Abstract
Direct assessment of forest regeneration from remote sensing data is a previously little-explored problem. This is due to several factors which complicate object detection of small trees in the understory. Most existing studies are based on airborne laser scanning (ALS) data, which often has insufficient point densities in the understory forest layers. The present study uses plot-based terrestrial laser scanning (TLS) and the survey design was similar to traditional forest inventory practices. Furthermore, a framework of methods was developed to solve the difficulties of detecting understory trees for quantifying regeneration in temperate montane forest. Regeneration is of special importance in our montane study area, since large parts are declared as protection forest against alpine natural hazards. Close to nature forest structures were tackled by separating 3D tree stem detection from overall tree segmentation. In support, techniques from 3D mathematical morphology, Hough transformation and state-of-the-art machine learning were applied. The methodical framework consisted of four major steps. These were the extraction of the tree stems, the estimation of the stem diameters at breast height (DBH), the image segmentation into individual trees and finally, the separation of two groups of regeneration. All methods were fully automated and utilized volumetric 3D image information which was derived from the original point cloud. The total amount of regeneration was split into established regeneration, consisting of trees with a height > 130 cm in combination with a DBH < 12 cm and unestablished regeneration, consisting of trees with a height < 130 cm. Validation was carried out against field-based expert estimates of percentage ground cover, differentiating seven classes that were similar to those used by forest inventory. The mean absolute error (MAE) of our method for established regeneration was 1.11 classes and for unestablished regeneration only 0.27 classes. Considering the metrical distances between the class centres, the MAE amounted 8.08% for established regeneration and 2.23% for unestablished regeneration.
1. Introduction
Forest regeneration is an important component of forest environments, since it fulfils various functions. These are for example its essential role for vital canopy succession and forest ecosystems, it constitutes habitats for wildlife and ensures stand stability in protection forests against natural hazards [1,2,3,4,5,6,7]. In the context of this study, we refer to regeneration as small trees of a specific maximum size and do not refer to processes or origin. Despite its importance, remote sensing-based approaches for direct detection and quantification of regeneration are underrepresented and less effective compared to monitoring the overstory canopy and mature trees [8,9].
Reasons for this discrepancy are on the one hand the little direct financial value of understory trees [2], but most important are the technical difficulties in surveying small trees occluded by larger trees [10]. Passive optical remote sensing data has been rarely used in this context, since it is limited to explanatory variables from the canopy surface [3]. One of the few examples for using optical imagery is the study of Nuanmu et al., who derived understory vegetation using phenology metrics from a time series of satellite images [10]. The limitations of optical remote sensing are overcome by laser scanning (LiDAR) data, which provides direct information of the 3D forest structure. Most studies which attempted to estimate regeneration or understory use airborne laser scanning (ALS), since it enables the coverage of large and continuous areas [2,3,8,11,12,13,14]. In the technical context of remote sensing methodology, the terms regeneration, juvenile trees and understory trees are often used for similar approaches and objectives.
The general disadvantage of ALS is, that point density decreases in the lower forest layers, since large proportions of the laser beam are reflected while penetrating the upper canopy [14,15]. This led to a predominance of approaches which estimated understory coverage indirectly, for example from statistical characteristics of the reflection distribution. Latifi et al. compared different regression models to derive understory density [8]. They undertook a detailed analysis of a large number of height-based variables. Hill and Broughton estimated the understory from last return reflections while considering an approximated crown depth of the overstory canopy based on height percentiles [13]. Criticism on their approach includes the requirement of seasonal timing for data acquisition with respect to the vegetation period and its dependence on additional ancillary data. Lindberg et al. estimated understory as part of the total vegetation structure [16]. They derived the vegetation volume by subdividing the vertical extent of their test plots into horizontal slices of 10 cm thickness. Their approach took advantage of direct usage of waveform information within the height intervals, but proved to be inaccurate for suppressed trees in heights below 10 m. Morsdorf et al. classified ALS return heights in combination with intensity values, in order to quantify forest height layers [17]. While achieving good results for dominant layers, the ground coverage of lower forest layers was clearly underestimated. Singh et al. determined understory in urban forests by classifying height and intensity features derived from the point distribution [18]. In comparison to other studies they additionally included optical satellite imagery. Their overall success was strongly supported by the sparse structure of the urban forest canopy, the contrasting dense structure of homogeneous understory vegetation as well as the data acquisition in leaf-off state. Wing et al. combined height distribution metrics with intensity data to derive a new explanatory variable for predicting understory vegetation [3]. The forest in their test area was characterized by a distinct height difference between overstory and understory layers which simplified the separation. Korpela et al. also estimated understory tree coverage by height metrics [14]. To increase the number of reflections in lower forest layers, they developed a model for compensating transmission losses of the LiDAR beam.
Relatively few approaches tried to overcome the indirect estimation of understory cover by aiming at direct object detection of the individual understory trees. Caused by the airborne perspective, the success of this task is generally limited, since only a fraction of the laser beams penetrates to the forest ground layers. A transitional solution leading from height distribution analysis to tree object detection was presented by Hamraz et al. [2]. They first separated the point cloud into multiple layers which were afterwards used individually for tree segmentation. Even if this improved the understory tree detection, the sparse point density in lower layers remained the limiting factor. Duncanson et al. distinguished over- and understory forest layers from statistical height distributions on their test site and computed canopy height models (CHM) for each [19]. The results of the subsequent watershed segmentation showed similar drawbacks like the previous approach. The method introduced by Ferraz et al. operated in reverse order [20]. They first clustered individual vegetation objects and then assigned them to forest layers. The same methodical order was followed by Amiri et al. [11], who applied a previously developed two-stage segmentation of forest trees. From the resulting tree objects, the regeneration was selected by a height threshold.
Despite the limitations of ALS for understory tree detection, terrestrial laser scanning (TLS) has not explicitly been explored for this objective so far. Due to its ground-based perspective, it generally provides higher relative reflection densities in lower forest layers. In addition, the higher total point densities, caused by short scan distances and system characteristics, support a more accurate recording of small and suppressed trees. Therefore, it can be assumed that object detection in the understory would be more successful with TLS than with ALS data. A few studies exist which used TLS data to retrieve non-object related information for understory vegetation. Hancock et al. used TLS data to calibrate ALS-derived 3D vegetation structure [21]. Gupta et al. computed point distribution statistics from TLS data to estimate changes in the understory structure [22]. Existing TLS-based studies which focus on stem detection in dense and multilayered forest come closest to the task of understory tree detection. Xia et al. isolated tree stems from a dense and heterogenous forest [23]. However, they only used data from a single scan position and validated only those tree stems that were visible from this single perspective. They did not distinguish between regeneration and mature trees. Brolly et al. mapped tree stems on forest regeneration patches [24]. Because the selected plots contained no mature or overstory trees, a discrimination of regeneration was not carried out. Lin et al. conducted a fundamental study on the technical suitability of mobile laser scanning (MLS) for characterizing understory trees [9]. While the general capability of these systems could be confirmed, there exist specific differences between MLS and TLS.
The present study refers to the research gap of TLS-based quantification of forest regeneration. It overcomes the concept of distinguishing visible height layers in the point cloud and defines regeneration by both stem diameter and crown top height. This is in accordance with the definitions of the Swiss national forest inventory (NFI) and of highly practical value. The test plots in our study area were located in temperate montane forest and mostly contained all-aged trees of several indistinct layers. Since large parts of the study area are declared as protection forest against alpine natural hazards, reliable information about the regeneration is of special local importance [4,5]. This challenging application scenario was approached by constructing a framework of four individual methods which complemented each other.
2. Study Area and Data
2.1. Study Area
For this study two locations in the Swiss Alps were used as study areas. Both were situated in the canton of Grison and comprised the local regions of Herrschaft and Schanfigg. The terrain consisted of mountain slopes, gullies and hilly plains, at altitudes between 1000 and 1900 m above sea level. The timberline in this region is at an altitude around 2000 m and the study area mainly contained temperate montane forest. Large parts of the forest are declared as protection forest against alpine natural hazards. To preserve the protective function of the forest over long and consistent periods of time, stable forest regeneration plays an important role. The management regime of the forest in this area is close to nature silviculture, which only allows cuts of single trees or small groups of trees and prefers natural regeneration. In many places the forest is horizontally and vertically strongly structured. Multiple height layers are often not clearly separable, since trees of different heights and ages are mixed and have interlocking crowns. The main tree species in our study area are Norway spruce (Picea abies L. H. Karst), silver fir (Abies alba Mill.), Scots pine (Pinus sylvestris) and L. European larch (Larix decidua Mill.).
On this study area 14 plots were evenly distributed and centred orthogonally on the inventory points of the Swiss NFI. The plots were of square shape and had an edge length of 25 m. Each of the plots was split into four quadrants which resulted in 56 sample areas for our study. They covered diverse forest stands and many of them were characterized by a dense and interlocking distribution of trees. This structure of concentrated biomass often caused visual occlusion effects, which challenged the detection of tree objects from the laser scanning data. Coniferous trees which had branches close to the ground or grew in clusters often appeared optically dense. This resulted in reduced laser reflections and made data analysis more demanding.
2.2. TLS Data
TLS surveys were conducted on each of the sample plots using a FARO Focus 3D S120 scanner system [25]. The scanner was configured with a point spacing of 6.14 mm at 10 m distance and a planar scan size of 10,310 × 4268 points. The phase shift technology recorded one reflection per emitted beam. Figure 1 schematically illustrates the setup of the scan positions on a sample plot.
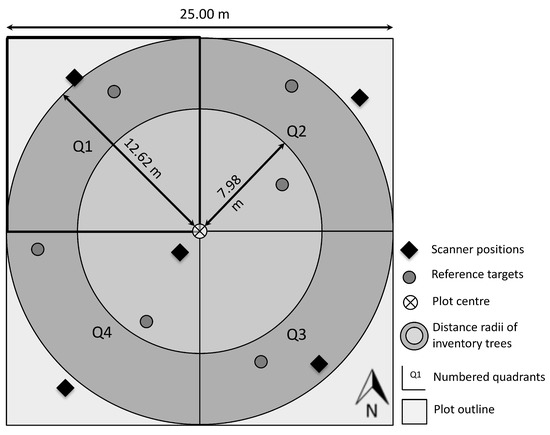
Figure 1.
Schema of the sample plot setup for scanning and reference data. The square shape of the sample plot was centred on the inventory point. Five TLS positions were oriented towards the corners and the centre and six reference targets were distributed over the plot area. The two concentric circles indicate the distance radii at which trees of different DBH are measured by the NFI. The plot was subdivided into four quadrants (Q1–Q4). For each quadrant ground cover of the regeneration was estimated in the field.
2.3. Reference Data
A so-called corner scan setup was chosen to minimize occlusion effects on the sample plots [26,27]. Four scans were positioned in direction of the corners of the plot and one towards the centre. Depending on the local conditions, slight variations were possible. In addition, six reference targets were evenly distributed on the plot, so that at least three were visible from each scan position. These targets consisted of reflective spheres mounted on poles of varying length and were used for registration of the single scans. The positions of the targets were determined relatively to one long-term GNSS measurement. After differential correction the average positional accuracy of these measurements amounted ±10 cm.
The preprocessing of the LiDAR data consisted of multiple steps. Firstly, the individual scans were registered and georeferenced using the Software FARO Scene 5.3 [28]. Then, the single scans were merged, clipped to the plot extend, thinned to 1 point/cm and normalized to ground height. In order to eliminate disturbances from ground vegetation like grass, all reflection below 50 cm height were removed. Finally, the point cloud of each plot was split into quadrants and transferred into a binary voxel grid. In the binary voxel grid a cell was defined as vegetation, if it contained at least one reflection. The quadrants matched the shape and size of the reference data for regeneration ground coverage. For the voxel grids a cell size of 10 cm edge length was determined empirically and was well suitable for the irregular point distribution, for preserving a relative high image resolution and for computational efficiency. Bienert et al. determined the same optimal voxel size for TLS data in forest environments [29].
All references have been collected together with the laser scans during a field campaign in 2015. The type of available reference data differed between plots, since the original field work intended to serve various interests. For the previous underlying studies on tree stem detection, DBH estimation and individual tree segmentation different reference data and plots were available then for the present regeneration detection. Details for the assessment of these framework components can be found in the corresponding publications [30,31,32].
For validating the regeneration coverage, suitable references were available on 14 spatially independent plots. For each of the four quadrants on these plots the ground cover of all regeneration trees was estimated. This resulted in a total of 56 reference quadrants. In accordance with the practices of the Swiss NFI [33], the regeneration was further subdivided into two types. The first type was the unestablished regeneration which bears a relatively high risk of browsing and other damages. It consisted of all trees with a maximum crown top height of 130 cm. The second class was the established regeneration and included all trees with a crown top height > 130 cm and with a DBH < 12 cm. Field-based estimation of ground cover was made visually by experienced forest inventory practitioners and was oriented both on the principles described by Braun-Blanquet as well as on the field survey manual of the Swiss NFI [33,34]. The percentage of crown cover was estimated in relation to the total ground surface for all trees belonging to the same type of regeneration. Overall, seven classes of percentage were discriminated: 0%, 1–5%, 6–15%, 16–25%, 26–50%, 51–75% and 76–100%. Table 1 provides an overview of the main characteristics of the reference plots and their single quadrants. Besides a general plot description, it contains two descriptions of the regeneration. The first one lists the ground cover of the regeneration for the complete plot and is separated by species. The second one provides the total ground cover for each quadrant of the plot for all species combined. The latter information was used as reference for validation of our method. Both types of descriptions are separated for unestablished and established regeneration.

Table 1.
Overview of the characteristics of each sample plot. The table lists general structural forest information as well as characteristics of the regeneration cover on the plots and the single quadrants.
3. Method
3.1. Outline
The overall framework for detecting and quantifying forest regeneration consisted of a set of individual submethods, of which each produced intermediate results. These intermediate results were used again by other submethods of the framework. Finally, they were combined to extract the regeneration and to measure its quantity. In the following sections the submethods were only outlined, because detailed descriptions were already provided in the individual underlying publications [30,31,32]. If not stated differently, the same parametrization as in the original publications were used. The focus of the present study was on the new application of regeneration estimation, which required a specific combination of the submethods. Figure 2 illustrates the major steps of this workflow, based on example data from a plot quadrant. Image (a) shows the original normalized point cloud which was overlaid with RGB information for visualisation. After preprocessing of the point cloud the voxel grid in image (b) was received. It is a binary vegetation image with ground information being removed. This 3D image was the basis for extracting the tree stems in part (c). The individual stem objects were used to derive the DBH as shown in illustration (d). The tree stems also functioned as priors for segmentation of the whole vegetation volume into individual tree objects in part (e). Finally, the tree objects were used together with the DBH information to identify the regeneration. In image (f) the regeneration is highlighted in light grey from the other forest trees.
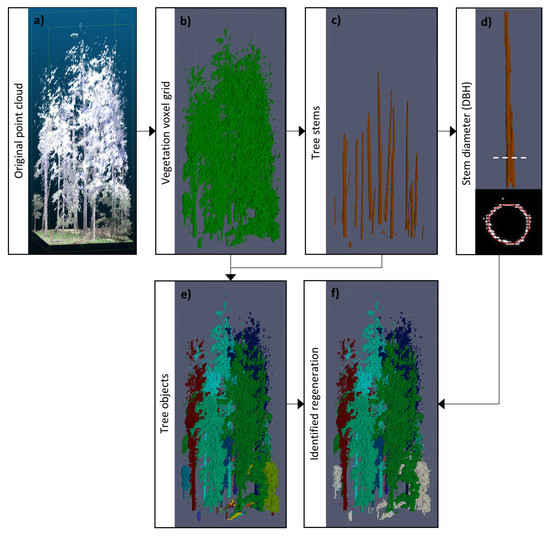
Figure 2.
Illustration of the intermediate results of the overall workflow for an example plot quadrant. Image (a) shows the original normalized point cloud which was overlaid with RGB information for visualisation. Image (b) depicts the binary vegetation information after removing ground points and converting the data to a voxel grid. From the vegetation volume the tree stems were detected in image (c). The DBH was directly derived from the tree stems in image (d). The tree stems were also used as priors for segmentation of the vegetation volume into individual tree clusters in image (e). Image (f) shows the regeneration in light grey which was identified from the individual tree clusters by using DBH information and tree height.
3.2. Tree Stem Detection
The detection of the tree stems was the first important submethod in our framework concept. The core part of this method is based on techniques from 3D mathematical morphology and has been described in detail in the publication from Heinzel and Huber [30].
As a first step we applied a series of 3D opening operations on the binary vegetation image. The opening operations produced a set of candidate segments which were likely to resemble tree stems or parts of tree stems. The idea behind was that tree stems were considered to be vertical linear structures. Therefore, a straight vertical line functioned as a simplified template and was used as structuring element (SE). The line was centred on each voxel and the voxel was kept, if the SE fitted into the binary vegetation image at this position. The formal definition of morphological opening is described in Equation (1) in terms of set theory. Using the notation of Soille [35], an opening of a set X is defined as an erosion of X with the SE B, followed by a dilation with the reflected SE . The underlying basic operators erosion and dilation are defined in Equations (2) and (3), where denotes a point of the set X, is the SE centred at and ⌀ is an empty set.
In effect, this opening procedure removed all voxels that were not likely to be part of a stem. Since tree stems naturally can be inclined, the original SE was tilted up to a certain degree and rotated around its vertical axis. This resulted in a series of 17 SEs which were iteratively used for opening of the vegetation volume. For all plots and quadrants a SE with a length of 21 voxels was used, after empirically testing different sizes. By summing the opening images of a quadrant, the stem candidate segments were received in form of connected components.
In a second step the stem candidate segments were filtered and combined. While some stems already consisted of a single segment, many stems were split into multiple fragments. The main reason for multiple stem segments were occlusion effects caused by dense foliage and branches. These fragments were combined by following logical processing rules as explained by Heinzel and Huber [30]. An important criterion was the vertical alignment of segments, which indicated the probability for representing the same tree stem. Sometimes other non-stem vegetation parts with a high density of laser reflections caused errors in the set of candidate segments. These erroneous segments could be identified by their atypical shape or based on their positional offset in relation to other stem segments. Many stems could not be extracted with their full length, since the upper parts of the trees often produced too little reflections on the stems caused by occlusion effects.
3.3. DBH Derivation
The second submethod of our framework was the derivation of the DBH from the tree stems. This submethod directly used the previously extracted stem objects and was applied as described in the publication of Heinzel and Huber [31]. The main concept of the DBH estimation relied on the Hough transformation of stem cross-section images into a suitable parameter space. General principles of Hough transform for image analysis and circle fitting have been described by Davies, Yuen et al. and Duda et al. [36,37,38]. For the present application, image slices of the single stem objects were taken at 130 cm height and with a thickness of 20 cm. With support of the original point cloud, the cross-section images were resampled to a higher pixel resolution of 1 cm. The individual cross-sections for each stem were then probed with a set of circles with n radii r. Each circle was centred on all x and y positions of the image. Hough transform allowed the optimization of the three parameters r, x, and y by accumulating all cells of the binary image at distances ri with i = {1, 2, …, n} from the centre positions x and y. The highest accumulation value indicated the optimal circle radius and the optimal centre position for fitting the stem cross-section. For our study, we probed all cross-section images with 50 different radii at 1 cm intervals. The determined optimal circle radius was doubled and represented the searched DBH.
3.4. Individual Tree Segmentation
The third submethod segmented the original vegetation volume into individual tree objects. We adopted a graph-based approach for spectral clustering which consisted of three steps. Input for the individual tree segmentation was the binary 3D vegetation image and the extracted tree stems. While the segmentation was carried out on the vegetation volume image, the tree stems were used as priors for the clustering process. A detailed description of the whole segmentation process was provided by Heinzel and Huber [32].
During the first step, a similarity graph was built from the 3D image data. Therefore, the weights were computed between each pair of n image voxels at the positions i and j. The weights model the similarity between two voxels based on distance metrics. The computation of the distance weights was oriented on the findings of Kamavar et al. and Reitberger et al. [39,40], but it was adapted for the inclusion of 3D stem priors and high-resolution TLS data. Accordingly, the total weight between two voxels was computed as the product of the horizontal distance , the vertical distance and the 3D-distance from the closest tree stem . In order to reduce the amount of 3D weight computations, distances between voxel pairs were only considered within a limited ellipsoidal space around each image cell. Equation (4) defines the weight computation based on the distance factors.
A first simple incorporation of prior information was directly implemented into the distance matrix. In cases where both positions i and j were located within different or the same stem object, the weights were set to 0 or 1, respectively. In all other cases the regular distance weights were used.
After constructing the weight matrix, the representation of the data points was changed for effective clustering. For similar purposes, general solutions directly derive m eigenvectors with the smallest eigenvalues from the normalized graph Laplacian , as described by von Luxburg [41]. In order to yield a better separation of trees in dense forest structures, we additionally incorporated prior information by using the tree stems. Based on the works of Hu et al. [42], the constraint clustering problem could be expressed as an optimization of the term in Equation (5).
is the label indicator matrix which has to be optimized, is the identity matrix, is the degree matrix of and encoded the voxels of the stem priors with one stem per column. is the correlation factor used in the constraint for the stem priors. After optimization, the columns of the matrix X described weighted combinations of the eigenvectors. The row vectors represented the transformed data points.
Afterwards, the row vectors were finally clustered by minimizing the energy of a Markov Random Field (MRF). The MRF included two types of costs. The unary label costs referred to the assignment of a label to the voxel i and the pairwise costs to the assignment of the labels and to the voxel pair . To solve this optimization problem the MRF framework described by Komodakis et al. and Komodakis and Tziritas was applied [43,44]. The derived labels defined the individual tree objects. An automated postprocessing was conducted in order to correct erroneously labelled tree fragments at the crown tops and at the image border.
3.5. Regeneration Estimation
The final and principal component of the overall processing framework bundles the intermediate results of all previous submethods in order to estimate the regeneration ground cover. Figure 3 presents the general workflow, starting with the individual tree objects and finishing with the validated ground cover classes. In a first step, the tree objects were classified into established and unestablished regeneration and separated from the remaining mature trees. Part (a) of Figure 4 illustrates the original tree objects on an example plot quadrant and part (b) shows the separated two types of regeneration. Criteria for this classification were the tree crown top height and the DBH while using the thresholds described in Section 2.3. The tree crown top height was determined from the maximum z coordinate of all voxels belonging to the same tree object. The DBH was automatically determined by the submethod in Section 3.3. At this point, the wrong allocation of only a single tree to either regeneration or non-regeneration could already lead to considerable error in ground coverage. Since Heinzel and Huber showed that for a few specific cases relatively large outliers might occur for DBH estimation [31], a separate check for potential DBH outliers was included. Therefore, we computed the overall ratio between tree height and DBH from forest inventory data of whole Switzerland, independent from species or other tree characteristics. The resulting height-DBH proportional factor of 0.014 was a rough value, but suitable to be used with a tolerant range of allowed deviation. Only estimated DBH values that were less than half or more than double of the theoretical height-derived DBH were considered to be outliers. In such cases the height-derived DBH was used instead.
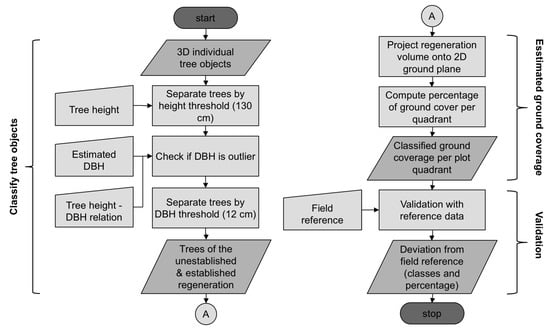
Figure 3.
Flowchart explaining the estimation of regeneration ground cover. The process was separated into three parts which are indicated with braces at the side of the chart. Firstly, trees belonging to the unestablished and established regeneration were determined from the set of individual tree objects. Secondly, the percentage of ground cover was calculated from the 2D projection of the regeneration. Finally, the ground cover classes were validated using field reference data.
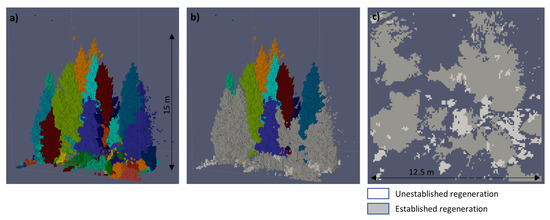
Figure 4.
Exemplary visualisation of the results from the regeneration ground cover estimation. Image (a) shows the previously isolated trees within a plot quadrant. From these tree objects two classes of regeneration have been extracted in image (b). For the ground cover computation, the regeneration has been vertically projected onto a 2D surface. Image (c) maps the footprints of both regeneration classes.
After determining those trees which belonged to the same type of regeneration, the volumetric image of these objects was projected onto the horizontal 2D plane of the normalized ground surface. Part (c) of Figure 4 illustrates this ground projection for the two types of regeneration. From this binary ground cover image the percentage of coverage in relation to the total size of the plot quadrant was computed and classified into seven classes as described in Section 2.3. These classes of percentage ground cover were oriented on the practices of the Swiss NFI. Finally, we received the classes of percentage ground cover for both unestablished and established regeneration within each plot quadrant.
3.6. Accuracy Assessment
The accuracy assessment was carried out by comparing the automatically determined ground cover classes against the field reference. Since the seven classes of percentage ground cover were in metrical order, the deviation between reference and predicted ground cover was denoted as number of classes. In addition, we used the percentage values at the class centres to describe the deviation as percentage of ground cover. This was useful, since the classes used by the Swiss NFI were of unequal and increasing size (see Section 2.3). From both, the deviation given in number of classes and the deviation given as percentage of ground cover, the mean bias error (MBE) and the mean absolute error (MAE) were computed according to the explanations of Willmot et al. and Willmott and Matsuura [45,46]. Both metrics are defined in Equations (6) and (7).
In above equations is the predicted ground cover of the i-th plot quadrant, is the reference ground cover of quadrant i and n is the total number of plot quadrants. All accuracy metrics were provided for unestablished and established regeneration.
4. Results
The results of the accuracy assessment are summarized in form of two confusion matrices in Table 2. Table 2a presents the results for the unestablished regeneration and Table 2b for the established regeneration. Both matrices contain the counts of occurrences for each individual combination of predicted and reference class. Altogether 56 quadrants from 14 plots have been assessed. The results of the individual plot quadrants are visualised in the two charts of Figure 5. The quadrants are sorted by plot number with four quadrants per plot. Figure 5a displays the predicted and reference classes of each quadrant for the unestablished regeneration. The same chart structure is used in Figure 5b for the established regeneration.

Table 2.
Confusion matrices summarizing the results of the accuracy assessment. The two subtables separately describe the results for (a) unestablished and (b) established regeneration. The matrices contain the counts of occurrences for each combination of reference and predicted class. All class combinations outside the matrix diagonal are highlighted in light grey, while the matrix diagonal is in dark grey. GC = ground cover.
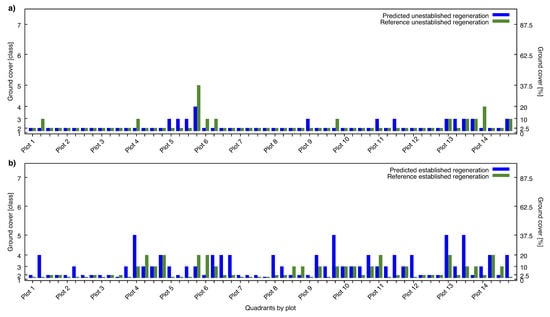
Figure 5.
Charts showing the predicted ground cover (blue) versus the field reference (green) for each individual quadrant. The quadrants are sorted by plot as indicated on the x-axis. The marking of the y-axis on the left side indicates the classes of ground cover. The marking on the right side indicates percentage of ground cover, given as the centre values of each class. Part (a) covers the unestablished regeneration and part (b) the established regeneration.
The unequal range of ground cover which is spanned by the individual classes, complicates the interpretation of the results. This characteristic of varying class sizes is visualised by the different units used for the y-axes on the left and the right side of the charts in Figure 5. The higher the class number indicated by the y-axis on the left side, the larger the spanned range of ground cover indicated by the y-axis on the right side. The individual sizes of the classes, as detailed in Section 2.3, are also provided as additional information next to the class numbers in Table 2a,b.
In order to circumvent the problem of varying class sizes, a different presentation of the confusion matrices is provided in Table 3 which uses the percentage of ground cover at the class centres. The intention was to describe the error as percent ground cover instead of counts of misclassifications. Table 3a gives a general overview of the percentage difference of ground cover between all possible combinations of classes. For this calculation, the percentage values at the class centres have been used. This matrix does not contain validation results, but serves as an interpretation aid for the following subtables. Table 3b,c multiply the percentage difference of each class combination with the related count in Table 2. This already provides a percentage-based view on the accuracy assessment, since the different class sizes are considered. Table 3b contains the results for the unestablished regeneration and Table 3c for the established regeneration. Finally, Table 3d,e provide the normalized differences. Therefore, the cells of Table 3b,c have been scaled with the L1-norm which was computed from all entries in a table. The absolute values of these normalized results sum up to 100% for each type of regeneration. Each normalized value contains the proportional share of the total error of the respective regeneration type. This representation of the confusion matrix, in combination with the class-based counts in Table 2, allows meaningful interpretation of the results.

Table 3.
Matrices describing the accuracy assessment results by using the differences between the percentage values at the class centres. Subtable (a) gives an overview of the general differences between percentage values at the class centres for all class combinations. In subtable (b) the percentage difference between each class combination was multiplied by the related count in the confusion matrix of Table 2a. Subtable (c) shows for the same for the established regeneration and uses the confusion matrix in Table 2b. In subtable (d) the differences from subtable (b) have been scaled with their L1-norm. Subtable (e) scales the respective differences for the established regeneration. UR = unestablished regeneration, ER = established regeneration, GC = ground cover.
Besides the confusion matrices, overall error metrics have been computed and detailed in Table 4. Table 4a shows the overall values measured in number of classes and Table 4b the values measured in percentage at the class centres. Both tables contain the MBE and the MAE for established and unestablished regeneration.

Table 4.
Overview of the overall accuracies for unestablished and established regeneration. Subtable (a) measures the mean error as number of classes. Subtable (b) relates to the same assessment but expresses the error as percentage ground cover, calculated from the ground cover values at the class centres. GC = ground cover.
5. Discussion
The described method allows the quantification of forest regeneration using TLS data. In comparison to airborne data, TLS data can capture understory trees more accurately and completely. The high point density in combination with the specific processing framework were able to differentiate two types of regeneration, even in dense forest constellations. In order to allow practical applicability, the design of the method was oriented on practices of the Swiss NFI. The plot-based data sets, the definition of regeneration types as well as the classes of ground cover were similar to those used by field-based forest inventory. The range and the gradation of the classes proved to minimize field estimation errors during long-term use by foresters. However, field-based visual estimation of regeneration ground cover, is generally prone for inaccuracies. Since the available reference data consisted of such estimations, potential deviations from the true values must be considered when interpreting the results.
5.1. Methodological Approach
The overall processing framework was constructed as a specific combination of individual submethods. The submethods were based on previously published studies which were developed using TLS data from the same field campaign but with a different set of sample plots [30,31,32]. From all methods, the detection of the tree stems played a key role, since the stems were subsequently used in the workflow for DBH estimation and as priors for single-tree isolation. An important characteristic of the stem detection is the volumetric approach, which extracts 3D stem objects directly from the image. The derived stem objects only consisted of the scanned parts of the stems and modelling of occluded parts was avoided. This restriction allows accurate DBH estimation and supports the 3D segmentation of tree objects even in difficult scenarios. The most complex component of the processing framework was the single-tree segmentation which is a composition of methods itself. It combines several techniques into an effective machine learning workflow. All components of this workflow were adapted and improved for usage with high TLS point densities and for 3D structures. The main concepts for single-tree segmentation were based on graph theory, spectral learning and Markov random fields. Each of the submethods of the overall framework was used with the same parameter values as described in the original publications. The parameters were determined empirically and used constantly for all plots.
The whole framework for regeneration estimation was automated and required no user interaction. The most difficult scenario were groups of small coniferous trees growing in dense clusters. Since these trees often had branches down to the ground, they worked as a barrier for the laser beam and allowed relatively few reflections from inside the tree cluster. These occlusion effects complicated the detection of tree stems and the segmentation into individual trees. Therefore, very dense clusters of small coniferous trees could not be segmented and used as individual objects instead. This topic has been discussed in detail by Heinzel and Huber [32]. While this phenomenon related to the true number of individual small trees, it did not affect the amount of ground coverage of the regeneration.
5.2. Interpretation of the Results
The evaluation of the automated quantification of forest regeneration showed good agreement with the field records. For the unestablished regeneration we achieved automated results that were almost identical with the field reference. The mean bias was close to zero and the MAE was only marginal with 0.27 classes or 2.23% of ground cover. The confusion matrices in Table 2a and Table 3d as well as the chart in Figure 5a reveal that the majority of the quadrants belonged to reference class two and were correctly classified.
For established regeneration, the mean bias indicated a weak trend towards overestimation of less than one class. The mean error was significantly higher compared to unestablished regeneration, but still amounted only little more than one class or 8.08% of ground cover. From the confusion matrices in Table 2b and Table 3e it can be seen that the number of misclassifications increases with smaller classes. This trend can at least partly be explained by the small range of ground cover, spanned by the lower classes. Small class sizes increase the impact of relatively small estimation errors. However, due to the unequal class sizes, the error of percentage ground cover needs not necessarily be higher for misclassifications with many counts than with few. As an example, it can be seen that for the established regeneration in Table 2b, six quadrants of reference class one have erroneously been predicted as class three. At the same time, three quadrants of reference class three have been predicted as class five. Even if the first case counts two times more misclassifications, Table 3e shows that the summed and normalized error in percentage ground cover is smaller. This is caused by the different distances between the class centres, as listed in Table 3a. A similar effect can be seen from the individual quadrants in Figure 5. In Figure 5a, the fourth quadrant of plot five seems to underestimate the reference ground cover by about the half. However, the deviation of predicted and reference class amounts only one class, while the percentage difference of ground cover at the class centres is relatively high. The unequal range spanned by the classes becomes clear from the different markings of the y-axis on the left and on the right side of the chart.
Comparing the distribution of ground cover classes, it can be seen that for unestablished regeneration class two is dominating. The low ground cover of this class, which is between 1% and 5%, often originates from small tree sizes. Even if several small trees exist within a quadrant, the percentage of ground cover is often less than 5%. For established regeneration the distribution of ground cover classes is more balanced. However, the number of counts decreases for higher classes and the maximum occurring reference class is class five with up to 50% ground cover.
Two aspects have to be considered when interpreting the accuracies. Firstly, deviations between reference and estimated ground cover of only one class difference could derive from only small mistakes in ground cover estimation. If the true regeneration cover of a plot quadrant is close to a class border, it could easily happen that different classes were estimated in the field and by automated TLS data processing. Depending on the application, errors of up to one class could therefore be considered as tolerable. Secondly, it should be considered that the field reference is not necessarily the true ground data. Visual estimation of ground cover in the field, even if done by experienced practitioners, is always a source of failure. In addition, it is prone to bias between different observers. Besides the availability for this study, the intention to use this kind of field data was that it is in accordance with the long-term practices of the Swiss NFI. Therefore, the good agreement of automated results and field references indicates practical usability of the present TLS-based method.
5.3. Comparison to other Studies
The comparison of the present method with other studies, both from a methodical and from a result’s point of view, is not straight forward. To our best knowledge, there is no other study on object-based quantification of forest regeneration based on TLS data. Furthermore, existing studies which estimate regeneration with different technical approaches and different types of data, often do not report results in consistent and comparable ways.
To give at least an impression of how our approach relates to others, we compare it to the small number of understory-related studies which also use object-based methods but work with airborne data. Generally, these comparisons show that the most apparent advantage of our TLS-based method is the higher information content in lower forest layers. Especially when aiming on understory information, all studies which use airborne datasets report limitations in detection of small trees due to sparse point densities [2,19,20,47]. Profound investigations on this phenomenon were carried out by Korpela et al. and Kükenbrink et al. [14,15]. The good results of the present TLS-based approach confirm our initial assumption of perspective-related advantages. Besides the perspective, terrestrial scanners also produce higher point densities due to shorter distances between scanner and objects. Both aspects support the correct detection of small trees. A disadvantage of TLS compared to airborne data is the limitation to cover only relatively small areas.
The methods used by most other studies on understory tree detection are different from our approach. From the relatively small number of available studies, the approaches of Duncanson et al. as well as Hamraz et al. are most different to ours. Both studies tried to apply methods based on digital surface models (DSM) in a way that allowed the extraction of understory trees. Therefore, they computed DSMs for different forest layers that were visible in the point cloud. Duncanson et al. subsequently applied a classic watershed method to the overstory and the understory CHMs [19]. Hamraz et al. used DSMs from multiple layers and applied his own DSM-based segmentation method [2,48]. In comparison to these DSM-based methods, the approach of Ferraz et al. comes closer to ours [20]. They directly clustered the 3D information of the point cloud by using the mean shift algorithm. This allowed the separation of overstory and understory trees, but still had problems to detect small trees that were overtopped by larger trees. Therefore, their approach is less suitable for quantification of regeneration. The approach of Amiri et al. also used mean shift clustering, but combined it with normalized cut segmentation [11]. The method is based on the ALS-based single-tree detection approaches of Reitberger et al. and Yao et al. [40,49], but focused on the estimation of regeneration coverage. Compared to other studies, their ALS-based concept is more similar to our approach, but differentiates in several methodical aspects. Besides the improved and prior-constrained 3D clustering, the present method uses volumetric stem information which can only be derived from TLS data. Furthermore, we estimate regeneration more differentiated by considering both DBH and height metrics and provide results for two types of regeneration.
Concerning the comparison of accuracies, the only object-based approach which reported regeneration cover is from Amiri et al. [11]. They achieved an average of 60% detected regeneration cover and on single plots they reached up to 70%. This refers to errors of 40% and 30% which are a relatively small when considering the use of airborne data. However, our method could strongly improve the accuracy, since the largest mean error in the case of established regeneration amounted only 8.08%. In order to compare our results to a broader range of studies, we also considered non-object-based methods, like height-based point distribution approaches. For these types of methods Wing et al. reported outstanding good results with an error of ±22% and a bias of 0%. The disadvantage of their approach is, that they cannot differentiate individual tree objects of the regeneration. This means that their method requires clearly separated forest layers. Hill and Broughton report correspondences of detected understory tree cover with field data of 77% in leaf-on state and 72% in leaf-off state [13]. The good results were achieved by seasonal timing and by considering all trees below the overstory canopy. Therefore, their method was not restricted to regeneration trees of defined size limits. Similarly, Morsdorf et al. estimated the vegetation cover of subdominant layers from ALS data with an accuracy of 48% [17]. Generally, the strongly varying definitions and thresholds used for understory estimation, which mostly do not match the size limits of forest regeneration, makes comparisons difficult. Independent from differences between approaches, it can be seen that the new method presented in this study achieved the best results. This demonstrates the advantage of using TLS data for regeneration estimation as well as the effectivity of our methodical framework.
6. Conclusions
The present study described a new method for estimating ground coverage of forest regeneration. In comparison to other studies, it used TLS instead of ALS data and introduced a processing framework which handled high data densities in combination with 3D object-based image analysis. Until now, this combination of properties was not used before and the accuracy of the results was better than earlier approaches. Methodically, the direct analysis of volumetric 3D data avoided to fall back to DSM-based methods or height distribution metrics of the point cloud. The use of TLS data allowed more precise and complete detection of suppressed trees, but it is not suitable for continuous wide area surveys. Instead, we tested our method on the basis of sample plots and adapted the analytical concept to the practices of the Swiss NFI. This supports potential integration into practical application and sample-based inventories. Since our method was developed on datasets in temperate montane forest which has a protective function against alpine natural hazards, additional tests with other types of forest should be carried out in the future. An interesting extension would be, to investigate the applicability of our method for unmanned aerial vehicle LiDAR, where the sensors are mounted on drone platforms. This could be an intermediate solution for surveying continuous areas on regional scale, since higher point densities and higher scan angles can be reached compared to classical ALS data.
Author Contributions
J.H. conceived and developed all methods, designed the fieldwork, and acquired and processed the data. C.G. conceived and defined the main research objectives, designed the fieldwork, and managed the project. The manuscript was written by Johannes Heinzel with input from C.G.
Funding
This study was conducted within the framework of the Swiss National Forest Inventory, a joint research program of the Swiss Federal Office for the Environment (FOEN) and the Swiss Federal Institute for Forest, Snow and Landscape Research (WSL). No further external funding was received.
Acknowledgments
The authors would like to thank Christoph Fischer for forestry related advise, Björn Dreier and Natalia Rehush for their support during the fieldwork and Susette Haegi for registration of the scans. Special thanks go to Markus Huber, who previously managed the research project. The forest administration of the canton Grisons, Switzerland, gave important assistance to this project.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ALS | airborne laser scanning |
| TLS | terrestrial laser scanning |
| MLS | mobile laser scanning |
| LiDAR | light detection and ranging |
| DBH | diameter at breast height |
| CHM | crown height model |
| DSM | digital surface model |
| NFI | national forest inventory |
| SE | structuring element |
| MRF | Markov random field |
| MAE | mean absolute error |
| MBE | mean bias error |
References
- Ishii, H.T.; Tanabe, S.I.; Hiura, T. Exploring the relationships among canopy structure, stand productivity, and biodiversity of temperate forest ecosystems. For. Sci. 2004, 50, 342–355. [Google Scholar]
- Hamraz, H.; Contreras, M.A.; Zhang, J. Vertical stratification of forest canopy for segmentation of understory trees within small-footprint airborne LiDAR point clouds. ISPRS J. Photogramm. Remote Sens. 2017, 130, 385–392. [Google Scholar] [CrossRef]
- Wing, B.M.; Ritchie, M.W.; Boston, K.; Cohen, W.B.; Gitelman, A.; Olsen, M.J. Prediction of understory vegetation cover with airborne lidar in an interior ponderosa pine forest. Remote Sens. Environ. 2012, 124, 730–741. [Google Scholar] [CrossRef]
- O’Hara, K.L. Multiaged forest stands for protection forests: concepts and applications. For. Snow Landsc. Res. 2006, 80, 45–55. [Google Scholar]
- Dorren, L.; Berger, F.; Jonsson, M.; Krautblatter, M.; Mölk, M.; Stoffel, M.; Wehrli, A. State of the art in rockfall—Forest interactions. Schweiz. Z. Forstwes. 2007, 158, 128–141. [Google Scholar] [CrossRef]
- Antos, J. Understory plants in temperate forests. In Forests and Forest Plants; Owens, J.N., Lund, H.G., Eds.; Eolss Publishers Co., Ltd.: Oxford, UK, 2009; pp. 262–279. [Google Scholar]
- Jules, M.J.; Sawyer, J.O.; Jules, E.S. Assessing the relationships between stand development and understory vegetation using a 420-year chronosequence. For. Ecol. Manag. 2008, 255, 2384–2393. [Google Scholar] [CrossRef]
- Latifi, H.; Heurich, M.; Hartig, F.; Müller, J.; Krzystek, P.; Jehl, H.; Dech, S. Estimating over- and understorey canopy density of temperate mixed stands by airborne LiDAR data. Forestry 2015, 89, 69–81. [Google Scholar] [CrossRef]
- Lin, Y.; Holopainen, M.; Kankare, V.; Hyyppa, J. Validation of Mobile Laser Scanning for Understory Tree Characterization in Urban Forest. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2014, 7, 3167–3173. [Google Scholar] [CrossRef]
- Tuanmu, M.N.; Viña, A.; Bearer, S.; Xu, W.; Ouyang, Z.; Zhang, H.; Liu, J. Mapping understory vegetation using phenological characteristics derived from remotely sensed data. Remote Sens. Environ. 2010, 114, 1833–1844. [Google Scholar] [CrossRef]
- Amiri, N.; Yao, W.; Heurich, M.; Krzystek, P.; Skidmore, A.K. Estimation of regeneration coverage in a temperate forest by 3D segmentation using airborne laser scanning data. Int. J. Appl. Earth Observ. Geoinf. 2016, 52, 252–262. [Google Scholar] [CrossRef]
- Fleming, S.; Cottin, A.; Woodhouse, I. The first spectral map of a forest understory from multispectral lidar. Lidar News 2015, 5, 26–30. [Google Scholar]
- Hill, R.; Broughton, R. Mapping the understorey of deciduous woodland from leaf-on and leaf-off airborne LiDAR data: A case study in lowland Britain. ISPRS J. Photogramm. Remote Sens. 2009, 64, 223–233. [Google Scholar] [CrossRef]
- Korpela, I.; Hovi, A.; Morsdorf, F. Understory trees in airborne LiDAR data—Selective mapping due to transmission losses and echo-triggering mechanisms. Remote Sens. Environ. 2012, 119, 92–104. [Google Scholar] [CrossRef]
- Kükenbrink, D.; Schneider, F.D.; Leiterer, R.; Schaepman, M.E.; Morsdorf, F. Quantification of hidden canopy volume of airborne laser scanning data using a voxel traversal algorithm. Remote Sens. Environ. 2017, 194, 424–436. [Google Scholar] [CrossRef]
- Lindberg, E.; Olofsson, K.; Holmgren, J.; Olsson, H. Estimation of 3D vegetation structure from waveform and discrete return airborne laser scanning data. Remote Sens. Environ. 2012, 118, 151–161. [Google Scholar] [CrossRef]
- Morsdorf, F.; Mårell, A.; Koetz, B.; Cassagne, N.; Pimont, F.; Rigolot, E.; Allgöwer, B. Discrimination of vegetation strata in a multi-layered Mediterranean forest ecosystem using height and intensity information derived from airborne laser scanning. Remote Sens. Environ. 2010, 114, 1403–1415. [Google Scholar] [CrossRef]
- Singh, K.K.; Davis, A.J.; Meentemeyer, R.K. Detecting understory plant invasion in urban forests using LiDAR. Int. J. Appl. Earth Observ. Geoinf. 2015, 38, 267–279. [Google Scholar] [CrossRef]
- Duncanson, L.; Cook, B.; Hurtt, G.; Dubayah, R. An efficient, multi-layered crown delineation algorithm for mapping individual tree structure across multiple ecosystems. Remote Sens. Environ. 2014, 154, 378–386. [Google Scholar] [CrossRef]
- Ferraz, A.; Bretar, F.; Jacquemoud, S.; Gonçalves, G.; Pereira, L.; Tomé, M.; Soares, P. 3-D mapping of a multi-layered Mediterranean forest using ALS data. Remote Sens. Environ. 2012, 121, 210–223. [Google Scholar] [CrossRef]
- Hancock, S.; Anderson, K.; Disney, M.; Gaston, K.J. Measurement of fine-spatial-resolution 3D vegetation structure with airborne waveform lidar: Calibration and validation with voxelised terrestrial lidar. Remote Sens. Environ. 2017, 188, 37–50. [Google Scholar] [CrossRef]
- Gupta, V.; Reinke, K.; Jones, S.; Wallace, L.; Holden, L. Assessing Metrics for Estimating Fire Induced Change in the Forest Understorey Structure Using Terrestrial Laser Scanning. Remote Sens. 2015, 7, 8180–8201. [Google Scholar] [CrossRef]
- Xia, S.; Wang, C.; Pan, F.; Xi, X.; Zeng, H.; Liu, H. Detecting stems in dense and homogeneous forest using single-scan TLS. Forests 2015, 6, 3923–3945. [Google Scholar] [CrossRef]
- Brolly, G.; Király, G.; Czimber, K. Mapping Forest Regeneration from Terrestrial Laser Scans. Acta Silv. Lign. Hung. 2013, 9, 135–146. [Google Scholar] [CrossRef]
- FARO Technologies Inc. FARO Laser Scanner Focus3D Manual; FARO Technologies Inc.: Lake Mary, FL, USA, 2013; p. 79. [Google Scholar]
- Yao, T.; Yang, X.; Zhao, F.; Wang, Z.; Zhang, Q.; Jupp, D.; Lovell, J.; Culvenor, D.; Newnham, G.; Ni-Meister, W.; et al. Measuring forest structure and biomass in New England forest stands using Echidna ground-based lidar. Remote Sens. Environ. 2011, 115, 2965–2974. [Google Scholar] [CrossRef]
- Zande, D.V.d.; Jonckheere, I.; Stuckens, J.; Verstraeten, W.W.; Coppin, P. Sampling design of ground-based lidar measurements of forest canopy structure and its effect on shadowing. Can. J. Remote Sens. 2008, 34, 526–538. [Google Scholar] [CrossRef]
- FARO Technologies Inc. FARO SCENE 5.1 Users Manual; FARO: Lake Mary, FL, USA, 2014. [Google Scholar]
- Bienert, A.; Queck, R.; Schmidt, A.; Bernhofer, C.; Maas, H. Voxel space analysis of terrestrial laser scans in forests for wind field modelling. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2010, 38, 92–97. [Google Scholar]
- Heinzel, J.; Huber, M. Detecting Tree Stems from Volumetric TLS Data in Forest Environments with Rich Understory. Remote Sens. 2017, 9, 9. [Google Scholar] [CrossRef]
- Heinzel, J.; Huber, M. Tree Stem Diameter Estimation From Volumetric TLS Image Data. Remote Sens. 2017, 9, 614. [Google Scholar] [CrossRef]
- Heinzel, J.; Huber, M. Constrained Spectral Clustering of Individual Trees in Dense Forest Using Terrestrial Laser Scanning Data. Remote Sens. 2018, 10, 1056. [Google Scholar] [CrossRef]
- Düggelin, C.; Keller, M. (Eds.) Schweizerisches Landesforstinventar, Feldaufnahme-Anleitung 2017; Eidg. Forschungsanstalt für Wald, Schnee und Landschaft: Birmensdorf, Schweiz, 2017; p. 220. [Google Scholar]
- Braun-Blanquet, J. Pflanzensoziologie. Grundzüge der Vegetationskunde; Springer: Berlin, Germany, 1928. [Google Scholar]
- Soille, P. Morphological Image Analysis: Principles and Applications; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2004; p. 302. [Google Scholar]
- Davies, E. A modified Hough scheme for general circle location. Pattern Recognit. Lett. 1988, 7, 37–43. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E. Use of the Hough transformation to detect lines and curves in pictures. Commun. ACM 1972, 15, 11–15. [Google Scholar] [CrossRef]
- Yuen, H.K.; Princen, J.; Dlingworth, J.; Kittler, J. A Comparative Study of Hough Transform Methods for Circle Finding. In Proceedings of the Alvey Vision Conference 1989, British Machine Vision Association and Society for Pattern Recognition, Reading, UK, 25–28 September 1989. [Google Scholar] [CrossRef]
- Kamvar, K.; Sepandar, S.; Klein, K.; Dan, D.; Manning, M.; Christopher, C. Spectral learning. In Proceedings of the 18th international joint conference on Artificial Intelligence International, Acapulco, Mexico, 9–15 August 2003. [Google Scholar]
- Reitberger, J.; Schnörr, C.; Krzystek, P.; Stilla, U. 3D segmentation of single trees exploiting full waveform LIDAR data. ISPRS J. Photogramm. Remote Sens. 2009, 64, 561–574. [Google Scholar] [CrossRef]
- Von Luxburg, U. A tutorial on spectral clustering. Stat. Comput. 2007, 17, 395–416. [Google Scholar] [CrossRef]
- Hu, H.; Feng, J.; Yu, C.; Zhou, J. Multi-class constrained normalized cut with hard, soft, unary and pairwise priors and its applications to object segmentation. IEEE Trans. Image Process. 2013, 22, 4328–4340. [Google Scholar] [CrossRef]
- Komodakis, N.; Tziritas, G.; Paragios, N. Performance vs computational efficiency for optimizing single and dynamic MRFs: Setting the state of the art with primal-dual strategies. Comput. Vis. Image Underst. 2008, 112, 14–29. [Google Scholar] [CrossRef]
- Komodakis, N.; Tziritas, G. Approximate Labeling via Graph Cuts Based on Linear Programming. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 1436–1453. [Google Scholar] [CrossRef] [PubMed]
- Willmott, C.J.; Matsuura, K.; Robeson, S.M. Ambiguities inherent in sums-of-squares-based error statistics. Atmos. Environ. 2009, 43, 749–752. [Google Scholar] [CrossRef]
- Willmott, C.J.; Matsuura, K. Advantages of the mean absolute error (MAE) over the root mean square error (RMSE) in assessing average model performance. Clim. Res. 2005, 30, 79–82. [Google Scholar]
- Lahivaara, T.; Seppanen, A.; Kaipio, J.P.; Vauhkonen, J.; Korhonen, L.; Tokola, T.; Maltamo, M. Bayesian Approach to Tree Detection Based on Airborne Laser Scanning Data. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2690–2699. [Google Scholar] [CrossRef]
- Hamraz, H.; Contreras, M.A.; Zhang, J. A robust approach for tree segmentation in deciduous forests using small-footprint airborne LiDAR data. Int. J. Appl. Earth Observ. Geoinf. 2016, 52, 532–541. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree species classification and estimation of stem volume and DBH based on single tree extraction by exploiting airborne full-waveform LiDAR data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).