An Intelligent Management System and Advanced Analytics for Boosting Date Production
Abstract
1. Introduction
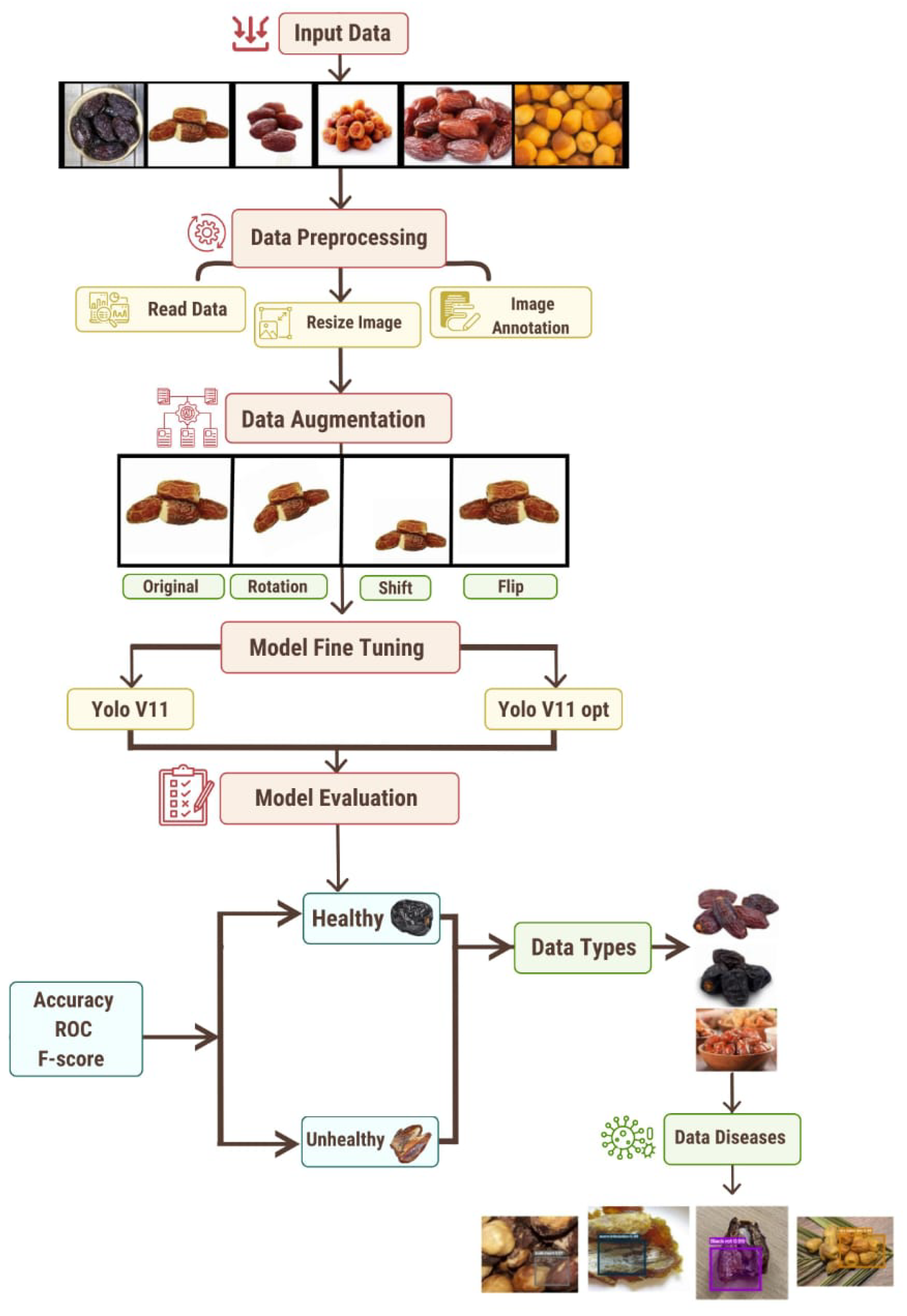
2. Proposed Model
- Input Data: This study is grounded on a curated dataset comprising 2482 high-resolution images of date fruits, encompassing a wide spectrum of phenotypes including healthy, diseased, and pathogen-affected samples. Such diversity in sample representation is critical for achieving generalizable feature extraction and robust model learning across heterogeneous conditions.To support systematic model development, the dataset was partitioned into three distinct subsets: training, validation, and testing. The training subset, containing 1737 images (70% of the dataset), was employed to facilitate the learning of discriminative patterns across various visual attributes. To fine-tune hyperparameters and monitor overfitting behavior, a validation set consisting of 372 samples (15%) was reserved. The final 373 samples (15%) were held back for the independent testing phase, ensuring an unbiased evaluation of the model’s generalization capabilities.This stratified data division preserved the class distribution across subsets and provided a rigorous foundation for training, optimizing, and assessing the proposed deep learning framework.
- Data Preprocessing: The data preparation pipeline was designed to ensure uniformity, compatibility with deep learning architectures, and robustness to environmental variability. The following steps were implemented:
- –
- Image Resizing: All images were resized to 640 × 640 pixels using bilinear interpolation. This standardized input size ensures compatibility with the YOLOv-based detection architecture, while preserving spatial structure.
- –
- Color Space and Channel Handling: Images were loaded in RGB format. When necessary, conversions from OpenCV’s default BGR to RGB were performed.
- –
- Pixel Normalization: Pixel intensity values were scaled to a [0, 1] range by dividing each channel by 255.
- –
- Channel-Wise Standardization: Images were normalized using ImageNet-derived means [0.485, 0.456, 0.406] and standard deviations [0.229, 0.224, 0.225] to align with pretrained CNN backbones and enhance training stability.
- –
- Dataset Partitioning: To ensure representative sampling and balanced class distribution, the dataset was proportionally segmented into training (70%), validation (15%), and testing (15%) subsets using stratified allocation.
- Data Augmentation: To enhance model generalization under real-world variability, a diverse augmentation strategy was implemented using the Albumentations library (https://github.com/albumentations-team/albumentations (accessed on 1 May 2025)). Each original image was augmented to create five variants with randomized transformations, preserving semantic consistency while introducing noise and distortion. The following augmentation techniques were applied:
- –
- Horizontal and vertical flipping to simulate camera orientation variability.
- –
- Rotation (±30°), scaling, and shifting to replicate changes in viewpoint and framing.
- –
- Brightness and contrast adjustments to mimic natural lighting changes, including overexposure and shadows.
- –
- Gaussian blur and additive Gaussian noise to reflect dust interference or sensor imperfections.
- –
- Coarse dropout to model occlusions caused by leaves, tools, or partial obstructions.
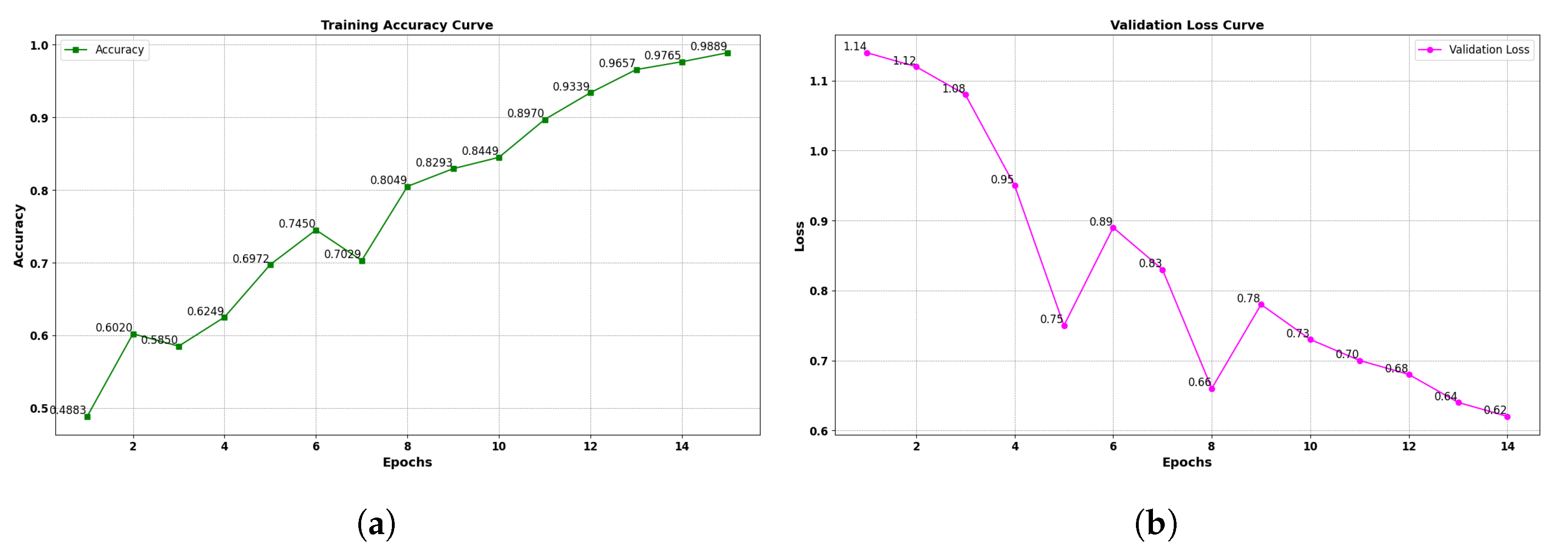
All augmented images were saved in high-quality JPEG format to retain spatial fidelity. This structured preprocessing pipeline contributes to improved model robustness and generalizability in non-ideal deployment scenarios. - Model Fine-Tuning: The proposed framework employs two deep-learning-based object detection architectures: the standard YOLOv11 and an optimized variant, YOLOv11-opt. Both models were fine-tuned using the preprocessed and augmented dataset to capture domain-specific features relevant to date fruit classification and disease detection. The fine-tuning process involved adjusting the model weights through transfer learning, allowing the architectures to learn subtle variations in visual attributes such as texture, color distribution, morphological patterns, and disease symptoms. This targeted adaptation enhances the models’ sensitivity to inter-class distinctions among healthy, diseased, and varietal date types, thereby improving detection precision and classification accuracy within the agricultural domain.
- –
- Stage 1—Health-Based Classification: In the initial classification stage, the fine-tuned models categorize the input date fruit images into two primary classes: healthy and unhealthy. This binary classification serves as the foundational step, enabling a structured hierarchical analysis. For images identified as healthy, the model classifies them into specific date varieties based on morphological and phenotypic characteristics. In contrast, images categorized as unhealthy are directed toward further pathological assessment.
- –
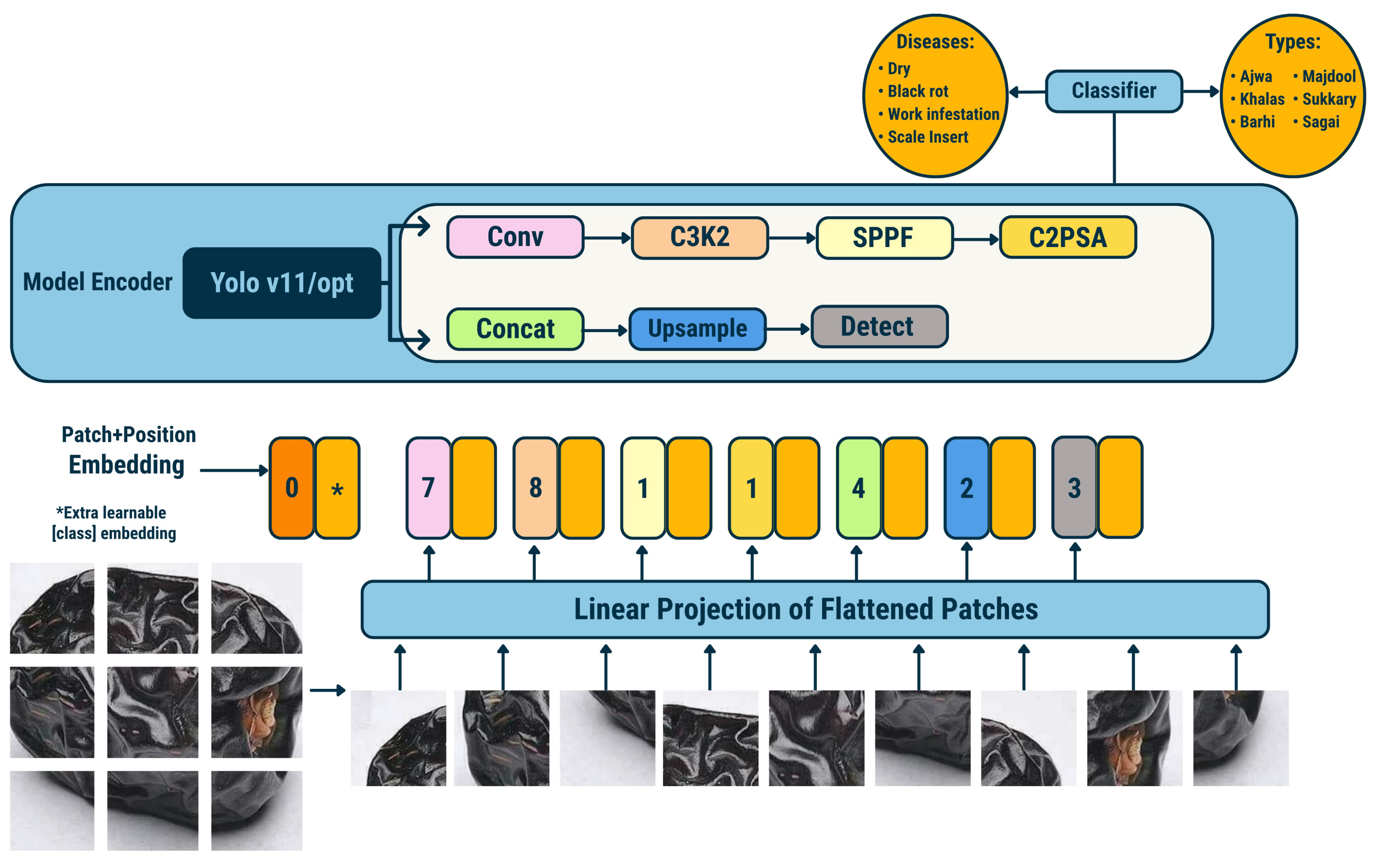
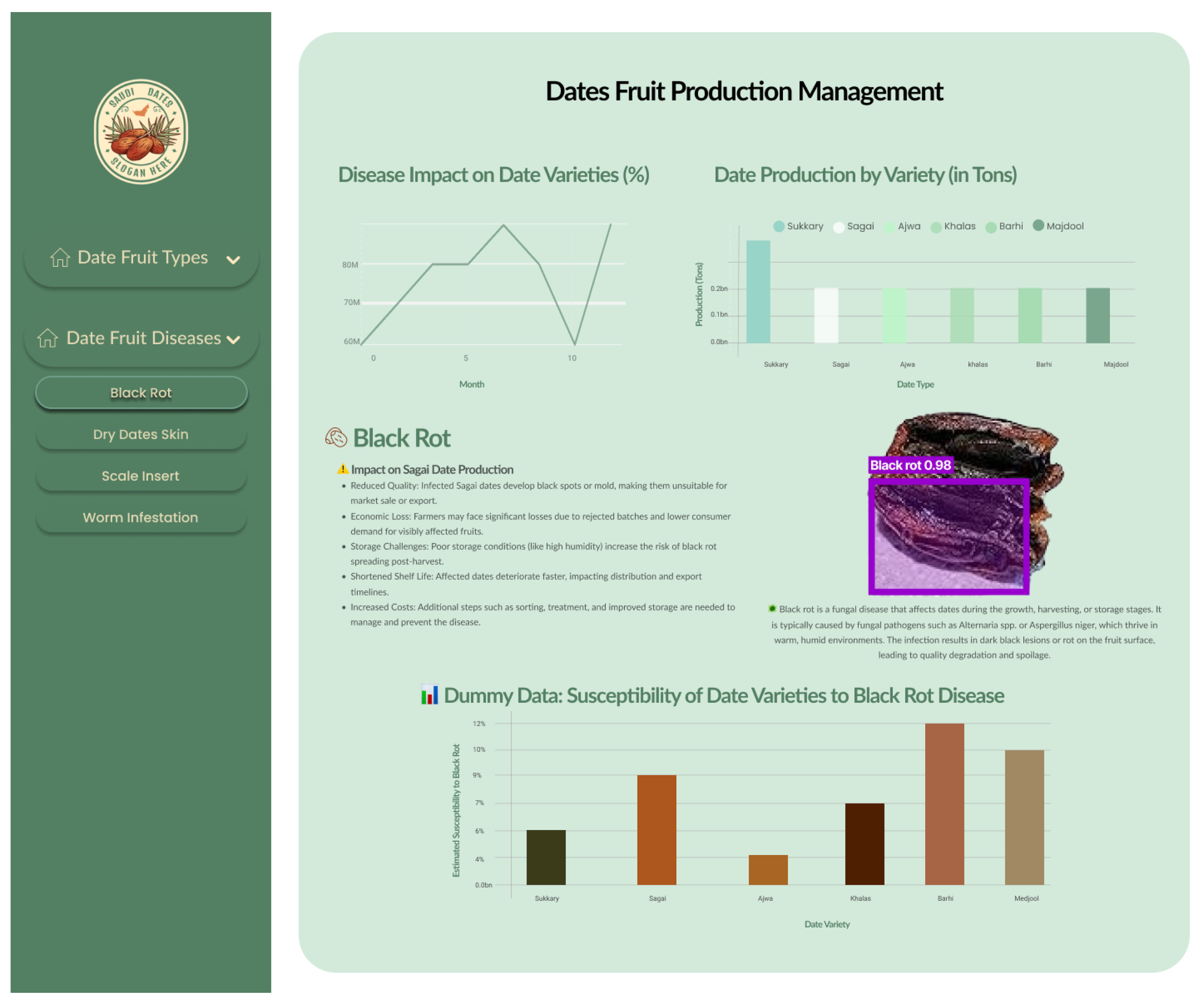
- Stage 2—Varietal and Disease-Specific Classification: Following the health-based categorization, a secondary, more detailed classification is performed. Healthy date fruits undergo varietal classification, in which the system distinguishes among different types, such as Ajwa, Barhi, Khalas, and others. Conversely, unhealthy dates are subjected to disease classification, where the model identifies and categorizes specific pathological conditions such as black rot, worm infestation, scale insect damage, or dry skin. This two-tiered decision-making framework facilitates both quality control for healthy produce and early detection of disease in defective samples, thereby supporting enhanced agricultural management and market optimization.
- Model Evaluation: Once trained, the models were evaluated using several performance metrics. These included accuracy, which quantifies the proportion of correct predictions; the ROC curve, which evaluates performance across different classification thresholds; and the F-score, which balances precision and recall, providing insight into the model’s robustness.
2.1. YOLOv11 Model
2.1.1. YOLOv11 Architecture Overview
- Backbone: Extracts hierarchical features via convolutional operations, enhanced with Cross-Stage Partial (CSP) structures and SiLU activations for efficient representation and gradient flow.
- –
- Encodes low- and high-level features.
- –
- Applies spatial downsampling.
- –
- Outputs semantically rich feature maps.
- Neck: Refines and fuses multi-scale features using a BiFPN++ structure, improving detection across object sizes [46].
- –
- Aggregates spatial and contextual information.
- –
- Boosts detection performance on varied object scales.
- Head: The head is designed to output the final predictions of the model, including bounding boxes, class scores, objectness confidence, and, optionally, segmentation masks. YOLOv11 utilizes a decoupled head architecture that separates classification and localization into distinct branches. This separation enables the network to focus on each task independently, thereby improving accuracy and interpretability, particularly in complex object detection scenarios [47].
- –
- Utilizes a decoupled head architecture for specialized learning.
- –
- Includes
- *
- Classification head—assigns class probabilities.
- *
- Segmentation head—generates masks for segmentation tasks (optional).
Layer-Wise Functional Overview
2.1.2. YOLOv11 Training Optimization for Date Type and Disease Classification
- Exponential Moving Average (EMA): Integrated EMA tracking to smooth parameter updates, stabilizing model weights during training and improving inference performance.
- Augmentations: Combined advanced augmentation techniques including Mosaic, MixUp, CutMix, CopyPaste, HSV jitter, and Random Affine transformations to increase data diversity and improve generalization.
- Dynamic Label Assignment: Strategies such as SimOTA++ and TaskAligned assignment were employed to dynamically match predictions with targets, enhancing the effectiveness of supervision during training.
- Input Resolution: A fixed input image size of was selected to optimize the trade-off between detection precision and real-time performance, particularly for small lesions and fine-grained date textures.
- Learning Rate Scheduling: A cosine annealing schedule with warm-up epochs was adopted to gradually reduce the learning rate, promoting stable convergence.
- Loss Weight Balancing: The weights of classification, objectness, and box regression loss components were fine-tuned to address class imbalance and improve detection accuracy.
- Batch Accumulation: A batch size of 32 was used alongside gradient accumulation (2×) to accommodate GPU memory constraints, while maintaining stable gradients.
- Object Confidence Threshold Tuning: The confidence threshold (conf_threshold) was empirically adjusted to 0.25 to reduce false positives without compromising recall.
- Patch and Position Embedding (Layer 1): The input image is divided into non-overlapping patches, each linearly projected and embedded with positional encodings and a learnable classification token.
- Linear Projection (Layer 2): Flattened patch embeddings are mapped into a latent feature space via a linear projection layer.
- YOLOv11/opt Backbone (Layers 3–9):
- –
- Conv (Layer 3, ×7): Convolutional layers for low- and mid-level hierarchical feature extraction.
- –
- C3K2 (Layer 4, ×6): Cross-Stage Partial blocks with residual connections to enhance feature reuse and learning efficiency.
- –
- SPPF (Layer 5, ×1): Spatial Pyramid Pooling Fast module for multi-scale receptive field aggregation.
- –
- C2PSA (Layer 6, ×1): Cross-Channel and Parallel Self-Attention module for channel-wise and spatial feature refinement.
- –
- Concat (Layer 7, ×1): Concatenation of multi-resolution feature maps to preserve diverse spatial information.
- –
- Upsample (Layer 8, ×1): Upsampling operation to restore spatial resolution before detection.
- –
- Detect (Layer 9, ×1): Detection head for bounding box regression and objectness prediction.
- Classification Module (Layer 10): Final classification of detected instances into predefined categories, such as specific fruit types or disease types.
| Algorithm 1 Optimized YOLOv11 Object Detection and Fine-Tuning Framework |
| Require: Pretrained YOLOv11 model , labeled dataset Ensure: Optimized model for real-time object detection
|
3. Experimental Results
3.1. Experimental Settings
3.1.1. Work Environment
3.1.2. Parameter Settings
3.1.3. Evaluation Metrics
3.2. Comparative Analysis
3.2.1. Yolov11 Performance Results Across Different Date Types
3.2.2. Date Disease Detection Performance
3.2.3. YOLOv11-Opt Performance in Disease Detection
3.2.4. Object Detection Performance Based on mAP and IoU Thresholds
3.3. Visual Analytics for Strategic Management of Date Production
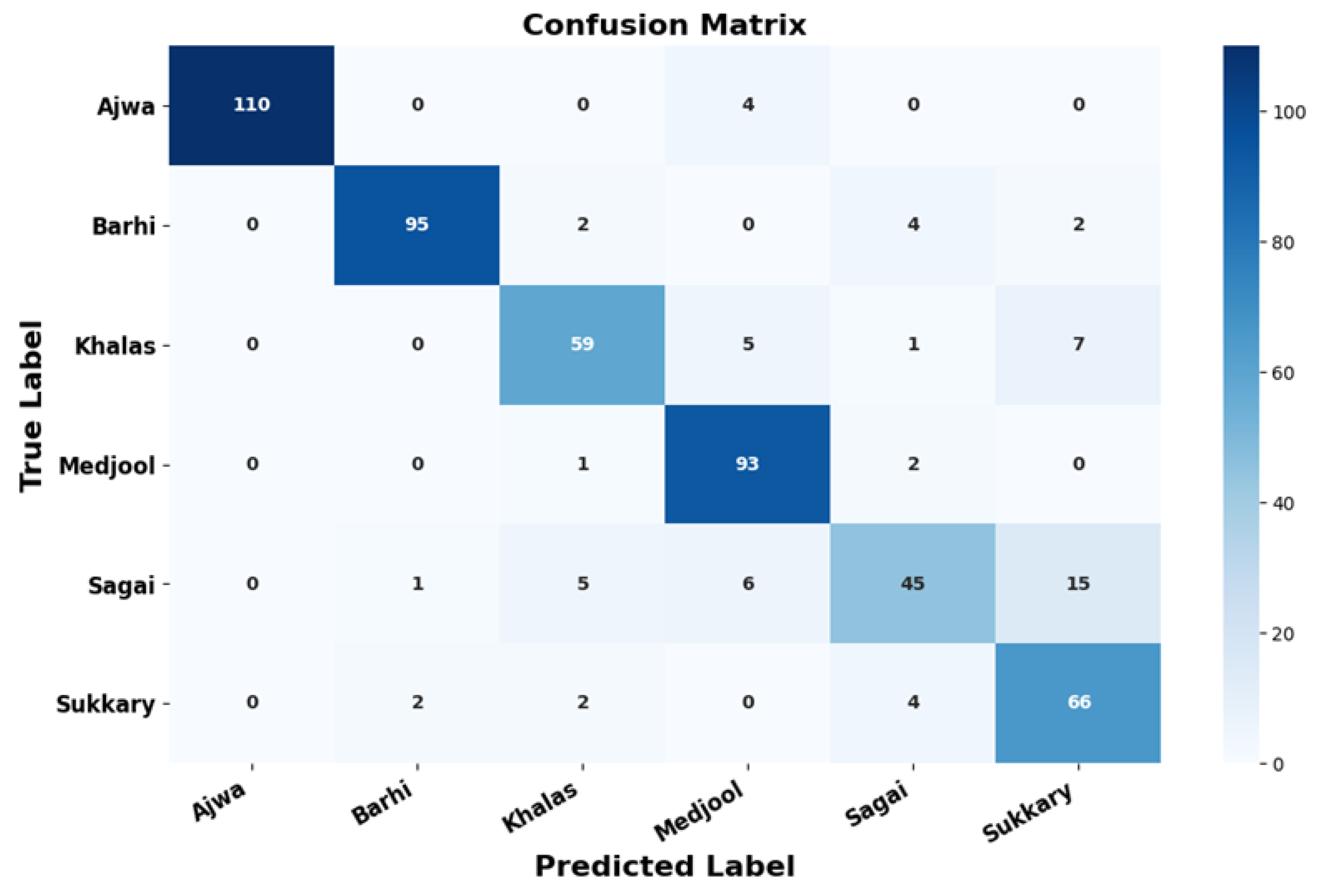
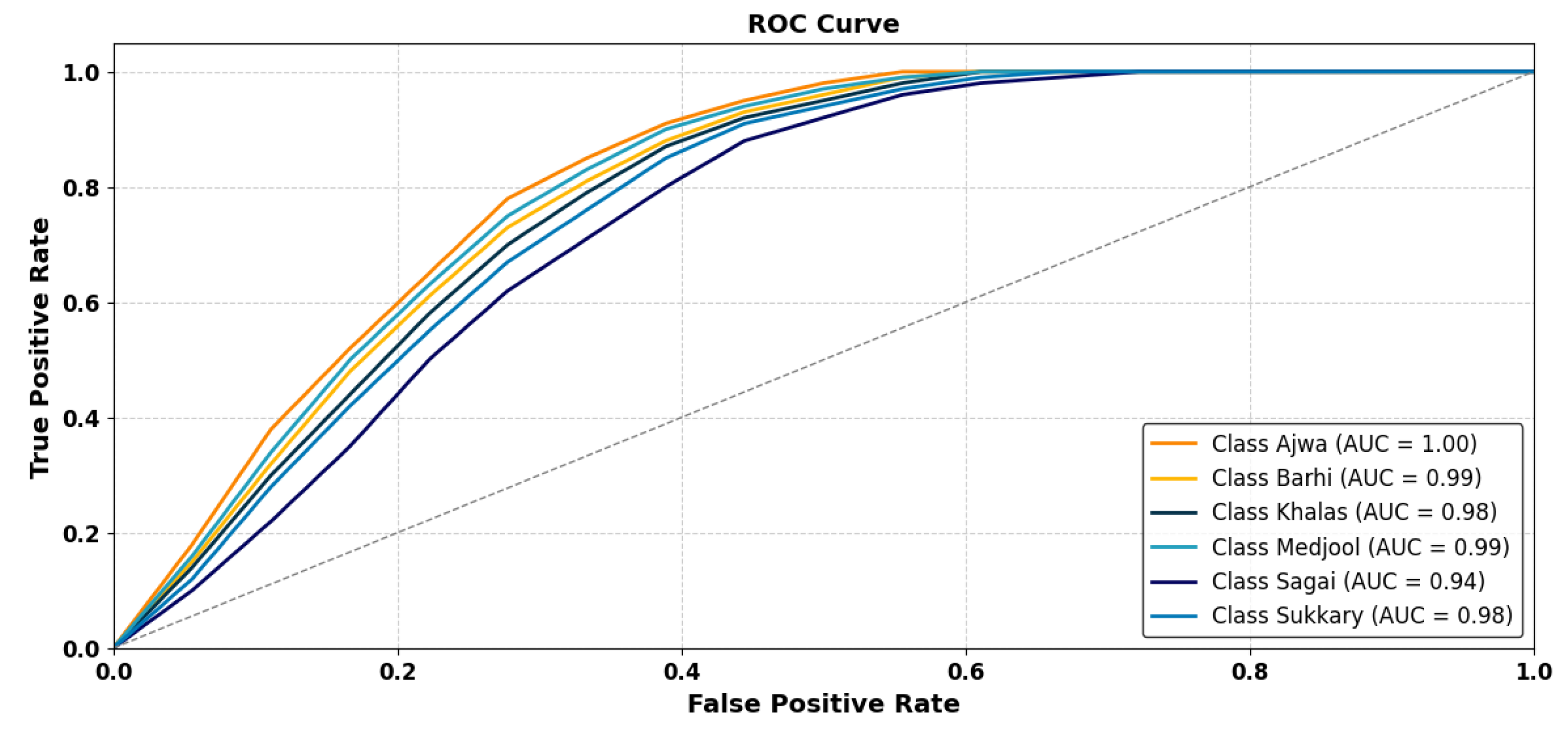
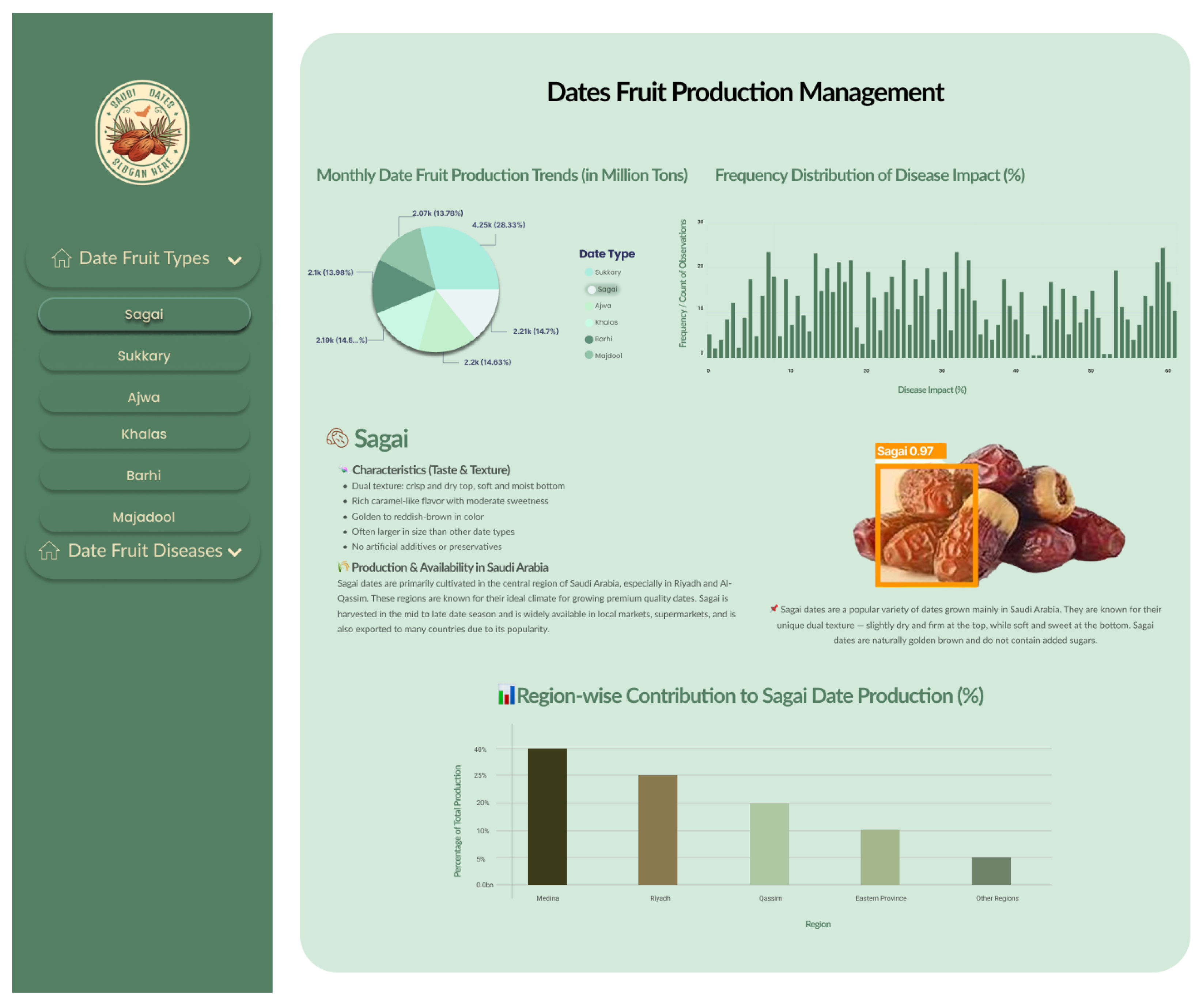
- Date Type Analytics—Figure 11: This visualization integrates categorical and temporal indicators to present the distribution of dominant date varieties, inter-seasonal yield variability, and comparative productivity levels. It facilitates identification of high-yield cultivars and production imbalances, offering a foundational reference for strategic resource allocation, genetic selection, and market-driven planning. The YOLOv11-Opt model achieved an average classification accuracy of 99.04% and ROC-AUC of 0.9961 across six major varieties (Ajwa, Barhi, Khalas, Medjool, Sagai, and Sukkary), demonstrating highly reliable predictive performance in varietal recognition.
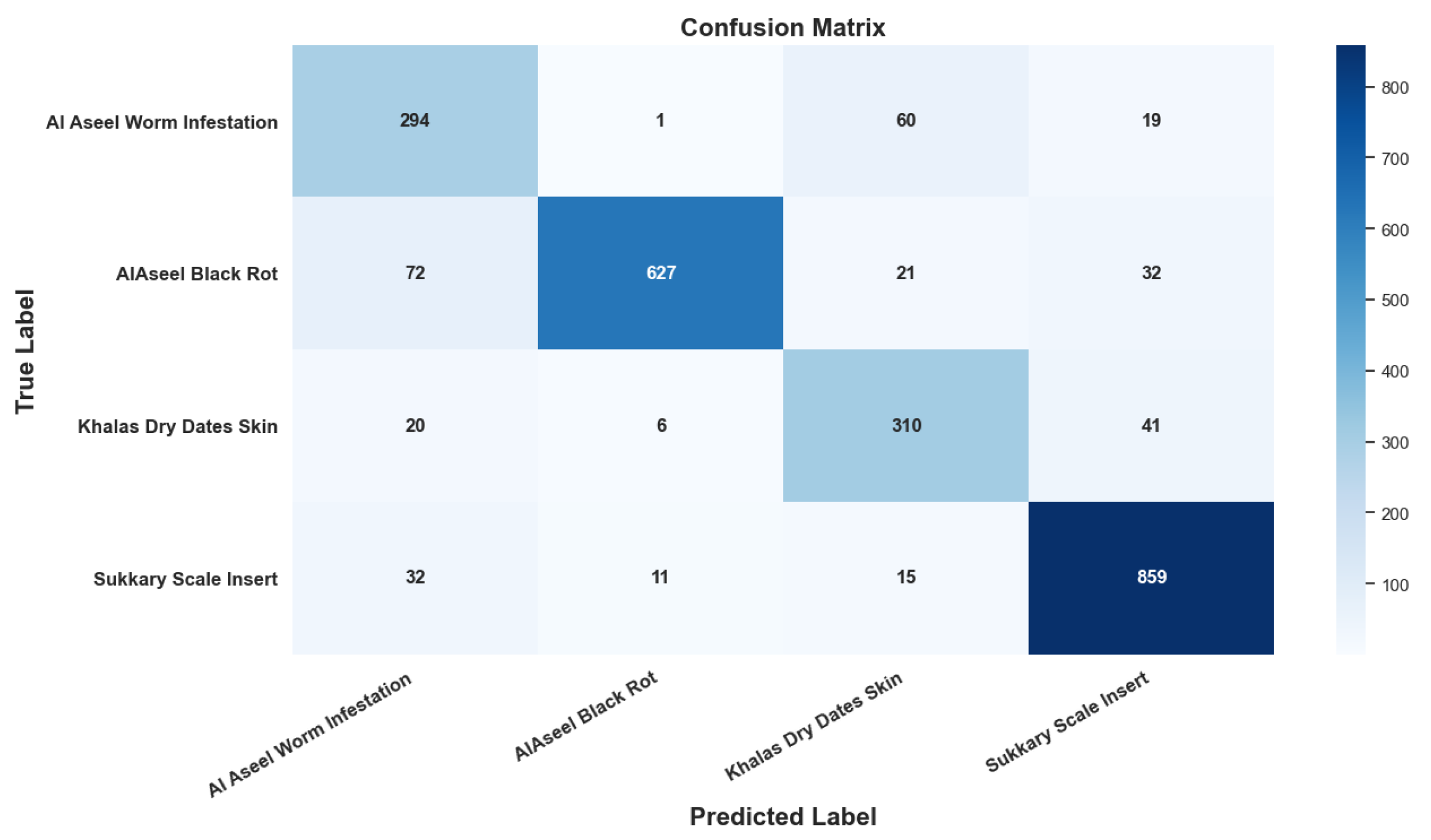
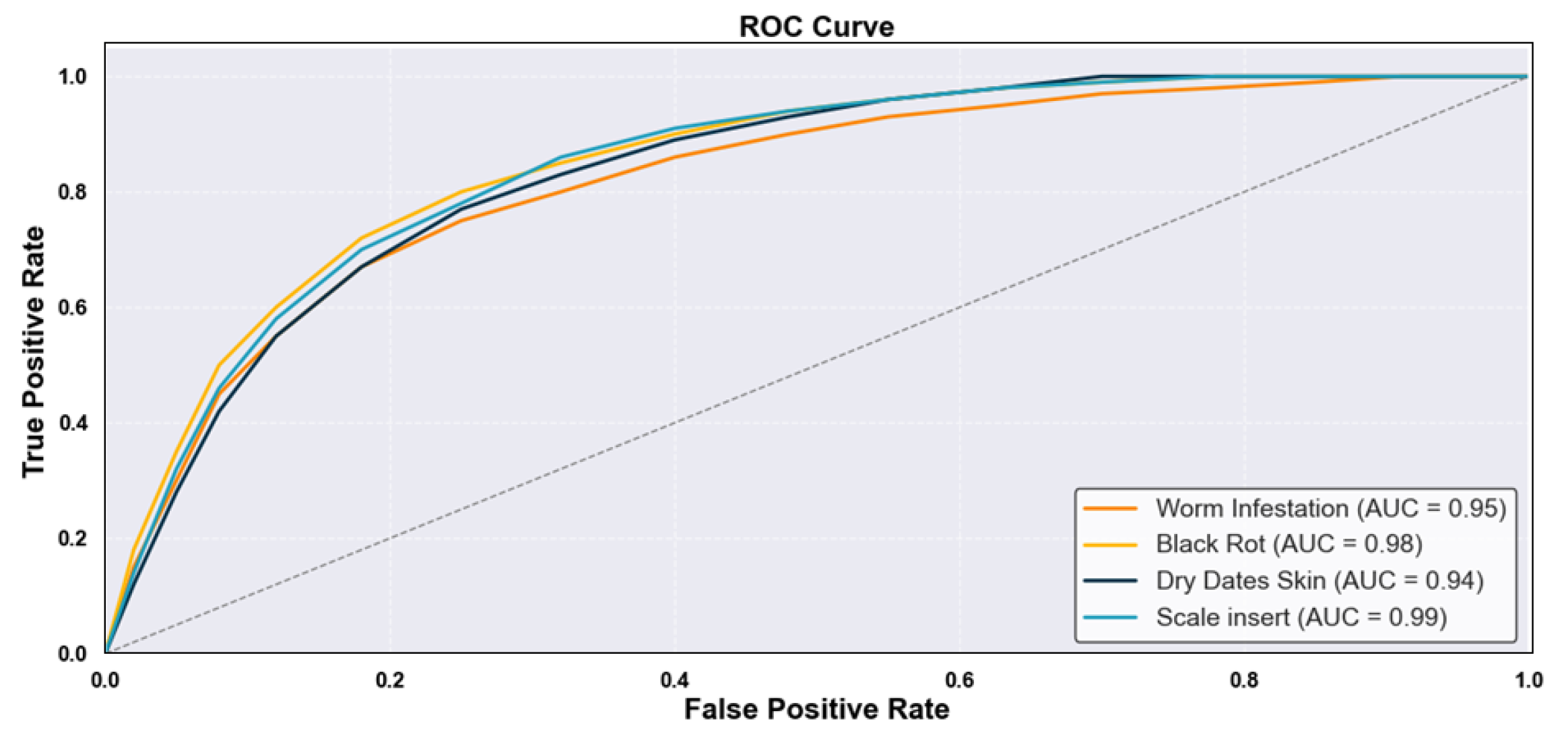
- Disease Monitoring—Figure 12: The analysis captures disease incidence and severity levels across recorded samples. By highlighting both frequent and sporadic outbreaks, the visualization aids in prioritizing phytosanitary measures and aligning early warning systems with real-time agronomic data. The insights serve as a decision-support layer for surveillance, treatment scheduling, and long-term health risk mitigation. The system demonstrated an average disease detection accuracy of 99.69% and an F1-score of 99.83% across conditions such as Worm Infestation, Black Rot, Scale Insert, and Dry Date Skin, confirming the robustness and precision of the proposed framework in real-world agricultural environments.
4. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Badr, A.; Allam, R.; Hassan, E. Advances in Date Palm Production: Challenges and Opportunities. Agric. Food Secur. 2019, 8, 20. [Google Scholar]
- Abdullah, M.; Al-Farsi, S. Nutritional Benefits and Health Effects of Dates: A Comprehensive Review. J. Food Sci. 2017, 82, 223–232. [Google Scholar] [CrossRef]
- Qazi, I.; Ali, Q.S.; Saad, M.; Ahmad, Z.; Khanb, M.U.; Rabbi, M.K.; ur Rahman, M. Explore the Economic Significance of the Dhakki Date Industry in the Local Region, Including its Contribution to Employment, Income Generation, and Overall Economic Development. Indus J. Biosci. Res. 2023, 1, 1–7. [Google Scholar]
- Zaid, A. Date Palm Cultivation; Food & Agriculture Organization: Rome, Italy, 2024. [Google Scholar]
- Alkatheri, A.H.; Alkatheeri, M.S.; Cheng, W.H.; Thomas, W.; Lai, K.S.; Lim, S.H.E. Innovations in extractable compounds from date seeds: Farms to future. AIMS Agric. Food 2024, 9, 256–281. [Google Scholar] [CrossRef]
- Rahman, M.; Al-Suhaibani, A.; Alghamdi, S. Water Management Strategies for Date Palm Production: A Review of Sustainable Practices. Desalin. Water Treat. 2020, 187, 156–163. [Google Scholar] [CrossRef]
- Mohamadizadeh, M.; Dehghan, P.; Azizi-Soleiman, F.; Maleki, P. Effectiveness of date seed on glycemia and advanced glycation end-products in type 2 diabetes: A randomized placebo-controlled trial. Nutr. Diabetes 2024, 14, 37. [Google Scholar] [CrossRef]
- Kiesler, R.; Franke, H.; Lachenmeier, D.W. A comprehensive review of the nutritional composition and toxicological profile of date seed coffee (Phoenix dactylifera). Appl. Sci. 2024, 14, 2346. [Google Scholar] [CrossRef]
- Al-Wajid, A.; Zawawi, M.; Siddique, M. Therapeutic Potential of Date Palm Seeds and Their Extracts. Pharmacogn. Res. 2018, 10, 289–296. [Google Scholar] [CrossRef]
- Majid, M.; Al-Khayri, J.M.; Jain, S.M. Anatomical Assessment of Skin Separation in Date Palm (Phoenix dactylifera L.) Fruit. Agriculture 2024, 13, 38. [Google Scholar]
- Himanshu; Kumar, N.; Khangwal, I.; Upadhyay, A. Assessment of nutritional composition, phytochemical screening, antioxidant, and antibacterial activities of date palm (Phoenix dactylifera) seeds. Discover Food 2024, 4, 151. [Google Scholar] [CrossRef]
- Karimi, E.; Dehghan, P.; Azizi-Soleiman, F.; Mohamadizadeh, M. Date seed (Phoenix dactylifera) supplementation modulates oxidative DNA damage, lipid peroxidation, and cardiometabolic risk factors in type 2 diabetes: A triple-blinded randomized placebo-controlled trial. J. Funct. Foods 2024, 117, 106226. [Google Scholar] [CrossRef]
- Elkeilani, M.; Al-Kayal, A. Biofuel Production from Date Palm By-Products: Current Status and Future Prospects. Renew. Sustain. Energy Rev. 2020, 132, 110037. [Google Scholar] [CrossRef]
- Al-Farsi, M.; Lee, C.Y. Nutritional and functional properties of dates: A review. Crit. Rev. Food Sci. Nutr. 2008, 48, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Gunnars, K. Medjool Dates: Nutrition, Benefits, and Uses. Healthline. 2024. Available online: https://www.healthline.com/nutrition/medjool-dates (accessed on 7 April 2025).
- Al-Shahib, W.; Marshall, R.J. Date fruits (Phoenix dactylifera L.): An overview. Food Res. Int. 2003, 36, 999–1013. [Google Scholar]
- Shabani, L.; Rezaee, M.; Azarpazhooh, E.; Ghaffari, H. Phytochemicals and biological activities of Phoenix dactylifera L. (date palm): A comprehensive review. J. Ethnopharmacol. 2022, 285, 114914. [Google Scholar]
- Chandio, F.A.; Liu, W.; Shah, S.H. Precision Agriculture and Data-Driven Decision Support for Sustainable Crop Management. J. Agric. Inform. 2021, 12, 1–12. [Google Scholar]
- Alawadhi, T.; Alfaris, M.; Alzahrani, M. Artificial Intelligence in Agriculture: A Case Study on Date Palm Farming. J. Agric. Eng. 2022, 8, 232–248. [Google Scholar]
- Alaoui, A.O.; Boutaleb Joutei, A. Date Palm Scale and Their Management. World J. Agric. Soil Sci. 2024, 9, WJASS.MS.ID.000710. [Google Scholar]
- Aziz, D.; Rafiq, S.; Saini, P.; Ahad, I.; Gonal, B.; Rehman, S.A.; Rashid, S.; Saini, P.; Rohela, G.K.; Aalum, K.; et al. Remote sensing and artificial intelligence: Revolutionizing pest management in agriculture. Front. Sustain. Food Syst. 2025, 9, 1551460. [Google Scholar] [CrossRef]
- Mohanty, S.; Hughes, D.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2021, 12, 1419. [Google Scholar] [CrossRef]
- Ahmed, F.; Malik, H. YOLOv7 for real-time detection of tomato plant diseases in complex backgrounds. Plant Methods 2023, 19, 25. [Google Scholar]
- Liu, Y.; Zhang, H. Vision Transformers for plant disease classification: A comparative study. Sensors 2023, 23, 2755. [Google Scholar]
- Albarrak, K.M.; Sorour, S.E. Web-Enhanced Vision Transformers and Deep Learning for Accurate Event-Centric Management Categorization in Education Institutions. Systems 2024, 12, 475. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M. Deep residual learning for detection of wheat rust disease. Comput. Electron. Agric. 2022, 197, 106951. [Google Scholar]
- Islam, M.; Rahman, M. Fungal infection detection in rice using UAV imagery and EfficientNet. Precis. Agric. 2024, 25, 123–136. [Google Scholar]
- Chen, J.; Wang, H. MobileNet-based attention model for real-time apple leaf disease classification. Comput. Electron. Agric. 2023, 206, 107586. [Google Scholar]
- Khanam, R.; Ali, M. YOLOv11: A hybrid lightweight model for high-precision object detection in agriculture. IEEE Trans. Image Process. 2024, 33, 1125–1138. [Google Scholar]
- Zafar, A. AI-integrated IoT architecture for predictive pest control in precision farming. J. Agric. Inform. 2024, 15, 45–60. [Google Scholar]
- Ma, Y.; Chen, L. LSTM-based yield forecasting model for wheat using time-series meteorological data. Agric. Syst. 2022, 195, 103327. [Google Scholar]
- Karthik, R.; Manogaran, G. Hyperspectral deep learning model for early sugarcane disease detection. Comput. Electron. Agric. 2022, 194, 106705. [Google Scholar]
- Shahid, M.; Zubair, M. IoT-enabled real-time analytics platform for corn nitrogen optimization. Sensors 2023, 23, 1356. [Google Scholar]
- Al-Mssallem, I.; Al-Dous, E.; Al-Moammar, K.; Mimida, S.N.; Alothman, Z.; Islam, S.; Alkuraya, A.; Reitz, T.R.; Ahmed, I.; Mahfouz, M.M. Recent developments in date palm genomics and molecular breeding. Front. Genet. 2022, 13, 959266. [Google Scholar]
- Tjoa, E.; Guan, C. Explainable AI: A review of methods and applications. Inf. Fusion 2021, 77, 1–15. [Google Scholar]
- Zhang, Y.; Sun, W. SHAP-based model interpretation for soybean root rot prediction using ensemble learning. Biosyst. Eng. 2023, 229, 90–102. [Google Scholar]
- Khanam, R.; Hussain, M. Yolov11: An overview of the key architectural enhancements. arXiv 2024, arXiv:2410.17725. [Google Scholar]
- Jocher, G.; Qiu, J. Ultralytics YOLO11. 2024. Available online: https://github.com/ultralytics/ultralytics (accessed on 1 May 2025).
- Liao, Y.; Li, L.; Xiao, H.; Xu, F.; Shan, B.; Yin, H. YOLO-MECD: Citrus Detection Algorithm Based on YOLOv11. Agronomy 2025, 15, 687. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, L. Fruit ripeness classification using YOLOv4 and image augmentation on embedded edge devices. Agric. Syst. 2021, 187, 102988. [Google Scholar]
- Zhang, H.; Liu, Q.; Chen, W. Research on litchi image detection in orchard using UAV based on improved YOLOv5. Sensors 2023, 23, 1123. [Google Scholar] [CrossRef]
- Alahi, A.; Singh, V. A Lightweight YOLO-Based Architecture for Apple Detection on Embedded Systems. Comput. Electron. Agric. 2021, 192, 106573. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, L.; Xu, K. YOLOv1 to v8: Unveiling Each Variant—A Comprehensive Review of YOLO. Inf. Fusion 2023, 95, 101786. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Sorour, S.E.; Aljaafari, M.; Alarfaj, A.A.; AlMusallam, W.H.; Aljoqiman, K.S. Fine-tuned Vision Transformers and YOLOv11 for precise detection of pediatric Adenoid Hypertrophy. Alex. Eng. J. 2025, 128, 366–393. [Google Scholar] [CrossRef]
- Tan, M.; Pang, R.; Le, Q.V. EfficientDet: Scalable and efficient object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 10781–10790. [Google Scholar]
- Liu, T.; Bai, Q.; Torigian, D.A.; Tong, Y.; Udupa, J.K. VSmTrans: A hybrid paradigm integrating self-attention and convolution for 3D medical image segmentation. Med. Image Anal. 2024, 98, 103295. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Searching for activation functions. Nat. Commun. 2020, 11, 1–10. [Google Scholar]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. Scaled-YOLOv4: Scaling Cross Stage Partial Network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 13029–13038. [Google Scholar]
- Ding, X.; Zhang, X.; Han, J.; Ding, G. RepVGG: Making VGG-style ConvNets Great Again. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 13733–13742. [Google Scholar]
- Li, C.; Li, L.; Jiang, H.; Weng, K.; Geng, Y.; Li, L.; Ke, Z.; Li, Q.; Cheng, M.; Nie, W.; et al. YOLOv6: A single-stage object detection framework for industrial applications. arXiv 2022, arXiv:2209.02976. [Google Scholar]
- Sorour, S.E.; Mine, T.; Goda, K.; Hirokawa, S. A predictive model to evaluate student performance. J. Inf. Process. 2015, 23, 192–201. [Google Scholar] [CrossRef]
- Sorour, S.E.; Mine, T.; Goda, K.; Hirokawa, S. Predicting students’ grades based on free style comments data by artificial neural network. In Proceedings of the 2014 IEEE Frontiers in Education Conference (FIE) Proceedings, Madrid, Spain, 22–25 October 2014; pp. 1–9. [Google Scholar]
- De Medeiros, A.K.A.; Guzzo, A.; Greco, G.; Van der Aalst, W.M.; Weijters, A.; Van Dongen, B.F.; Saccà, D. Process mining based on clustering: A quest for precision. In Proceedings of the Business Process Management Workshops: BPM 2007 International Workshops, BPI, BPD, CBP, ProHealth, RefMod, semantics4ws, Brisbane, Australia, 24 September 2007; Springer: Berlin/Heidelberg, Germany, 2008; pp. 17–29. [Google Scholar]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Amigó, E.; Gonzalo, J.; Artiles, J.; Verdejo, F. A comparison of extrinsic clustering evaluation metrics based on formal constraints. Inf. Retr. 2009, 12, 461–486. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Diagnostic tests 1: Sensitivity and specificity. BMJ 1994, 308, 1552. [Google Scholar] [CrossRef] [PubMed]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef]
- Everingham, M.; Van Gool, L.; Williams, C.K.I.; Winn, J.; Zisserman, A. The Pascal Visual Object Classes (VOC) Challenge. Int. J. Comput. Vis. 2010, 88, 303–338. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Lawrence Zitnick, C. Microsoft COCO: Common objects in context. In Proceedings of the ECCV, Zurich, Switzerland, 6–12 September 2014; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]


| Layer | Functional Description |
|---|---|
| Conv + BN + SiLU | Implements convolutional filtering, followed by batch normalization and SiLU activation to facilitate nonlinear feature transformation, improve convergence stability, and enhance representational capacity [48]. |
| CSPBlock | Enhances feature reuse and network depth, while reducing computational complexity by partitioning feature maps into cross-stage paths, thus improving learning efficiency and inference speed [49]. |
| RepConv | Introduces a multi-branch convolutional structure optimized during training, which is re-parameterized into a single convolution during inference for speed and memory efficiency [50]. |
| SPPF+ | Aggregates multi-scale contextual features using an optimized Spatial Pyramid Pooling module, thereby improving detection of objects at varying scales and positions [44]. |
| BiFPN++ | Provides adaptive feature fusion across scales using a bidirectional weighted mechanism with learnable coefficients, facilitating robust feature propagation and alignment [46]. |
| Decoupled Head | Segregates the detection head into distinct branches for objectness scoring, bounding box regression, and class prediction, improving task specialization and detection accuracy [51]. |
| Parameter | Optimized Value/Strategy | Contribution/Effect |
|---|---|---|
| imgsz | 640 × 640 | Enhanced detail capture for small lesions |
| batch_size | 32 + gradient accumulation (×2) | Balanced memory and convergence |
| learning_rate | 0.01 (cosine annealing + warm-up) | Stable and effective convergence |
| weight_decay | 0.0005 | Reduced overfitting risk |
| momentum | 0.937 | Smoothed weight updates |
| optimizer | SGD with momentum | Effective in large-scale vision models |
| lr_scheduler | Cosine annealing (warm-up = 3 epochs) | Controlled decay, improved generalization |
| conf_threshold | 0.25 | Balanced precision-recall trade-off |
| box_loss_gain | 0.05 | Focused on localization accuracy |
| cls_loss_gain | 0.5 | Prioritized correct classification |
| obj_loss_gain | 1.0 | Balanced detection objectness |
| Component | Specification |
|---|---|
| Processor | Intel(R) Core(TM) i7-8750H @ 2.20 GHz (6 cores, 12 threads) |
| RAM | 16 GB |
| System Architecture | 64-bit, x64-based |
| GPU | NVIDIA GPU with CUDA support |
| Operating System | Windows + WSL2 (Windows Subsystem for Linux) |
| Library/Tool | Version | Purpose |
|---|---|---|
| Python | 3.9.16 | Primary programming language. |
| PyTorch | 1.12.1+cu113 | Deep learning framework with CUDA 11.3 support. |
| Torchvision | 0.13.1 | Pretrained models and image transforms for PyTorch. |
| Ultralytics YOLO | v8.0.112 | Model loading, training, and evaluation. |
| Albumentations | 1.3.1 | Data augmentation with field-variation simulation. |
| Albumentations.pytorch | ToTensorV2 | Tensor conversion with channel normalization. |
| OpenCV (cv2) | 4.8.0 | Image reading, resizing, and preprocessing. |
| scikit-learn | 1.2.2 | Data splitting and evaluation metrics. |
| NumPy | 1.24.2 | Numerical array manipulation. |
| Pandas | 1.5.3 | Handling metadata and analysis logs. |
| Matplotlib | 3.7.1 | Plotting training results and confusion matrices. |
| Seaborn | 0.12.2 | Statistical data visualization (e.g., heatmaps). |
| tqdm | 4.65.0 | Progress bar during processing loops. |
| glob (built-in) | - | Pattern-based file retrieval. |
| shutil (built-in) | - | File operations (copy, move). |
| os (built-in) | - | Path and environment management. |
| random (built-in) | - | Seed setting for reproducibility. |
| time (built-in) | - | Execution time benchmarking. |
| %matplotlib inline | - | Notebook-based inline plot rendering. |
| Parameter | Value |
|---|---|
| Data Augmentation | |
| Horizontal Flip | 0.5 |
| Vertical Flip | 0.2 |
| Rotation | 0.5 |
| Brightness & Contrast Adjustment | 0.3 |
| Gaussian Blur | 0.3 |
| Gaussian Noise | 0.3 |
| Shift-Scale-Rotate | 0.5 |
| Coarse Dropout | 0.3 |
| Convert to Tensor | 0.1 |
| Training Configuration | |
| Epochs | 100 |
| Batch Size | 16 |
| Optimizer | SGD |
| Learning Rate | 0.1 |
| Weight Decay | 0.0005 |
| Momentum | 0.9 |
| Device | CUDA |
| Model | Date Type | Accuracy | Precision | Recall | F-Score | Specificity | ROC |
|---|---|---|---|---|---|---|---|
| YOLOv11 | Ajwa | 0.987 | 0.985 | 0.983 | 0.985 | 0.982 | 0.986 |
| Barhi | 0.984 | 0.984 | 0.986 | 0.982 | 0.982 | 0.987 | |
| Khalas | 0.975 | 0.975 | 0.976 | 0.973 | 0.982 | 0.977 | |
| Medjool | 0.988 | 0.984 | 0.986 | 0.981 | 0.980 | 0.985 | |
| Sagai | 0.986 | 0.985 | 0.983 | 0.983 | 0.986 | 0.987 | |
| Sukkary | 0.985 | 0.983 | 0.980 | 0.987 | 0.988 | 0.981 | |
| Avg. Overall | 0.986 | 0.982 | 0.982 | 0.981 | 0.982 | 0.982 | |
| YOLOv11-Opt | Ajwa | 0.994 | 0.999 | 0.993 | 0.990 | 0.987 | 0.994 |
| Barhi | 0.995 | 0.994 | 0.997 | 0.994 | 0.995 | 0.996 | |
| Khalas | 0.996 | 0.996 | 0.993 | 0.994 | 0.995 | 0.996 | |
| Medjool | 0.995 | 0.9983 | 0.997 | 0.993 | 0.996 | 0.995 | |
| Sagai | 0.985 | 0.988 | 0.989 | 0.985 | 0.983 | 0.993 | |
| Sukkary | 0.987 | 0.991 | 0.989 | 0.991 | 0.993 | 0.992 | |
| Avg. Overall | 0.990 | 0.990 | 0.993 | 0.995 | 0.995 | 0.996 |
| Date Type | Disease | Accuracy | Precision | Recall | F1-Score | Specificity | ROC-AUC |
|---|---|---|---|---|---|---|---|
| Al Aseel | Worm Infestation | 0.983 | 0.985 | 0.980 | 0.983 | 0.982 | 0.989 |
| Black Rot | 0.984 | 0.980 | 0.986 | 0.988 | 0.989 | 0.985 | |
| Dry Date Skin | 0.980 | 0.982 | 0.986 | 0.980 | 0.985 | 0.982 | |
| Scale Insert | 0.987 | 0.985 | 0.986 | 0.986 | 0.985 | 0.986 | |
| Sukkary | Worm Infestation | 0.985 | 0.980 | 0.981 | 0.976 | 0.976 | 0.973 |
| Black Rot | 0.981 | 0.982 | 0.988 | 0.990 | 0.985 | 0.980 | |
| Dry Date Skin | 0.986 | 0.985 | 0.980 | 0.973 | 0.970 | 0.987 | |
| Scale Insert | 0.986 | 0.989 | 0.988 | 0.988 | 0.988 | 0.989 | |
| Khalas | Worm Infestation | 0.979 | 0.961 | 0.974 | 0.967 | 0.976 | 0.984 |
| Black Rot | 0.982 | 0.985 | 0.980 | 0.983 | 0.987 | 0.974 | |
| Dry Date Skin | 0.990 | 0.980 | 0.975 | 0.978 | 0.977 | 0.983 | |
| Scale Insert | 0.987 | 0.976 | 0.979 | 0.979 | 0.984 | 0.987 | |
| Average Overall | 0.985 | 0.986 | 0.985 | 0.987 | 0.988 | 0.986 | |
| Date Type | Disease | Accuracy | Precision | Recall | F1-Score | Specificity | ROC-AUC |
|---|---|---|---|---|---|---|---|
| Al Aseel | Worm Infestation | 0.990 | 0.996 | 0.999 | 0.997 | 0.998 | 0.993 |
| Black Rot | 0.998 | 0.895 | 0.999 | 0.992 | 0.995 | 0.993 | |
| Dry Date Skin | 0.992 | 0.993 | 0.994 | 0.999 | 0.997 | 0.993 | |
| Scale Insert | 0.995 | 0.997 | 0.993 | 0.993 | 0.996 | 0.995 | |
| Sukkary | Worm Infestation | 0.995 | 0.993 | 0.994 | 0.998 | 0.996 | 0.997 |
| Black Rot | 0.993 | 0.990 | 0.992 | 0.996 | 0.991 | 0.998 | |
| Dry Date Skin | 0.997 | 0.998 | 0.999 | 0.994 | 0.992 | 0.998 | |
| Scale Insert | 0.997 | 0.999 | 0.990 | 0.993 | 0.996 | 0.998 | |
| Khalas | Worm Infestation | 0.999 | 0.995 | 0.996 | 0.999 | 0.987 | 0.993 |
| Black Rot | 0.992 | 0.998 | 0.998 | 0.993 | 0.995 | 0.992 | |
| Dry Date Skin | 0.991 | 0.992 | 0.996 | 0.999 | 0.998 | 0.995 | |
| Scale Insert | 0.996 | 0.998 | 0.994 | 0.998 | 0.998 | 0.991 | |
| Average Overall | 0.997 | 0.994 | 0.994 | 0.998 | 0.993 | 0.996 | |
| Model | mAP@0.5 | mAP@0.5:0.95 |
|---|---|---|
| YOLOv11—Date Type | 0.979 | 0.974 |
| YOLOv11-Opt—Date Type | 0.989 | 0.995 |
| YOLOv11—Disease Detection | 0.981 | 0.987 |
| YOLOv11-Opt—Disease Detection | 0.993 | 1.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorour, S.E.; Alsayyari, M.; Alqahtani, N.; Aldosery, K.; Altaweel, A.; Alzhrani, S. An Intelligent Management System and Advanced Analytics for Boosting Date Production. Sustainability 2025, 17, 5636. https://doi.org/10.3390/su17125636
Sorour SE, Alsayyari M, Alqahtani N, Aldosery K, Altaweel A, Alzhrani S. An Intelligent Management System and Advanced Analytics for Boosting Date Production. Sustainability. 2025; 17(12):5636. https://doi.org/10.3390/su17125636
Chicago/Turabian StyleSorour, Shaymaa E., Munira Alsayyari, Norah Alqahtani, Kaznah Aldosery, Anfal Altaweel, and Shahad Alzhrani. 2025. "An Intelligent Management System and Advanced Analytics for Boosting Date Production" Sustainability 17, no. 12: 5636. https://doi.org/10.3390/su17125636
APA StyleSorour, S. E., Alsayyari, M., Alqahtani, N., Aldosery, K., Altaweel, A., & Alzhrani, S. (2025). An Intelligent Management System and Advanced Analytics for Boosting Date Production. Sustainability, 17(12), 5636. https://doi.org/10.3390/su17125636







