Precision Farming Multimodal Technologies Using Optical Sensors for the Detection of Citrus Tristeza Virus Endemics
Abstract
1. Introduction
2. Materials and Methods
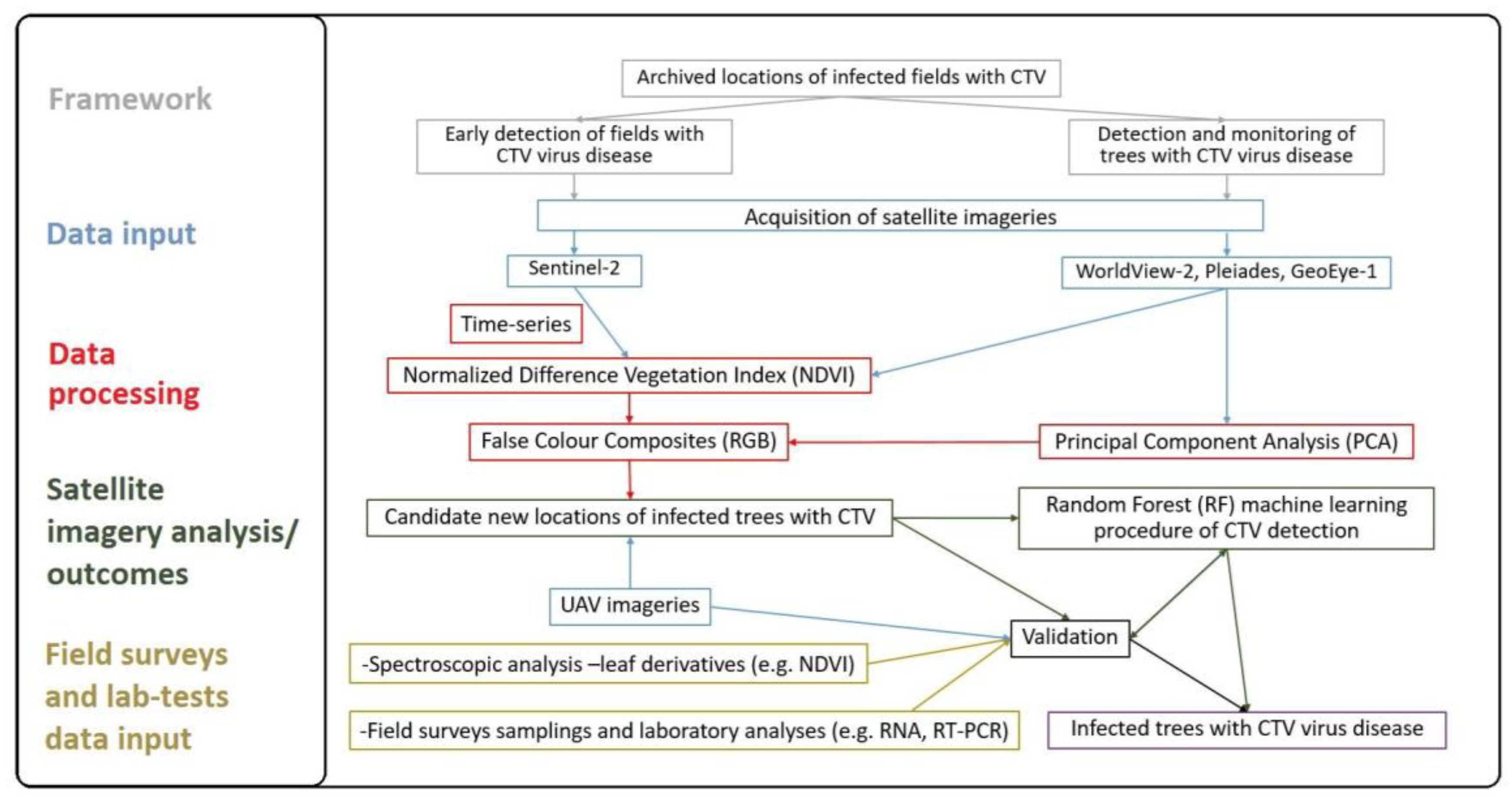
2.1. Early Detection of Fields with CTV Using Remote Sensing
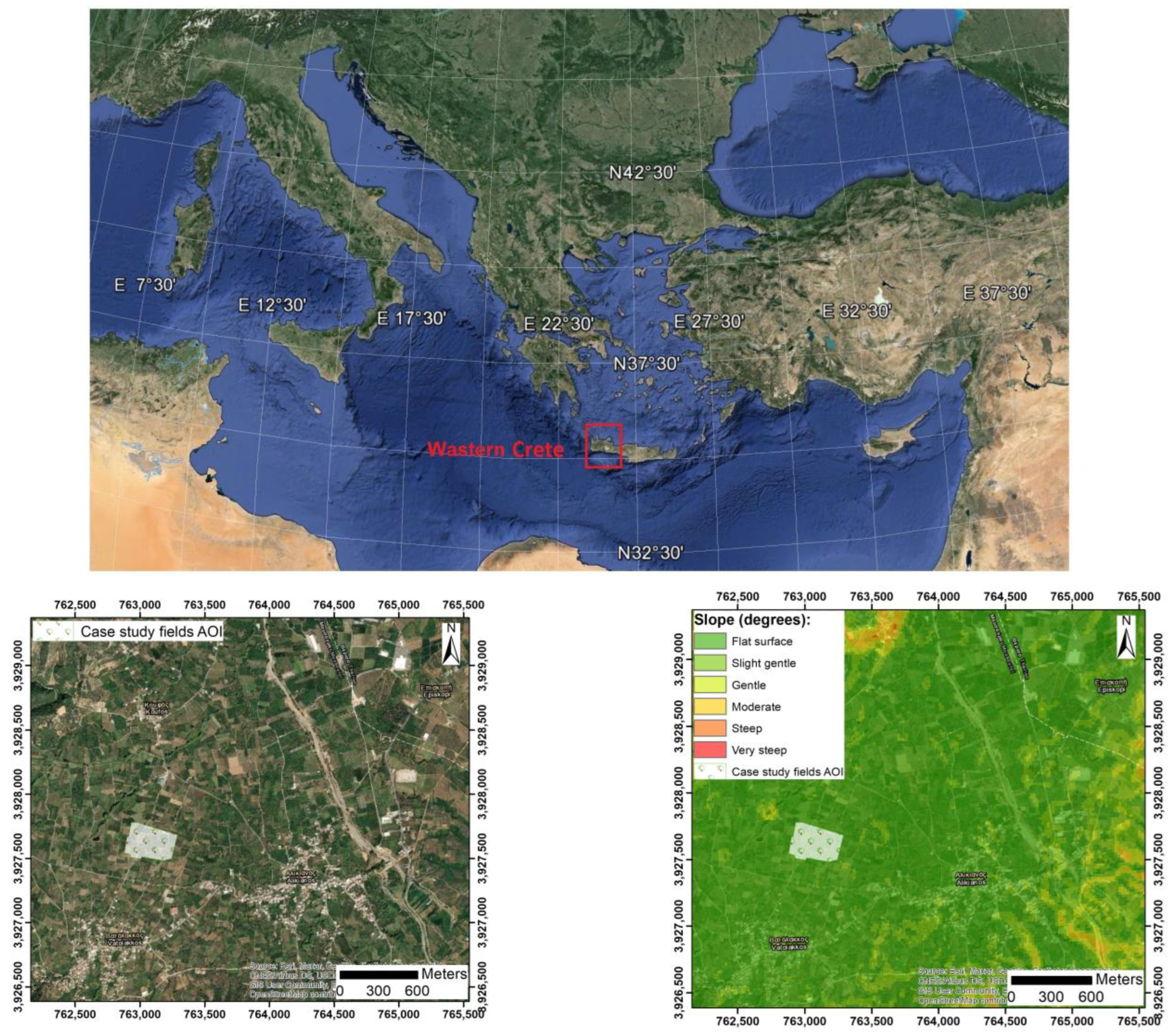
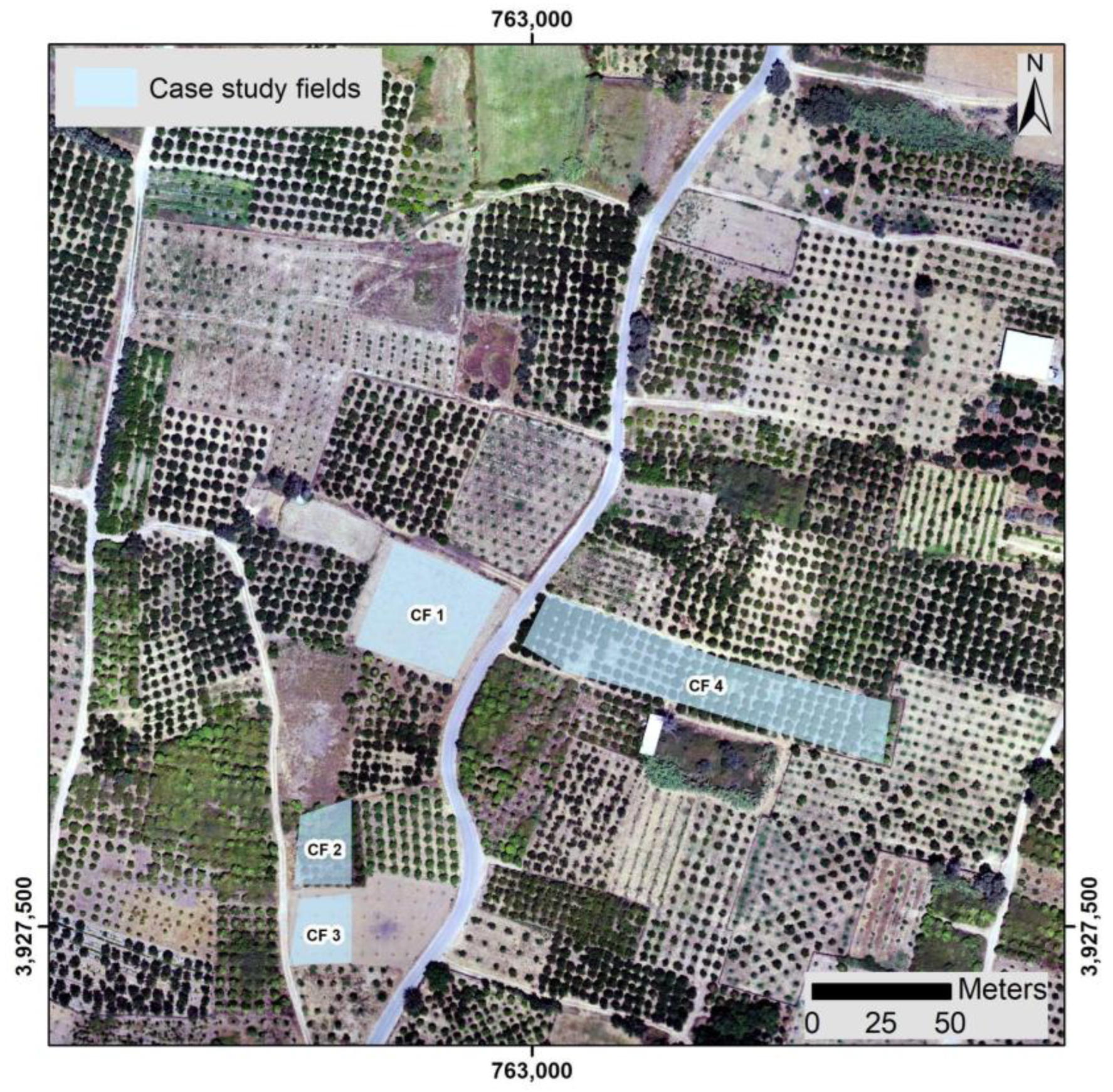
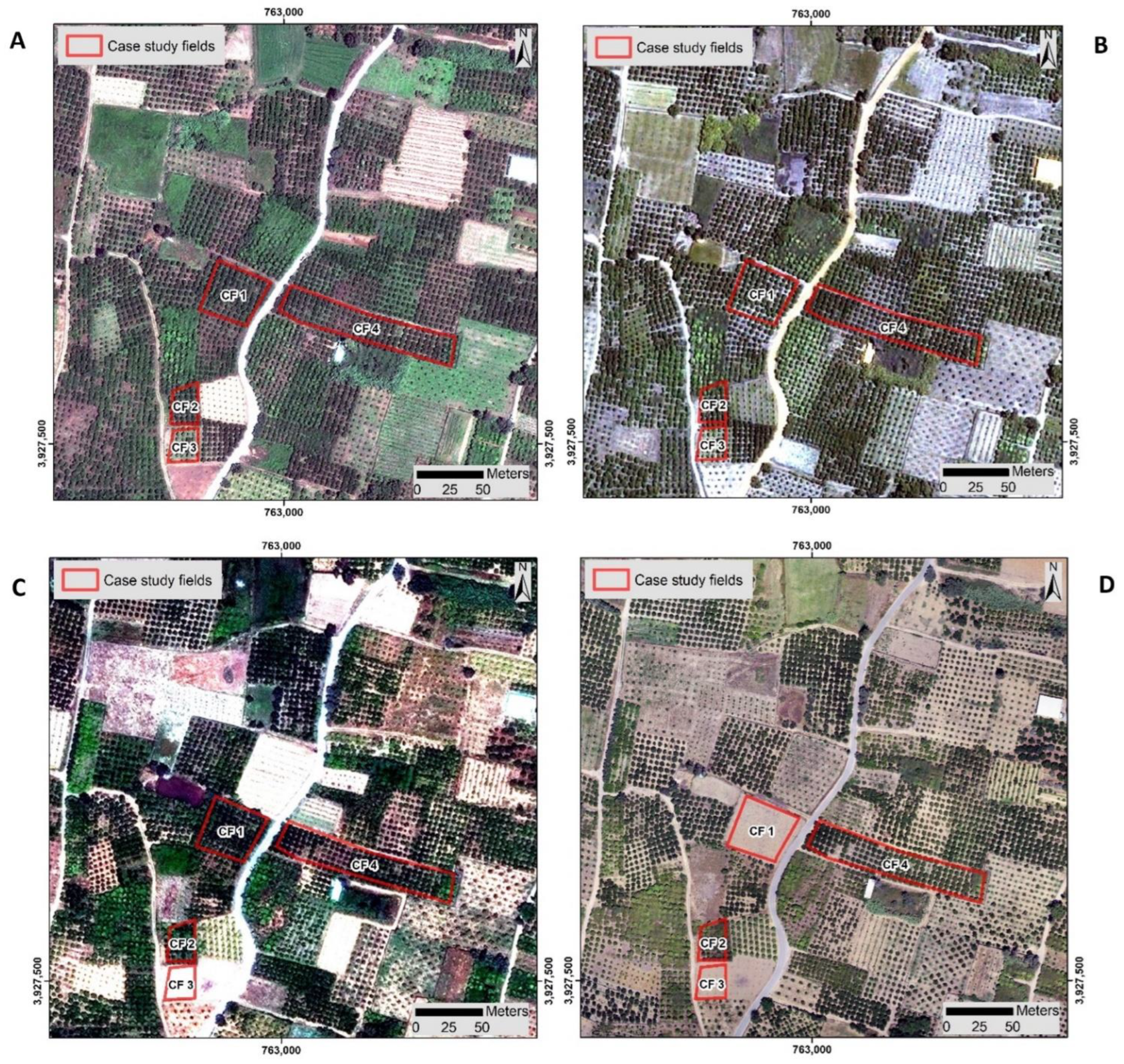
2.1.1. Case Study Infected Fields
2.1.2. Time-Series Visualization of CTV Infection Using Remote Sensing
2.2. Detection and Monitoring of Trees with CTV Using Remote Sensing
2.2.1. Satellite Imageries
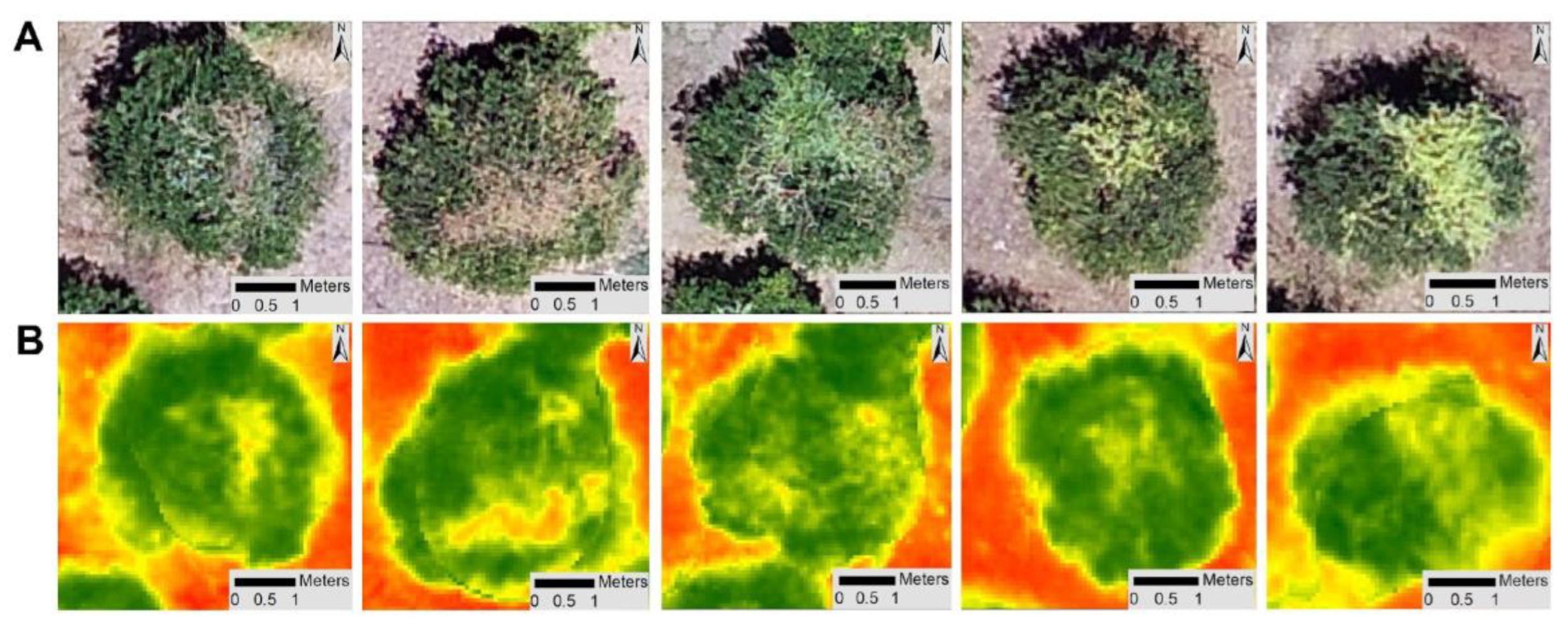
2.2.2. Unmanned Autonomous Vehicles (UAVs) Imageries
2.3. Validation
2.3.1. Field Surveys and CTV Detection
Leaf Sampling Collection
Virus Detection and Sequencing
Spectroscopy Analysis
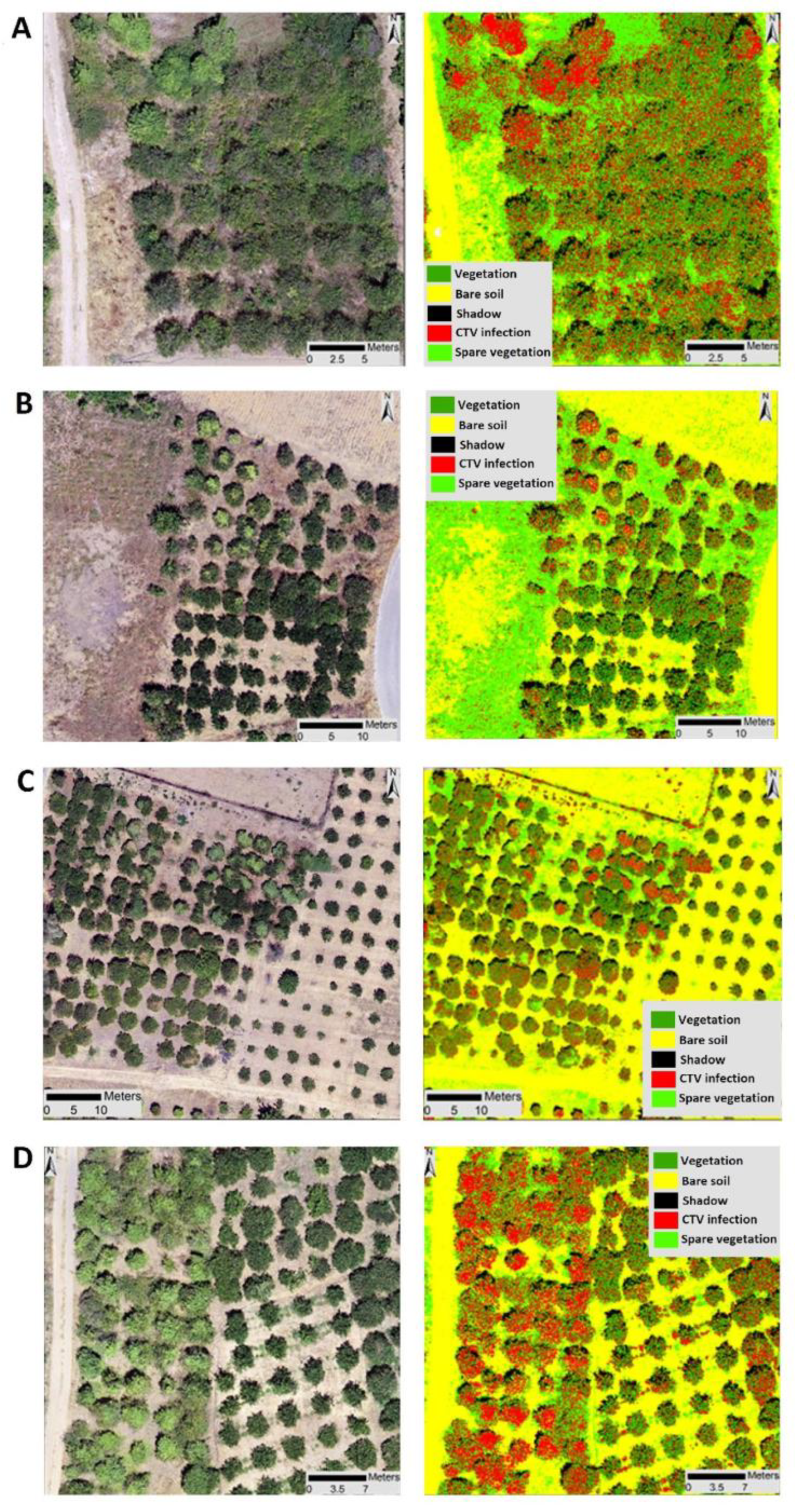
2.4. Semi-Automatic Machine Learning Procedure of CTV Detection and Accuracy Assessment
3. Results
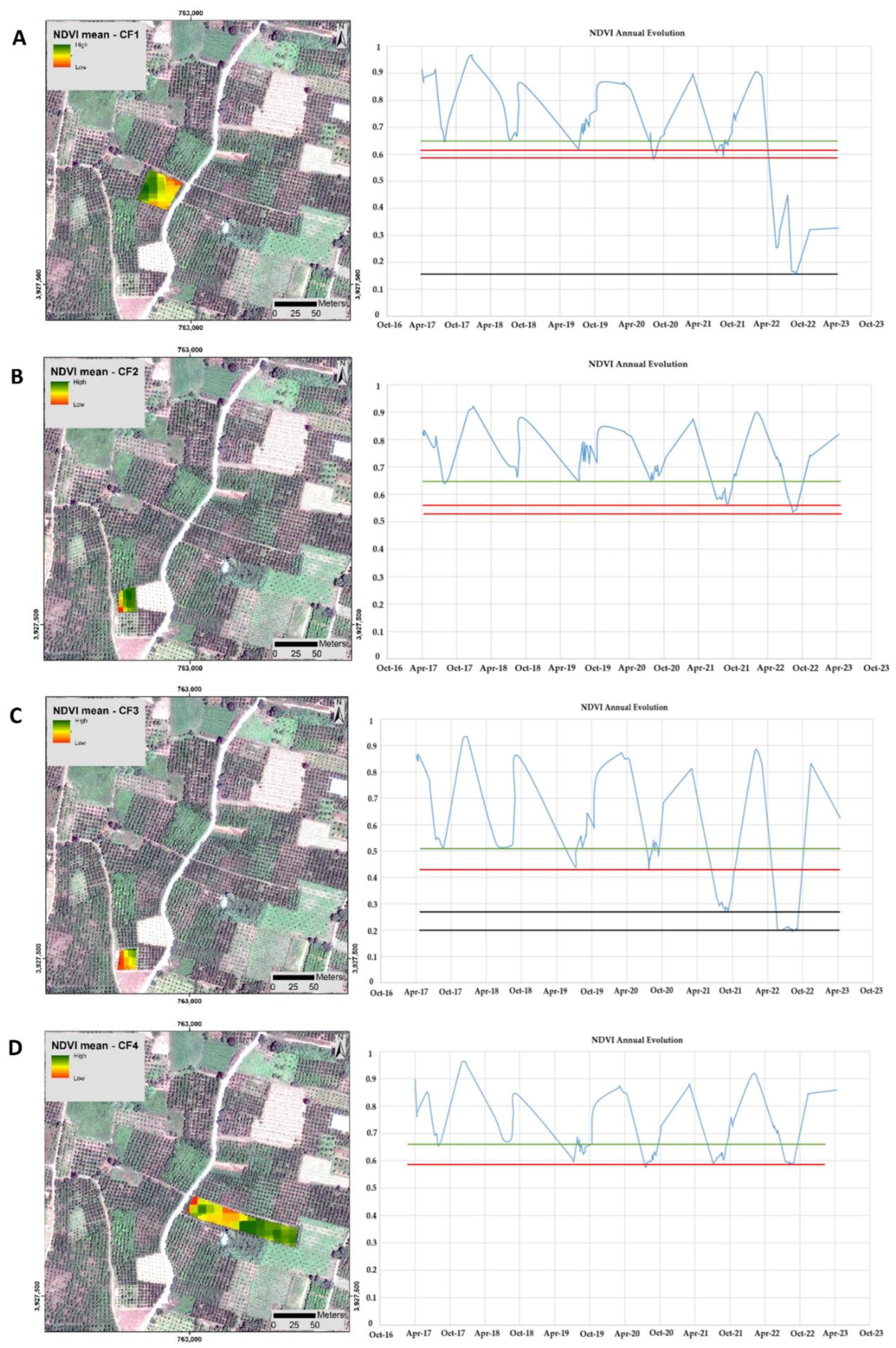
3.1. Time-Series Early Visualization of CTV and Epidemiology Using Remote Sensing
3.2. Detection and Monitoring of Trees with CTV Using Remote Sensing
3.3. Validation
3.4. Semi-Automatic Machine Learning Procedure of CTV Detection and Accuracy Assessment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A




| Candidate Field | Survey Year | Host | No. Collected Samples | 2021 | 2022 | ||
|---|---|---|---|---|---|---|---|
| Tissue-Print ELISA | RT-PCR | Tissue-Print ELISA | RT-PCR | ||||
| CdF1 | 2021 autumn | Orange | 3 | 0 | 0 | 0 | 0 |
| CdF2 | 2021 autumn | Orange | 3 | 1 (33.4%) | 1 (33.4%) | 2 (66.7%) | 2 (66.7%) |
| CdF3 | 2021 autumn | Orange | 2 | 0 | 0 | 1 (50%) | 1 (50%) |
| CdF4 | 2021 autumn | Orange | 3 | 2 (66.7%) | 2 (66.7%) | 3 (100%) | 3 (100%) |
| CdF5 | 2021 autumn | Orange | 3 | 1 (33.4%) | 3 (100%) | 2 (66.7%) | 3 (100%) |
| CdF6 | 2022 spring | Orange | 2 | n.a | n.a | 1 (50%) | 1 (50%) |
| CdF7 | 2022 spring | Orange | 6 | n.a | n.a | 4 (66.6%) | 5 (83.4%) |
| CdF8 | 2022 spring | Orange | 8 | n.a | n.a | 4 (50%) | 6 (75%) |
| CdF9 | 2022 spring | Orange | 5 | n.a | n.a | 2 (40%) | 3 (60%) |
| Percentage of candidate CTV-infected trees being infected based on laboratory analyses | 35 | 4/14 (28.5%) | 6/14 (43%) | 19/35 (54.3%) | 24/35 (68.57%) | ||
| WorldView-2 | Eigenvalues | % of covariance |
|---|---|---|
| PC 1 | 8984.86 | 66.58 |
| PC 2 | 4343.18 | 32.18 |
| PC 3 | 147.42 | 1 |
| PC 4 | 18.01 | 0.1 |
| Sum | 13,493.17 | 100 |
| Pleiades | Eigenvalues | % of covariance |
| PC 1 | 343,505.9 | 73.4 |
| PC 2 | 1,200,056.88 | 25.65 |
| PC 3 | 3994.88 | 0.85 |
| PC 4 | 372.2 | 0.08 |
| Sum | 467,928.88 | 100 |
| GeoEye-1 | Eigenvalues | % of covariance |
| PC 1 | 42,036.74 | 67 |
| PC 2 | 20,359.23 | 32.46 |
| PC 3 | 270.38 | 0.43 |
| PC 4 | 53.93 | 0.08 |
| Sum | 62,720.28 | 100 |
| A. WorldView-2 | ||||
|---|---|---|---|---|
| Factor loadings | PC 1 | PC 2 | PC 3 | PC 4 |
| Band 1 | 0.35 | 0.05 | −0.37 | 0.85 |
| Band 2 | 0.64 | 0.18 | −0.53 | −0.51 |
| Band 3 | 0.65 | 0.01 | 0.75 | 0.04 |
| Band 4 | −0.15 | 0.98 | 0.1 | 0.04 |
| B. Pleiades | ||||
| Factor loadings | PC 1 | PC 2 | PC 3 | PC 4 |
| Band 1 | 0.66 | −0.43 | −0.6 | 0.054 |
| Band 2 | 0.44 | −0.21 | 0.57 | −0.66 |
| Band 3 | 0.33 | −0.16 | 0.54 | 0.74 |
| Band 4 | 0.5 | 0.85 | −0.06 | 0.01 |
| C. GeoEye-1 | ||||
| Factor loadings | PC 1 | PC 2 | PC 3 | PC 4 |
| Band 1 | 0.12 | −0.46 | −0.51 | 0.7 |
| Band 2 | 0.16 | −0.47 | −0.5 | −0.7 |
| Band 3 | 0.12 | −0.71 | 0.69 | 0.012 |
| Band 4 | 0.97 | 0.22 | 0.063 | 0.03 |
| Confusion Matrix | Building | Vegetation | Bare Soil | Shadow | CTV-Infected Trees | Sparse Vegetation |
|---|---|---|---|---|---|---|
| Vegetation | 0 | 41 | 0 | 1 | 0 | 0 |
| Bare soil | 0 | 0 | 58 | 0 | 0 | 0 |
| Shadow | 0 | 0 | 0 | 30 | 0 | 0 |
| CTV infection | 0 | 7 | 6 | 0 | 139 | 13 |
| Sparse vegetation | 0 | 0 | 1 | 0 | 3 | 16 |
| Overall Accuracy (OA) | 0.897 | |||||
| Kappa statistics | 0.85 | |||||
| User’s accuracy | Producer’s accuracy | |||||
| Vegetation | 0.97 | Vegetation | 0.85 | |||
| Bare soil | 0.96 | Bare soil | 0.89 | |||
| Shadow | 1 | Shadow | 0.96 | |||
| CTV infection | 0.84 | CTV infection | 0.97 | |||
| Sparse vegetation | 0.8 | Sparse vegetation | 0.52 | |||
References
- FAO. FAOSTAT Database. 2020. Available online: http://www.fao.org/faostat/en/#data (accessed on 20 December 2023).
- Tennant, P.F.; Robinson, D.; Fisher, L.; Bennett, S.M.; Hutton, D.; Coates-Beckford, P.; McLaughlin, W. Diseases and pests of citrus (Citrus spp.). Tree For. Sci. Biotechnol. 2009, 3, 81–107. [Google Scholar]
- Bar-Joseph, M.; Marcus, R.; Lee, R.F. The continuous challenge of Citrus tristeza virus control. Annu. Rev. Phytopathol. 1989, 27, 291–316. [Google Scholar] [CrossRef]
- Moreno, P.; Ambrós, S.; Albiach-Martí, M.R.; Guerri, J.; Peña, L. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 2008, 9, 251–268. [Google Scholar] [CrossRef]
- Karasev, A.; Boyko, V.; Gowda, S.; Nikolaeva, O.; Hilf, M.; Koonin, E.; Niblett, C.; Cline, K.; Gumpf, D.; Lee, R.; et al. Complete Sequence of the Citrus Tristeza Virus RNA Genome. Virology 1995, 208, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Sabade, A.; López, C.; Rubio, L.; Flores, R.; Guerri, J.; Moreno, P. Polymorphism of a specific region in gene p23 of Citrus tristeza virus allows discrimination between mild and severe isolates. Arch. Virol. 2003, 148, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Sagheer, A.; Zhou, C.Y.; Zhou, Y.; Cao, M.J.; Wang, X.F. Distribution and research advances in Citrus tristeza virus. J. Integr. Agric. 2012, 11, 346–358. [Google Scholar]
- Bester, R.; Cook, G.; Maree, H.J. Citrus Tristeza Virus Genotype Detection Using High-Throughput Sequencing. Viruses 2021, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- ELSTAT. 2019. Available online: http://www.statistics.gr/el/statistics/agr (accessed on 15 January 2024).
- Beris, D.; Malandraki, I.; Vassilakos, N.; Theologidis, I.; Rampou, A.; Kektsidou, O.; Massart, S.; Varveri, C. Association of Citrus Virus A to Citrus Impietratura Disease Symptoms. Phytopathology 2021, 111, 1782–1789. [Google Scholar] [CrossRef]
- Mathioudakis, M.M.; Tektonidis, N.; Karagianni, A.; Mikalef, L.; Gómez, P.; Hasiów-Jaroszewska, B. Incidence and Epidemiology of Citrus Viroids in Greece: Role of Host and Cultivar in Epidemiological Characteristics. Viruses 2023, 15, 605. [Google Scholar] [CrossRef]
- Katsarou, K.; Chiumenti, M.; Kalantidis, K.; Mathioudakis, M.M. First Report of Citrus Viroids Infecting Persian (Tahiti) Lime in Greece. Plant Dis. 2020, 104, 998. [Google Scholar] [CrossRef]
- Kyriakopoulou, P.E. Virus and virus like diseases of citrus in Greece and the Greek certification program. Int. Organ. Citrus Virol. Conf. Proc. 2002, 15, 408–411. [Google Scholar] [CrossRef]
- Dimou, D.; Drossopoulou, J.; Moschos, E.; Varveri, C.; Bem, F. First report of Citrus tristeza virus in Greece. Plant Dis. 2002, 86, 329. [Google Scholar] [CrossRef] [PubMed]
- Owen, C.; Mathioudakis, Μ.M.; Gazivoda, A.; Gal, P.; Nol, N.; Kalliampakou, K.; Figas, A.; Bellan, A.; Iparaguirre, A.; Rubio, L.; et al. Evolution and molecular epidemiology of Citrus tristeza virus on Crete: Recent introduction of a severe strain. J. Phytopathol. 2014, 162, 839–843. [Google Scholar] [CrossRef]
- Chatzivassiliou, E.K.; Nolasco, G. Detection of a new variant of Citrus tristeza virus in Greek citrus crops. Phytopathol. Mediterr. 2015, 53, 140–147. [Google Scholar]
- Varveri, C.; Olmos, A.; Pina, J.A.; Marroquín, C.; Cambra, M. Biological and molecular characterization of a distinct Citrus tristeza virus isolate originating from a lemon tree in Greece. Plant Pathol. 2015, 64, 792–798. [Google Scholar] [CrossRef]
- Ghosh, D.K.; Kokane, S.B.; Gowda, S. Development of a reverse transcription recombinase polymerase based isothermal amplification coupled with lateral flow immunochromatographic assay (CTV-RT-RPA-LFICA) for rapid detection of Citrus tristeza virus. Sci. Rep. 2020, 10, 20593. [Google Scholar] [CrossRef]
- Wang, Y.M.; Ostendorf, B.; Gautam, D.; Habili, N.; Pagay, V. Plant Viral Disease Detection: From Molecular Diagnosis to Optical Sensing Technology—A Multidisciplinary Review. Remote Sens. 2022, 14, 1542. [Google Scholar] [CrossRef]
- Beck, P.S.; Martínez Sanchez, A.L.; Di Leo, M.; Chemin, Y.; Caudullo, G.; de la Fuente, B.; Zarco-Tejada, P.J. Remote Sensing in support of Plant Health Measures. In Findings from the Canopy Health Monitoring (CanHeMon) Project; EUR 29724 EN; Publications Office of the European Union: Luxembourg, 2019; ISBN 978-92-76-02051-6. JRC115403. [Google Scholar] [CrossRef]
- Terentev, A.; Dolzhenko, V.; Fedotov, A.; Eremenko, D. Current State of Hyperspectral Remote Sensing for Early Plant Disease Detection: A Review. Sensors 2022, 22, 757. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Pu, R.; Gonzalez-Moreno, P.; Yuan, L.; Wu, K.; Huang, W. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165, 104943. [Google Scholar] [CrossRef]
- Gualano, S.; Santoro, F.; Djelouah, K.; D’Onghia, A.M. Proximal and remote sensing in the monitoring of Citrus Tristeza Virus (CTV) infected trees: Preliminary results. In Proceedings of the XXVIII International Horticultural Congress on Science and Horticulture for People (IHC2010): International Symposium on the Challenge for a Sustainable Production, Protection and Consumption of Mediterranean Fruits and Nuts, Lisbon, Portugal, 22–27 August 2010; Article number: 940_88. pp. 641–646. [Google Scholar]
- Janssen, L.L.F.; Bakker, W.H. Principles of Remote Sensing: An Introductory Textbook, 2nd ed.; ITC Educational Textbook Series; International Institute for Aerospace Survey and Earth Sciences: Enschede, The Netherlands, 2001; p. 180. [Google Scholar]
- Afonso, A.M.; Guerra, R.; Cavaco, A.M.; Pinto, P.; Andrade, A.; Duarte, A.; Power, D.M.; Marques, N.T. Identification of asymptomatic plants infected with Citrus tristeza virus from a time series of leaf spectral characteristics. Comput. Electron. Agric. 2017, 141, 340–350. [Google Scholar] [CrossRef]
- Deng, X.; Zhu, Z.; Yang, J.; Zheng, Z.; Huang, Z.; Yin, X.; Wei, S.; Lan, Y. Detection of Citrus Huanglongbing Based on Multi-Input Neural Network Model of UAV HRS. Remote Sens. 2020, 12, 2678. [Google Scholar] [CrossRef]
- Golhani, K.; Balasundram, S.K.; Vadamalai, G.; Pradhan, B. Use of reflectance spectroscopy as a tool for screening viroid-inoculated oil palm seedlings. OAJAR 2017, 2, 1–4. [Google Scholar]
- Santoro, F.; Gualano, S.; Djelouah, K.; Guario, A.; D’Onghia, A.M. Remote sensing to support the monitoring of Citrus tristeza virus (CTV) infected areas. In Citrus Tristeza Virus and Toxoptera Citricidus: A Serious Threat to the Mediterranean Citrus Industry; D’Onghia, A.M., Djelouah, K., Roistacher, C.N., Eds.; Options Méditerranéennes: Série B. Etudes et Recherches; n. 65; CIHEAM: Bari, Italy, 2009; pp. 165–171. [Google Scholar]
- Selvaraja, S.; Balasundram, S.K.; Vadamalai, G.; Husni, M.H.A.; Khosla, R. Remote Sensing as a Tool to Assess Orange Spotting Disease in Oil Palm (Elaies guineensis). Mitt. Klosterneubg. 2014, 64, 12–26. [Google Scholar]
- Tian, X.; Fan, S.; Huang, W.; Wang, Z.; Li, J. Detection of early decay on citrus using hyperspectral transmittance imaging technology coupled with principal component analysis and improved watershed segmentation algorithms. Postharvest Biol. Technol. 2020, 161, 111071. [Google Scholar] [CrossRef]
- Vanella, D.; Consoli, S.; Ramírez-Cuesta, J.M.; Tessitori, M. Suitability of the MODIS-NDVI Time-Series for a Posteriori Evaluation of the Citrus Tristeza Virus Epidemic. Remote Sens. 2020, 12, 1965. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Camino, C.; Beck, P.S.A.; Calderon, R.; Hornero, A.; Hernandez-Clemente, R.; Kattenborn, T.; Montes-Borrego, M.; Susca, L.; Morelli, M.; et al. Previsual symptoms of Xylella fastidiosa infections revealed in spectral plant-trait alterations. Nat. Plants 2018, 4, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Chu, B.; Zhang, C.; Liu, F.; Jiang, L.; He, Y. Hyperspectral imaging for presymptomatic detection of tobacco disease with successive projections algorithm and machine-learning classifiers. Sci. Rep. 2017, 7, 4125. [Google Scholar] [CrossRef]
- Calderon, R.; Navas-Cortes, J.A.; Lucena, C.; Zarco-Tejada, P.J. High resolution airborne hyperspectral and thermal imagery for early detection of Verticillium wilt of olive using fluorescence, temperature and narrow-band spectral indices. Remote Sens. Environ. 2013, 139, 231–245. [Google Scholar] [CrossRef]
- Zhang, C.; Kovacs, J.M. The application of small unmanned aerial systems for precision agriculture: A review. Precis. Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Radoglou-Grammatikis, P.; Sarigiannidis, P.; Lagkas, T.; Moscholios, I. A compilation of UAV applications for precision agriculture. Comput. Netw. 2020, 172, 107148. [Google Scholar] [CrossRef]
- Shegani, M.; Tsikou, D.; Velimirovic, A.; Afifi, H.; Karayianni, A.; Gazivoda, A.; Manevska, K.; Manakos, I.; Livieratos, I.C. Citrus tristeza virus on the island of Crete: A survey and detection protocol applications. J. Plant Pathol. 2012, 94, 71–78. [Google Scholar]
- Argyriou, A.V.; Sarris, A.; Teeuw, R.M. Using geoinformatics and geomorphometrics to quantify the geodiversity of Crete, Greece. Int. J. Appl. Earth Obs. Geoinf. 2016, 51, 47–59. [Google Scholar] [CrossRef]
- Fletcher, R.S.; Skaria, M. Response of ratio vegetation indices to foot rot-infected citrus trees. Evaluating airborne normalized difference vegetation index imagery for citrus orchard surveys. Int. J. Remote Sens. 2004, 25, 3967–3972. [Google Scholar] [CrossRef]
- Singh, A. Principal components analysis in remote sensing. In Proceedings of the IGARSS 93—IEEE International Geoscience and Remote Sensing Symposium, Tokyo, Japan, 18–21 August 1993; Volume 4, pp. 1680–1682. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Wintz, P. Digital Image Processing; Addison-Wesley: Boston, MA, USA, 1977. [Google Scholar]
- Ortega-Terol, D.; Hernandez-Lopez, D.; Ballesteros, R.; Gonzalez-Aguilera, D. Automatic Hotspot and Sun Glint Detection in UAV Multispectral Images. Sensors 2017, 17, 2352. [Google Scholar] [CrossRef] [PubMed]
- Mathioudakis, Μ.Μ.; Saponari, M.; Hasiów-Jaroszewska, B.; Elbeaino, T.; Koubouris, G. Detection of viruses in olive cultivars in Greece, using a rapid and effective RNA extraction method, for certification of virus-tested propagation material. Phytopathol. Mediterr. 2020, 59, 203–211. [Google Scholar] [CrossRef]
- Nolasco, G.; Sequeira, Z.; Soares, C.; Mansinho, A.; Bailey, A.M.; Niblett, C.L. Asymmetric PCR ELISA: Increased sensitivity and reduced costs for the detection of plant viruses. Eur. J. Plant Pathol. 2002, 108, 293–298. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Hornero, A.; Beck, P.S.A.; Kattenborn, T.; Kempeneers, P.; Hernández-Clemente, R. Chlorophyll content estimation in an open-canopy conifer forest with Sentinel-2A and hyperspectral imagery in the context of forest decline. Remote Sens. Environ. 2019, 223, 320–335. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Belgiu, M.; Dragut, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Chan, J.C.W.; Paelinckx, D. Evaluation of Random Forest and Adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Feng, L.; Guo, M.; Wang, W.; Chen, Y.; Shi, Q.; Guo, W.; Lou, Y.; Kang, H.; Chen, Z.; Zhu, Y. Comparative Analysis of Machine Learning Methods and a Physical Model for Shallow Landslide Risk Modeling. Sustainability 2023, 15, 6. [Google Scholar] [CrossRef]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support vector machine versus random forest for remote sensing image classification: A meta-analysis and systematic review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Wei, X.; Johnson, M.A.; Langston, D.B., Jr.; Mehl, H.L.; Li, S. Identifying optimal wavelengths as disease signatures using hyperspectral sensor and machine learning. Remote Sens. 2021, 13, 2833. [Google Scholar] [CrossRef]
- Adelabu, S.; Mutanga, O.; Adam, E. Evaluating the impact of red-edge band from Rapideye image for classifying insect defoliation levels. ISPRS J. Photogramm. Remote Sens. 2014, 95, 34–41. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, H.; Duan, J.; He, L.; Yuan, X.; Gao, Y.; Liu, W.; Li, X.; Feng, W. Improved Spectral Detection of Nitrogen Deficiency and Yellow Mosaic Disease Stresses in Wheat Using a Soil Effect Removal Algorithm and Machine Learning. Remote Sens. 2023, 15, 2513. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Advances in hyperspectral remote sensing of vegetation and agricultural croplands. In Hyperspectral Remote Sensing of Vegetation; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; CRC Press: Boca Raton, FL, USA, 2011; pp. 3–35. [Google Scholar]
- Wang, D.; Vinson, R.; Holmes, M.; Seibel, G.; Bechar, A.; Nof, S.; Tao, Y. Early Detection of Tomato Spotted Wilt Virus by Hyperspectral Imaging and Outlier Removal Auxiliary Classifier Generative Adversarial Nets (OR-AC-GAN). Sci. Rep. 2019, 9, 4377. [Google Scholar] [CrossRef]
- Kouadio, L.; El Jarroudi, M.; Belabess, Z.; Laasli, S.-E.; Roni, M.Z.K.; Amine, I.D.I.; Mokhtari, N.; Mokrini, F.; Junk, J.; Lahlali, R. A Review on UAV-Based Applications for Plant Disease Detection and Monitoring. Remote Sens. 2023, 15, 4273. [Google Scholar] [CrossRef]
- Siebring, J.; Valente, J.; Franceschini, M.H.D.; Kamp, J.; Kooistra, L. Object-based image analysis applied to low altitude aerial imagery for potato plant trait retrieval and pathogen detection. Sensors 2019, 19, 5477. [Google Scholar] [CrossRef]
- Yamamoto, K.; Togami, T.; Yamaguchi, N. Super-resolution of plant disease images for the acceleration of image-based phenotyping and vigor diagnosis in agriculture. Sensors 2017, 17, 2557. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Qureshi, J.; Roberts, P. Laboratory and UAV-based identification and classification of tomato yellow leaf curl, bacterial spot, and target spot diseases in tomato utilizing hyperspectral imaging and machine learning. Remote Sens. 2020, 12, 2732. [Google Scholar] [CrossRef]


| Stage | Datasets | Methods | Outcomes |
|---|---|---|---|
| Early detection of fields with CTV using remote sensing | Sentinel-2 | Time-series of vegetation index NDVI | Vegetation dynamics related to CTV-infected fields |
| Detection and monitoring of trees with CTV using remote sensing | GeoEye-1 Pleiades WorldView-2 UAV |
| Detection of candidate CTV-infected trees with FCCs |
| Validation | Leaf samples |
| Positive candidate CTV-infected trees |
| Semi-automatic machine learning procedure of CTV detection and accuracy assessment | Training samples of CTV-infected trees |
| Semi-automatic detection of CTV-infected trees based on training samples |
| Satellite Sensor | Date of Acquisition | Satellite Sensor Spatial Resolution (in Meters) and Wavelengths | Coordinate System |
|---|---|---|---|
| GeoEye-1 | 2 June 2017 | 0.41 m in panchromatic 1.65 m in multispectral bands Blue: 450–510 nm, Green: 520–580 nm, Red: 655–690 nm, Near-infrared: 780–920 nm | WGS UTM Zone 34N |
| Pleiades | 28 July 2018 | 0.7 m in panchromatic 2.8 m in multispectral bands Blue: 450–530 nm, Green: 510–590 nm, Red: 620–700 nm, Near-infrared: 775–915 nm | WGS UTM Zone 34N |
| Worldview-2 | 21 May 2021 | 0.41 m in panchromatic 1.64 m in multispectral bands Blue: 450–510 nm, Green: 510–580 nm, Red: 630–690 nm, Near-infrared: 770–895 nm | WGS UTM Zone 34N |
| NDVI | Ground Truth | |||
|---|---|---|---|---|
| UAV July 2022-RGB321 | Spectroscopy | UAV | Standard Deviation | Infected by CTV |
 | 0.730 | 0.760 | 0.018 | No (healthy sample) |
 | 0.640 | 0.490 | 0.110 | Yes |
 | 0.620 | 0.700 | 0.056 | Yes |
 | 0.587 | 0.585 | 0.002 | Yes |
 | 0.520 | 0.580 | 0.040 | Yes |
 | 0.670 | 0.700 | 0.025 | Yes |
 | 0.630 | 0.660 | 0.022 | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Argyriou, A.V.; Tektonidis, N.; Alevizos, E.; Ferentinos, K.P.; Kourgialas, N.N.; Mathioudakis, M.M. Precision Farming Multimodal Technologies Using Optical Sensors for the Detection of Citrus Tristeza Virus Endemics. Sustainability 2024, 16, 5748. https://doi.org/10.3390/su16135748
Argyriou AV, Tektonidis N, Alevizos E, Ferentinos KP, Kourgialas NN, Mathioudakis MM. Precision Farming Multimodal Technologies Using Optical Sensors for the Detection of Citrus Tristeza Virus Endemics. Sustainability. 2024; 16(13):5748. https://doi.org/10.3390/su16135748
Chicago/Turabian StyleArgyriou, Athanasios V., Nikolaos Tektonidis, Evangelos Alevizos, Konstantinos P. Ferentinos, Nektarios N. Kourgialas, and Matthaios M. Mathioudakis. 2024. "Precision Farming Multimodal Technologies Using Optical Sensors for the Detection of Citrus Tristeza Virus Endemics" Sustainability 16, no. 13: 5748. https://doi.org/10.3390/su16135748
APA StyleArgyriou, A. V., Tektonidis, N., Alevizos, E., Ferentinos, K. P., Kourgialas, N. N., & Mathioudakis, M. M. (2024). Precision Farming Multimodal Technologies Using Optical Sensors for the Detection of Citrus Tristeza Virus Endemics. Sustainability, 16(13), 5748. https://doi.org/10.3390/su16135748








