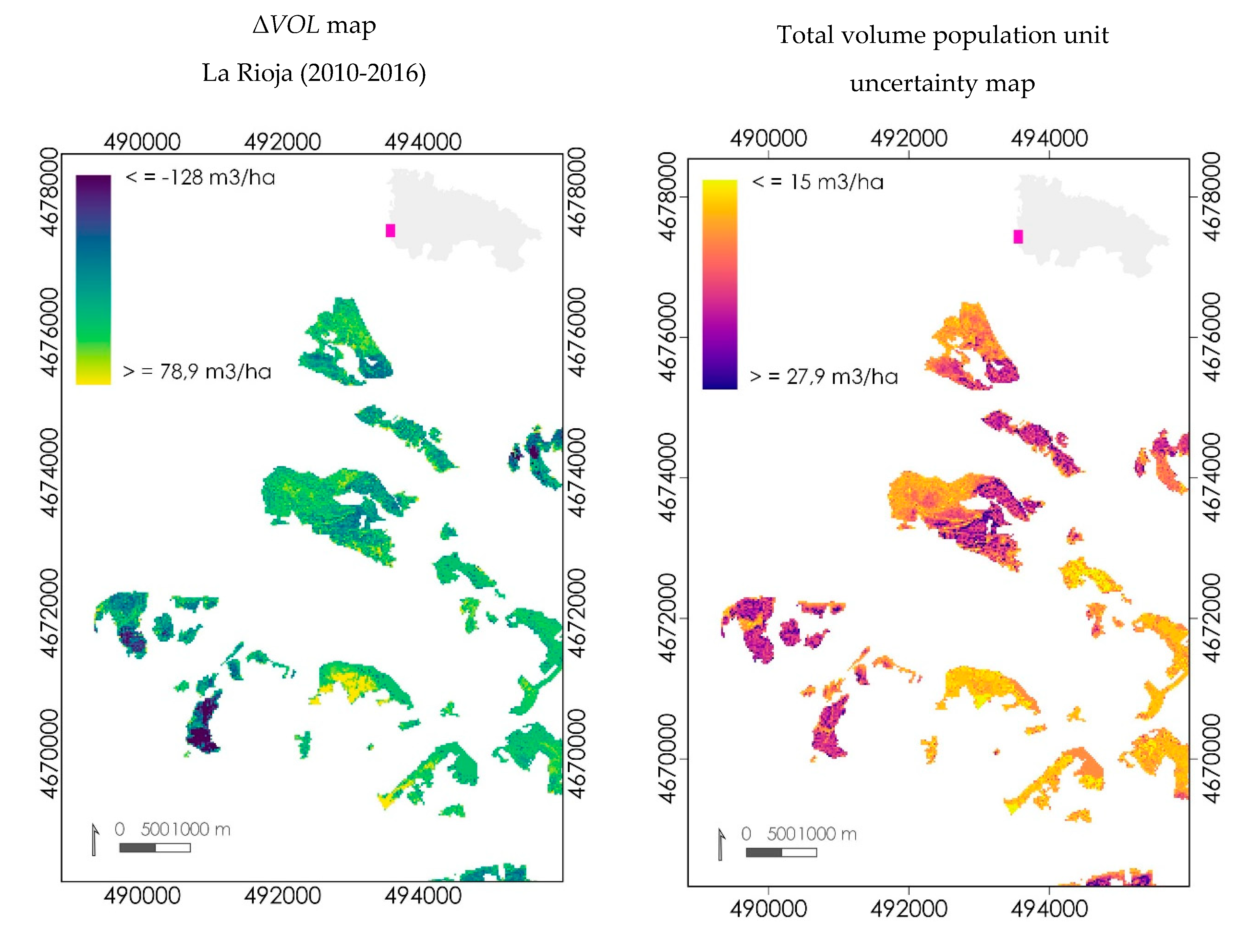
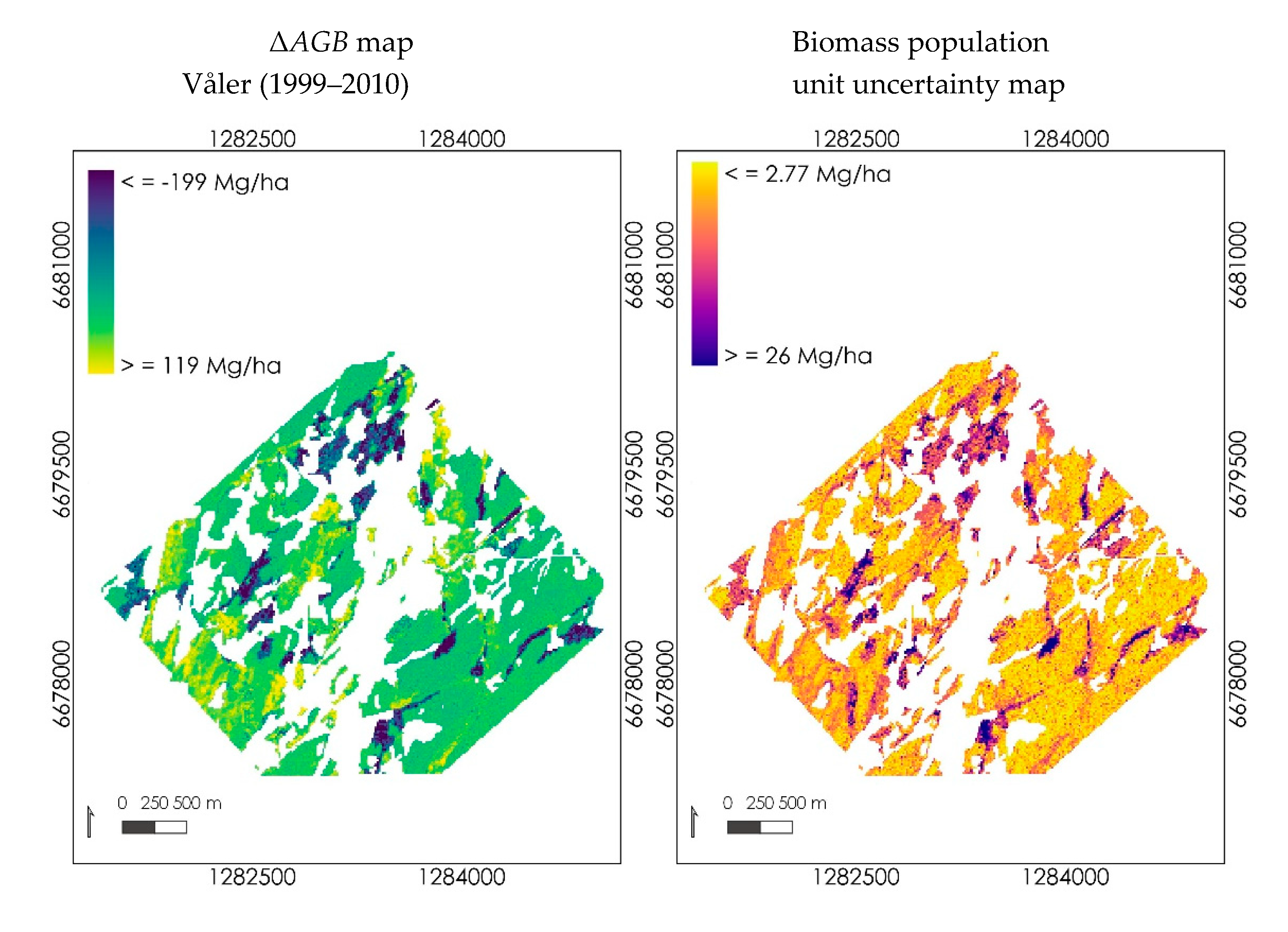
3.3.2. Mapping
4. Discussion
4.1. RF Optimitation: Landsat Variables
4.2. Statistical Inference and Bootstrap Techniques
4.3. Population Change
4.4. Data Considerations
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barrett, F.; McRoberts, R.E.; Tomppo, E.; Cienciala, E.; Waser, L.T. A questionnaire-based review of the operational use of remotely sensed data by national forest inventories. Remote Sens. Environ. 2016, 174, 279–289. [Google Scholar] [CrossRef]
- Kangas, A.; Astrup, R.; Breidenbach, J.; Fridman, J.; Gobakken, T.; Korhonen, K.T.; Maltamo, M.; Nilsson, M.; Nord-Larsen, T.; Næsset, E.; et al. Remote sensing and forest inventories in Nordic countries–roadmap for the future. Scand. J. For. Res. 2018, 33, 397–412. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Tomppo, E.O.; Næsset, E. Advances and emerging issues in national forest inventories. Scand. J. For. Res. 2010, 25, 368–381. [Google Scholar] [CrossRef]
- Tomppo, E.; Gschwantner, T.; Lawrence, M.; McRoberts, R.E. National Forest Inventories: Pathways for Common Reporting; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Maltamo, M.; Næsset, E.; Vauhkonen, J. (Eds.) Forestry Applications of Airborne Laser Scanning. Managed Forest Ecosystems; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Næsset, E. Estimating timber volume of forest stands using airborne laser scanner data. Remote Sens. Environ. 1997, 61, 246–253. [Google Scholar] [CrossRef]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Nӕsset, E. Airborne laser scanning as a method in operational forest inventory: Status of accuracy assessments accomplished in Scandinavia. Scand. J. For. Res. 2007, 22, 433–442. [Google Scholar] [CrossRef]
- Gobakken, T.; Næsset, E. Assessing effects of laser point density, ground sampling intensity, and field sample plot size on biophysical stand properties derived from airborne laser scanner data. Can. J. For. Res. 2008, 38, 1095–1109. [Google Scholar] [CrossRef]
- Holmgren, J. Prediction of tree height, basal area and stem volume in forest stands using airborne laser scanning. Scand. J. For. Res. 2004, 19, 543–553. [Google Scholar] [CrossRef]
- Maltamo, M.; Eerikäinen, K.; Packalén, P.; Hyyppä, J. Estimation of stem volume using laser scanning-based canopy height metrics. Forestry 2006, 79, 217–229. [Google Scholar] [CrossRef]
- Næsset, E.; Bollandsås, O.M.; Gobakken, T.; Gregoire, T.G.; Ståhl, G. Model-assisted estimation of change in forest biomass over an 11year period in a sample survey supported by airborne LiDAR: A case study with post-stratification to provide “activity data”. Remote Sens. Environ. 2013, 128, 299–314. [Google Scholar] [CrossRef]
- Noordermeer, L.; Bollandsås, O.M.; Gobakken, T.; Næsset, E. Direct and indirect site index determination for Norway spruce and Scots pine using bitemporal airborne laser scanner data. For. Ecol. Manag. 2018, 428, 104–114. [Google Scholar] [CrossRef]
- Zhao, K.; Londo, A.; Suarez, J.C.; Hu, T.; Garcia, M.; Wang, C. Utility of multitemporal lidar for forest and carbon monitoring: Tree growth, biomass dynamics, and carbon flux. Remote Sens. Environ. 2018, 204, 883–897. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T.; Bollandsås, O.M. Indirect and direct estimation of forest biomass change using forest inventory and airborne laser scanning data. Remote Sens. Environ. 2015, 164, 36–42. [Google Scholar] [CrossRef]
- Domingo, D.; Alonso, R.; de la Riva, J.; Lamelas, M.T.; Rodríguez, F.; Montealegre, A.L. Temporal Transferability of Pine Forest Attributes Modeling Using Low-Density Airborne Laser Scanning Data. Remote Sens. 2019, 11, 261. [Google Scholar] [CrossRef]
- Saarela, S.; Grafström, A.; Ståhl, G.; Kangas, A.; Holopainen, M.; Tuominen, S.; Nordkvist, K.; Hyyppä, J. Model-assisted estimation of growing stock volume using different combinations of LiDAR and Landsat data as auxiliary information. Remote Sens. Environ. 2015, 158, 431–440. [Google Scholar] [CrossRef]
- Packalén, P.; Suvanto, A.; Maltamo, M. A Two Stage Method to Estimate Species-specific Growing Stock. Photogramm. Eng. Remote Sens. 2009, 75, 1451–1460. [Google Scholar] [CrossRef]
- Ståhl, G.; Saarela, S.; Schnell, S.; Holm, S.; Breidenbach, J.; Healey, S.P.; Patterson, P.L.; Magnussen, S.; Næsset, E.; McRoberts, R.E.; et al. Use of models in large-area forest surveys: Comparing model-assisted, model-based and hybrid estimation. For. Ecosyst. 2016, 3, 5. [Google Scholar] [CrossRef]
- Saarela, S.; Holm, S.; Grafström, A.; Schnell, S.; Næsset, E.; Gregoire, T.G.; Nelson, R.F.; Ståhl, G. Hierarchical model-based inference for forest inventory utilizing three sources of information. Ann. For. Sci. 2016, 73, 895–910. [Google Scholar] [CrossRef]
- Puliti, S.; Saarela, S.; Gobakken, T.; Ståhl, G.; Næsset, E. Combining UAV and Sentinel-2 auxiliary data for forest growing stock volume estimation through hierarchical model-based inference. Remote Sens. Environ. 2018, 204, 485–497. [Google Scholar] [CrossRef]
- Ståhl, G.; Holm, S.; Gregoire, T.G.; Gobakken, T.; Næsset, E.; Nelson, R. Model-based inference for biomass estimation in a LiDAR sample survey in Hedmark County, Norway. Can. J. For. Res. 2011, 41, 83–95. [Google Scholar] [CrossRef]
- Chen, Q.; McRoberts, R.E.; Wang, C.; Radtke, P.J. Forest aboveground biomass mapping and estimation across multiple spatial scales using model-based inference. Remote Sens. Environ. 2016, 184, 350–360. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T. Inference for lidar-assisted estimation of forest growing stock volume. Remote Sens. Environ. 2013, 128, 268–275. [Google Scholar] [CrossRef]
- Andersen, H.-E.; Reutebuch, S.E.; Mcgaughey, R.J.; D’Oliveira, M.V.; Marcus, V.N.; Keller, M. Monitoring selective logging in western Amazonia with repeat lidar flights. Remote Sens. Environ. 2014, 151, 157–165. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T. Estimation for inaccessible and non-sampled forest areas using model-based inference and remotely sensed auxiliary information. Remote Sens. Environ. 2014, 154, 226–233. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Matasci, G.; Hermosilla, T.; Wulder, M.A.; White, J.C.; Coops, N.C.; Hobart, G.W.; Zald, H.S.J. Large-area mapping of Canadian boreal forest cover, height, biomass and other structural attributes using Landsat composites and lidar plots. Remote Sens. Environ. 2018, 209, 90–106. [Google Scholar] [CrossRef]
- Navarro, J.A.; Fernández-Landa, A.; Tomé, J.L.; Guillén-Climent, M.L.; Ojeda, J.C. Testing the quality of forest variable estimation using dense image matching: A comparison with airborne laser scanning in a Mediterranean pine forest. Int. J. Remote Sens. 2018, 39, 4744–4760. [Google Scholar] [CrossRef]
- Condés, S.; McRoberts, R.E. Updating national forest inventory estimates of growing stock volume using hybrid inference. For. Ecol. Manag. 2017, 400, 48–57. [Google Scholar] [CrossRef]
- Fortin, M.; Manso, R.; Schneider, R. Parametric bootstrap estimators for hybrid inference in forest inventories. Forestry 2018, 91, 354–365. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Magnussen, S.; Tomppo, E.O.; Chirici, G. Parametric, bootstrap, and jackknife variance estimators for the k-Nearest Neighbors technique with illustrations using forest inventory and satellite image data. Remote Sens. Environ. 2011, 115, 3165–3174. [Google Scholar] [CrossRef]
- Hou, Z.; McRoberts, R.E.; Ståhl, G.; Packalen, P.; Greenberg, J.A.; Xu, Q. How much can natural resource inventory benefit from finer resolution auxiliary data? Remote Sens. Environ. 2018, 209, 31–40. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T.; Chirici, G.; Condés, S.; Hou, Z.; Saarela, S.; Chen, Q.; Ståhl, G.; Walters, B.F. Assessing components of the model-based mean square error estimator for remote sensing assisted forest applications. Can. J. For. Res. 2018, 48, 642–649. [Google Scholar] [CrossRef]
- Alberdi, I.; Cañellas, I.; Vallejo Bombín, R. The Spanish National Forest Inventory: History, development, challenges and perspectives. Pesqui. Florest. Bras. 2017, 37, 361. [Google Scholar] [CrossRef]
- Álvarez-González, J.G.; Cañellas, I.; Alberdi, I.; Gadow, K.V.; Ruiz-González, A.D. National Forest Inventory and forest observational studies in Spain: Applications to forest modeling. For. Ecol. Manag. 2014, 316, 54–64. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Moser, P.; Zimermann Oliveira, L.; Vibrans, A.C. A general method for assessing the effects of uncertainty in individual-tree volume model predictions on large-area volume estimates with a subtropical forest illustration. Can. J. For. Res. 2014, 45, 44–51. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Westfall, J.A. Propagating uncertainty through individual tree volume model predictions to large-area volume estimates. Ann. For. Sci. 2016, 73, 625–633. [Google Scholar] [CrossRef]
- Valbuena, R.; Mauro, F.; Rodriguez-Solano, R.; Manzanera, J.A. Accuracy and precision of GPS receivers under forest canopies in a mountainous environment. Span. J. Agric. Res. 2013, 8, 1047–1057. [Google Scholar] [CrossRef]
- Mauro, F.; Valbuena, R.; Manzanera, J.A.; García-Abril, A. Influence of global navigation satellite system errors in positioning inventory plots for treeheight distribution studies. Can. J. For. Res. 2011, 41, 11–23. [Google Scholar] [CrossRef]
- Crecente-Campo, F.; Rojo-Alboreca, A.; Diéguez-Aranda, U. A merchantable volume system for Pinus sylvestris L. in the major mountain ranges of Spain. Ann. For. Sci. 2009, 66, 808. [Google Scholar] [CrossRef]
- McGaughey, R.; Forester, R.; Carson, W. Fusing LIDAR data, photographs, and other data using 2D and 3D visualization techniques. Proc. Terrain Data Appl. Vis. Connect. 2003, 28, 16–24. [Google Scholar]
- Vauhkonen, J.; Maltamo, M.; McRoberts, R.E.; Næsset, E. Chapter 1 Introduction to forestry applications of airborne laser scanning. In Forestry Applications of Airborne Laser Scanning, Concepts and Case Studies; Maltamo, M., Næsset, E., Vauhkonen, J., Eds.; Springer: Dordrecht, The Netherlands, 2014; p. 464. [Google Scholar]
- Banskota, A.; Kayastha, N.; Falkowski, M.J.; Wulder, M.A.; Froese, R.E.; White, J.C. Forest Monitoring Using Landsat Time Series Data: A Review. Can. J. Remote Sens. 2014, 40, 362–384. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T. Comparing the stock-change and gain–loss approaches for estimating forest carbon emissions for the aboveground biomass pool. Can. J. For. Res. 2018, 48, 1535–1542. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Royall, R.M.; Herson, J. Robust Estimation in Finite Populations I. J. Am. Stat. Assoc. 1973, 68, 880–889. [Google Scholar] [CrossRef]
- Valliant, R.; Dorfman, A.H.; Royall, R. Finite Population Sampling and Inference; Wiley: New York, NY, USA, 2000; p. 536. [Google Scholar]
- Särndal, C.-E.; Swensson, B.; Wretman, J. Model Assisted Survey Sampling; Springer: New York, NY, USA, 2019; p. 694. [Google Scholar]
- Breidt, F.; Opsomer, J.D. Local Polynomial Regression Estimators in Survey Sampling. Ann. Stat. 2000, 28, 1026–1053. [Google Scholar]
- Breidt, F.; Opsomer, J. Nonparametric and Semiparametric Estimation in Complex Surveys. In Handbook of Statistics—Sample Surveys: Inference and Analysis; Pfeffermann, D., Rao, C.R., Eds.; North-Holland: Amsterdam, The Netherlands, 2009; Volume 28, pp. 103–120. [Google Scholar]
- Breidt, F.J.; Opsomer, J.D. Model-Assisted Survey Estimation with Modern Prediction Techniques. Stat. Sci. 2017, 32, 190–205. [Google Scholar] [CrossRef]
- Lehtonen, R.; Särndal, C.-E.; Veijanen, A. Does the model matter? Comparing model-assisted and model-dependent estimators of class frequencies for domains. Stat. Transit. 2005, 7, 649–673. [Google Scholar]
- Särndal, C.-E. Combined inference in survey sampling. Pak. J. Stat. 2011, 27, 359–370. [Google Scholar]
- Zheng, H.; Little, R. Penalized spline nonparametric mixed models for inference about a finite population mean from two-stage samples. Surv. Methodol. 2004, 30, 209–218. [Google Scholar]
- Liu, R. Bootstrap Procedures under some Non-I.I.D. Models. Ann. Stat. 1988, 16, 1696–1708. [Google Scholar] [CrossRef]
- Flachaire, E. Bootstrapping heteroskedastic regression models: Wild bootstrap vs. pairs bootstrap. Comput. Stat. Data Anal. 2005, 49, 361–376. [Google Scholar] [CrossRef]
- Freedman, D.A. Bootstrapping Regression Models. Ann. Stat. 1981, 6, 1218–1228. [Google Scholar] [CrossRef]
- Diaconis, P.; Efron, B. Computer-Intensive Methods in Statistics. Sci. Am. 1983, 248, 116–130. [Google Scholar] [CrossRef]
- Carpenter, J.; Bithell, J. Bootstrap confidence intervals: When, which, what? A practical guide for medical statisticians. Stat. Med. 2000, 19, 1141–1164. [Google Scholar] [CrossRef]
- Ranalli, M.G.; Mecatti, F. Comparing Recent Approaches For Bootstrapping Sample Survey Data: A First Step Towards A Unified Approach. In Proceedings of the Joint Statistical Meeting (JSM), San Diego, CA, USA, 28 July–2 August 2012. [Google Scholar]
- Mentch, L.; Hooker, G. Quantifying Uncertainty in Random Forests via Confidence Intervals and Hypothesis Tests. J. Mach. Learn. Res. 2016, 17, 1–41. [Google Scholar]
- McRoberts, R.E. Probability- and model-based approaches to inference for proportion forest using satellite imagery as ancillary data. Remote Sens. Environ. 2010, 114, 1017–1025. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O. Evaluating the utility of the medium-spatial resolution Landsat 8 multispectral sensor in quantifying aboveground biomass in uMgeni catchment, South Africa. ISPRS J. Photogramm. Remote Sens. 2015, 101, 36–46. [Google Scholar] [CrossRef]
- Woodall, C.W.; Russell, M.; Andersen, H.-E.; Domke, G.M.; Deo, R.K.K. Evaluating the influence of spatial resolution of Landsat predictors on the accuracy of biomass models for large-area estimation across the eastern USA. Environ. Res. Lett. 2018, 13, 0550004. [Google Scholar]
- Fekety, P.A.; Falkowski, M.J.; Hudak, A.T.; Jain, T.B.; Evans, J.S. Transferability of Lidar-derived Basal Area and Stem Density Models within a Northern Idaho Ecoregion. Can. J. Remote Sens. 2018, 44, 131–143. [Google Scholar] [CrossRef]
- Tompalski, P.; White, J.C.; Coops, N.C.; Wulder, M.A. Demonstrating the transferability of forest inventory attribute models derived using airborne laser scanning data. Remote Sens. Environ. 2019, 227, 110–124. [Google Scholar] [CrossRef]
- Næsset, E. Effects of different sensors, flying altitudes, and pulse repetition frequencies on forest canopy metrics and biophysical stand properties derived from small-footprint airborne laser data. Remote Sens. Environ. 2009, 113, 148–159. [Google Scholar] [CrossRef]
- Holmgren, J.; Nilsson, M.; Olsson, H. Simulating the effects of lidar scanning angle for estimation of mean tree height and canopy closure. Can. J. Remote Sens. 2003, 29, 623–632. [Google Scholar] [CrossRef]
- Montaghi, A. Effect of scanning angle on vegetation metrics derived from a nationwide Airborne Laser Scanning acquisition. Can. J. Remote Sens. 2013, 39, S152–S173. [Google Scholar] [CrossRef]
- Hou, Z.; Xu, Q.; McRoberts, R.E.; Greenberg, J.A.; Liu, J.; Heiskanen, J.; Pitkänen, S.; Packalen, P. Effects of temporally external auxiliary data on model-based inference. Remote Sens. Environ. 2017, 198, 150–159. [Google Scholar] [CrossRef]
- Mauro, F.; Ritchie, M.; Wing, B.; Frank, B.; Monleon, V.; Temesgen, H.; Hudak, A. Estimation of changes of forest structural attributes at three different spatial aggregation levels in Northern California using multitemporal LiDAR. Remote Sens. 2019, 11, 923–954. [Google Scholar] [CrossRef]
- Nӕsset, E.; Gobakken, T.; Bollandsas, O.M.; Gregoire, T.G.; Nelson, R.; Stahl, G. Comparison of precision of biomass estimates in regional field sample surveys and airborne LiDAR-assisted surveys in Hedmark County, Norway. Remote Sens. Environ. 2013, 130, 108–120. [Google Scholar] [CrossRef]
- Økseter, R.; Bollandsås, O.M.; Gobakken, T.; Næsset, E. Modeling and predicting aboveground biomass change in young forest using multi-temporal airborne laser scanner data. Scand. J. For. Res. 2015, 30, 458–469. [Google Scholar] [CrossRef]
- Byrne, J.C.; Strand, E.K.; Eitel, J.U.H.; Martinuzzi, S.; Vierling, L.A.; Falkowski, M.J.; Hudak, A.T. Quantifying aboveground forest carbon pools and fluxes from repeat LiDAR surveys. Remote Sens. Environ. 2012, 123, 25–40. [Google Scholar]
- Tao, X.; Huang, C.; Zhao, F.; Schleeweis, K.; Masek, J.; Liang, S. Mapping forest disturbance intensity in North and South Carolina using annual Landsat observations and field inventory data. Remote Sens. Environ. 2019, 221, 351–362. [Google Scholar] [CrossRef]
- Pflugmacher, D.; Cohen, W.B.; Kennedy, R.E.; Yang, Z. Using Landsat-derived disturbance and recovery history and lidar to map forest biomass dynamics. Remote Sens. Environ. 2014, 151, 124–137. [Google Scholar] [CrossRef]
- Ahmed, O.S.; Franklin, S.E.; Wulder, M.A.; White, J.C. Characterizing stand-level forest canopy cover and height using Landsat time series, samples of airborne LiDAR, and the Random Forest algorithm. ISPRS J. Photogramm. Remote Sens. 2015, 101, 89–101. [Google Scholar] [CrossRef]
- Durante, P.; Martín-Alcón, S.; Gil-Tena, A.; Algeet, N.; Tomé, J.L.; Recuero, L.; Palacios-Orueta, A.; Oyonarte, C. Improving Aboveground Forest Biomass Maps: From High-Resolution to National Scale. Remote Sens. 2019, 11, 795. [Google Scholar] [CrossRef]
| Dataset | Variable | Number of Plots | Statistics | |||
|---|---|---|---|---|---|---|
| Mean | Min | Max | Stdev | |||
| La Rioja 2010 | VOL (m3/ha) | 155 | 234.74 | 2.53 | 548.68 | 127.8 |
| La Rioja 2016 | 49 | 236.53 | 5.21 | 517.15 | 135.39 | |
| Våler 1999 | AGB (Mg/ha) | 176 | 112.4 | 2.23 | 349.12 | 66.13 |
| Våler 2010 | 176 | 131.15 | 0 | 462.17 | 91.83 | |
| Estimate | La Rioja 2010 | La Rioja 2016 | Våler 1999 | Våler 2010 |
|---|---|---|---|---|
| 234.74 * | 236.53 a | 112.40 | 131.15 | |
| 10.26 * | 19.34 a | 5.00 | 6.94 | |
| 197.86 * | 183.26 a | 105.55 | 119.64 | |
| 4.33 * | 7.09 a | 2.04 | 2.6 | |
| 197.22 | 183.45 | 105.83 | 119.47 | |
| 200.15 | 190.41 | 113.45 | 131.35 | |
| 6.14 | 24.88 | 2.96 | 3.34 | |
| 3.5 | 7.44 | 1.92 | 2.25 | |
| 3.03 | 7.87 | 1.82 | 2.05 |
| Dataset | Measure | Auxiliary Variable | |||||||
|---|---|---|---|---|---|---|---|---|---|
| p25 | p99 | crr | lfcc | ||||||
| Sample | Pop | Sample | Pop | Sample | Pop | Sample | Pop | ||
| La Rioja 2010 | Mean | 8.28 | 7.03 | 14.80 | 13.19 | 0.54 | 0.49 | 79.93 | 64.67 |
| Min | 2.40 | 0.00 | 4.51 | 0.00 | 0.25 | 0.00 | 7.79 | 0.00 | |
| Max | 16.62 | 38.75 | 27.26 | 44.72 | 0.76 | 0.88 | 100 | 100 | |
| Range | 14.22 | 38.75 | 22.75 | 44.72 | 0.51 | 0.88 | 92.21 | 100 | |
| p25 | p99 | ndmi | lfcc | ||||||
| Sample | Pop | Sample | Pop | Sample | Pop | Sample | Pop | ||
| La Rioja 2016 | Mean | 9.61 | 7.52 | 16.42 | 14.59 | 0.29 | 0.29 | 69.21 | 65.36 |
| Min | 2.22 | 0.00 | 3.36 | 0.00 | 0.06 | -0.31 | 7.63 | 0.00 | |
| Max | 17.27 | 39.23 | 24.92 | 44.58 | 0.48 | 0.79 | 99.93 | 100 | |
| Range | 15.05 | 39.23 | 21.56 | 44.58 | 0.42 | 1.1 | 92.3 | 100 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esteban, J.; McRoberts, R.E.; Fernández-Landa, A.; Tomé, J.L.; Nӕsset, E. Estimating Forest Volume and Biomass and Their Changes Using Random Forests and Remotely Sensed Data. Remote Sens. 2019, 11, 1944. https://doi.org/10.3390/rs11161944
Esteban J, McRoberts RE, Fernández-Landa A, Tomé JL, Nӕsset E. Estimating Forest Volume and Biomass and Their Changes Using Random Forests and Remotely Sensed Data. Remote Sensing. 2019; 11(16):1944. https://doi.org/10.3390/rs11161944
Chicago/Turabian StyleEsteban, Jessica, Ronald E. McRoberts, Alfredo Fernández-Landa, José Luis Tomé, and Erik Nӕsset. 2019. "Estimating Forest Volume and Biomass and Their Changes Using Random Forests and Remotely Sensed Data" Remote Sensing 11, no. 16: 1944. https://doi.org/10.3390/rs11161944
APA StyleEsteban, J., McRoberts, R. E., Fernández-Landa, A., Tomé, J. L., & Nӕsset, E. (2019). Estimating Forest Volume and Biomass and Their Changes Using Random Forests and Remotely Sensed Data. Remote Sensing, 11(16), 1944. https://doi.org/10.3390/rs11161944






