Changes in Microbial and Metabolic Pathways of Solidifying Manganese and Removing Nitrogen from Electrolytic Manganese Residue by the Sulfate-Reducing Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. EMR, SRB, and Medium
2.2. Remediation Experiment of EMR by SRB
2.3. High-Throughput Sequencing
2.4. Data Analysis
3. Results
3.1. Changes in the Physical and Chemical Properties of Leachate
3.2. Remediation Mechanisms
3.3. Microorganism Community Diversity of Leachate
3.4. Analysis of Microbial Differences at Genus Level
3.5. Differences in Microbial Metabolic Function
3.6. Metabolism Pathway
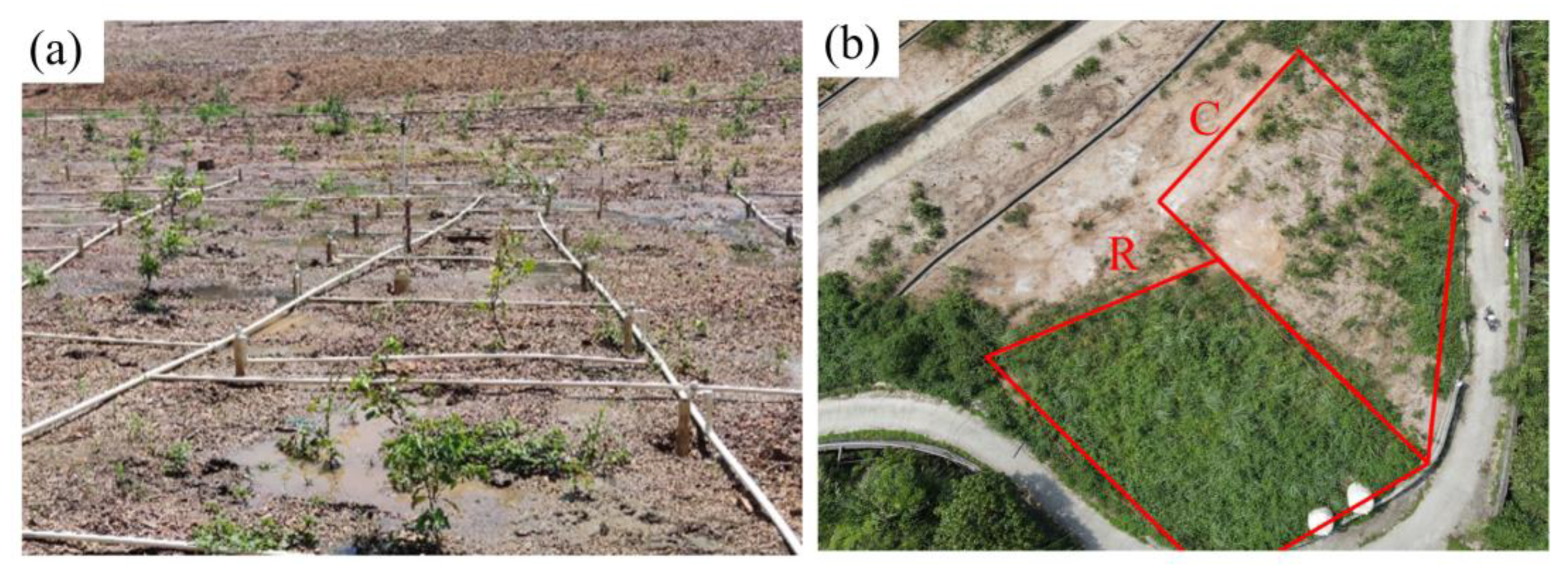
3.7. Vegetation Restoration
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, B.; Hou, D.; Duan, N.; Zhou, C.; Wang, J.; Dan, Z. Immobilization of high concentrations of soluble Mn(II) from electrolytic manganese solid waste using inorganic chemicals. Environ. Sci. Pollut. Res. Int. 2015, 22, 7782–7793. [Google Scholar] [CrossRef]
- Lan, J.; Zhang, S.; Mei, T.; Dong, Y.; Hou, H. Mechanochemical modification of electrolytic manganese residue: Ammonium nitrogen recycling, heavy metal solidification, and baking-free brick preparation. J. Clean. Prod. 2021, 329, 127727. [Google Scholar] [CrossRef]
- Chen, H.; Long, Q.; Zhang, Y.; Wang, S.; Deng, F. A novel method for the stabilization of soluble contaminants in electrolytic manganese residue: Using low-cost phosphogypsum leachate and magnesia/calcium oxide. Ecotox. Environ. Safe. 2020, 194, 110384. [Google Scholar] [CrossRef]
- Lan, J.R.; Sun, Y.; Chen, X.; Zhan, W.; Du, Y.; Zhang, T.C.; Ye, H.P.; Du, D.Y.; Hou, H.B. Bio-leaching of manganese from electrolytic manganese slag by Microbacterium trichothecenolyticum Y1: Mechanism and characteristics of microbial metabolites. Bioresource 2021, 319, 124056. [Google Scholar] [CrossRef]
- Shu, J.; Chen, M.; Wu, H.; Li, B.; Wang, B.; Li, B.; Liu, R.; Liu, Z. An innovative method for synergistic stabilization/solidification of Mn2+, NH4+-N, PO43− and F− in electrolytic manganese residue and phosphogypsum. J. Hazard. Mater. 2019, 376, 212–222. [Google Scholar] [CrossRef]
- Lan, J.R.; Sun, Y.; Guo, L.; Li, Z.M.; Du, D.Y.; Zhang, T.C. A novel method to recover ammonia, manganese and sulfate from electrolytic manganese residues by bio-leaching. J. Clean. Prod. 2019, 223, 499–507. [Google Scholar] [CrossRef]
- Tian, Y.; Shu, J.C.; Chen, M.J.; Wang, J.Y.; Wang, Y.; Luo, Z.G.; Wang, R.; Yang, F.H.; Xiu, F.R.; Sun, Z. Manganese and ammonia nitrogen recovery from electrolytic manganese residue by electric field enhanced leaching. J. Clean. Prod. 2019, 236, 8. [Google Scholar] [CrossRef]
- Aschner, M.; Erikson, K.M.; Dorman, D.C. Manganese dosimetry: Species differences and implications for neurotoxicity. Curr. Opin. Toxicol. 2005, 35, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Crossgrove, J.; Zheng, W. Manganese toxicity upon overexposure. NMR Biomed. 2004, 17, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.H.; Wu, S.S.; Liu, R.L.; Liu, Z.H.; Tao, C.Y. Efficient recovery of manganese and ammonia nitrogen from filter-pressing electrolytic manganese residue by organic complexation. J. Hydrol. 2022, 45, 102532. [Google Scholar] [CrossRef]
- Chen, H.; Long, Q.; Zhou, F.; Shen, M. Elec-accumulating behaviors of manganese in the electrokinetics-processed electrolytic manganese residue with carbon dioxide and oxalic acid. J. Electroanal. Chem. 2020, 865, 114162. [Google Scholar] [CrossRef]
- He, D.; Shu, J.; Zeng, X.; Wei, Y.; Chen, M.; Tan, D.; Liang, Q. Synergistic solidification/stabilization of electrolytic manganese residue and carbide slag. Sci. Total Environ. 2022, 810, 152175. [Google Scholar] [CrossRef]
- He, D.J.; Shu, J.C.; Wang, R.; Chen, M.J.; Wang, R.; Gao, Y.S.; Liu, R.L.; Liu, Z.H.; Xu, Z.H.; Tan, D.Y.; et al. A critical review on approaches for electrolytic manganese residue treatment and disposal technology: Reduction, pretreatment, and reuse. J. Hazard. Mater. 2021, 418, 126235. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, J.; Mercier, G.; Morel, J.L.; Charbonnier, P.; Blais, J.F.; Simonnot, M.O. Recovery of zinc and manganese from pyrometallurgy sludge by hydrometallurgical processing. J. Clean. Prod. 2017, 168, 311–321. [Google Scholar] [CrossRef]
- Shu, J.; Liu, R.; Liu, Z.; Chen, H.; Du, J.; Tao, C. Solidification/stabilization of electrolytic manganese residue using phosphate resource and low-grade MgO/CaO. J. Hazard. Mater. 2016, 317, 267–274. [Google Scholar] [CrossRef]
- Cameselle, C. Enhancement of Electro-Osmotic Flow During The electrokinetic treatment of a contaminated soil. Electrochim. Acta 2015, 181, 31–38. [Google Scholar] [CrossRef]
- Lee, D.-J.; Liu, X.; Weng, H.-L. Sulfate and organic carbon removal by microbial fuel cell with sulfate-reducing bacteria and sulfide-oxidising bacteria anodic biofilm. Bioresour. Technol. 2014, 156, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.; Sasaki, K.; Hiroyoshi, N.; Tsunekawa, M.; Hirajima, T. The Effect of Mn2+ Concentration on Mn Removal by a Sulfate Reducing Bacteria Bioreactor. Mater. Trans. 2004, 45, 2429–2434. [Google Scholar] [CrossRef]
- GB 8978-1996; Integrated wastewater discharge standard. State Environmental Protection Administration: Beijing, China, 1996.
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Chang, Y.; Hung, C.; Lee, J.; Liao, H.; Chou, H. Microbial community analysis of anaerobic bio-corrosion in different ORP profiles. Int. Biodeterior. Biodegrad. 2014, 95, 93–101. [Google Scholar] [CrossRef]
- Shen, H.; Zhang, Q.; Zhang, X.; Jiang, X.; Xiong, Z. In situ effects of biochar field-aged for six years on net N mineralization in paddy soil. Soil Tillage Res. 2021, 205, 104766. [Google Scholar] [CrossRef]
- Liang, F.; Wang, R.; Hongzhong, X.; Yang, X.; Zhang, T.; Hu, W.; Mi, B.; Liu, Z. Investigating pyrolysis characteristics of moso bamboo through TG-FTIR and Py-GC/MS. Bioresour. Technol. 2018, 256, 53–60. [Google Scholar] [CrossRef]
- Dan, M.; Zhang, Q.; Yu, S.; Prakash, A.; Lin, Y.; Zhou, Y. Noble-metal-free MnS/In2S3 composite as highly efficient visible light driven photocatalyst for H2 production from H2S. Appl. Catal. B 2017, 217, 530–539. [Google Scholar] [CrossRef]
- Guo, L.; Chen, A.; Li, C.; Wang, Y.; Yang, D.; He, N.; Liu, M.D. Solution chemistry mechanisms of exogenous silicon influencing the speciation and bioavailability of cadmium in alkaline paddy soil. J. Hazard. Mater. 2022, 438, 129526. [Google Scholar] [CrossRef]
- Shu, J.; Cai, L.; Zhao, J.; Feng, H.; Chen, M.; Zhang, X.; Wu, H.; Yang, Y.; Liu, R. A low cost of phosphate-based binder for Mn2+ and NH4+-N simultaneous stabilization in electrolytic manganese residue. Ecotoxicol. Environ. Saf. 2020, 205, 111317. [Google Scholar] [CrossRef]
- Sun, M.; Liu, H.; Liu, Y.; Qu, J.; Li, J. Graphene-based transition metal oxide nanocomposites for the oxygen reduction reaction. Nanoscale 2015, 7, 1250–1269. [Google Scholar] [CrossRef]
- Kamarisima; Miyanaga, K.; Tanji, Y. The utilization of aromatic hydrocarbon by nitrate- and sulfate-reducing bacteria in single and multiple nitrate injection for souring control. Biochem. Eng. J. 2018, 143, 75–80. [Google Scholar] [CrossRef]
- Xia, Z.; Wang, Q.; She, Z.; Gao, M.; Zhao, Y.; Guo, L.; Jin, C. Nitrogen removal pathway and dynamics of microbial community with the increase of salinity in simultaneous nitrification and denitrification process. Sci. Total Environ. 2019, 697, 134047. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Biswal, B.K.; Chen, G.H.; Wu, D. Sulfidogenic anaerobic digestion of sulfate-laden waste activated sludge: Evaluation on reactor performance and dynamics of microbial community. Bioresour. Technol. 2020, 297, 122396. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, H.; Ueno, Y.; Sekiguchi, Y.; Takii, S.; Hanada, S.; Matsuura, K.; Ibe, A. Dethiosulfatibacter aminovorans gen. nov., sp. nov., a novel thiosulfate-reducing bacterium isolated from coastal marine sediment via sulfate-reducing enrichment with Casamino acids. Int. J. Syst. Evol. Microbiol. 2007, 57 (Pt 10), 2320–2326. [Google Scholar]
- Lima, A.T.; Ottosen, L. Recovering rare earth elements from contaminated soils: Critical overview of current remediation technologies. Chemosphere 2021, 265, 129163. [Google Scholar] [CrossRef] [PubMed]
- Wiessner, A.; Kappelmeyer, U.; Kuschk, P.; Kästner, M. Sulphate reduction and the removal of carbon and ammonia in a laboratory-scale constructed wetland. Water Res. 2005, 39, 4643–4650. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Sun, Q.; Zhao, C.; Wen, D.; Tang, X. Quinoline biodegradation and its nitrogen transformation pathway by a Pseudomonas sp. strain. Biodegradation 2010, 21, 335–344. [Google Scholar] [CrossRef]
- Chang, M.; Wang, Y.; Pan, Y.; Zhang, K.; Zhu, T. Nitrogen removal from wastewater via simultaneous nitrification and denitrification using a biological folded non-aerated filter. Bioresour. Technol. 2019, 289, 121696. [Google Scholar] [CrossRef]
- Ouyang, Y.; Norton, J.M.; Stark, J.M.; Reeve, J.R.; Habteselassie, M.Y. Ammonia-oxidizing bacteria are more responsive than archaea to nitrogen source in an agricultural soil. Soil. Biol. Biochem. 2016, 96, 4–15. [Google Scholar] [CrossRef]
- Serrato, R.V.; Sassaki, G.L.; Gorin, P.; Cruz, L.M.; Pedrosa, F.O.; Choudhury, B.; Carlson, R.W.; Iacomini, M. Structural characterization of an acidic exoheteropolysaccharide produced by the nitrogen-fixing bacterium Burkholderia tropica. Carbohydr. Polym. 2008, 73, 564–572. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276.42. [Google Scholar] [CrossRef]
- Cai, J.; Jiang, J.X.; Zheng, P. Isolation and identification of bacteria responsible for simultaneous anaerobic ammonium and sulfate removal. Sci. China Chem. 2010, 53, 645–650. [Google Scholar] [CrossRef]
- Aranda-Tamaura, C.; Estrada-Alvarado, M.I.; Texier, A.C.; Cuervo, F.; Gomez, J.; Cervantes, F.J. Effects of different quinoid redox mediators on the removal of sulphide and nitrate via denitrification. Chemosphere 2007, 69, 1722–1727. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.J.; Liu, L.H.; Sun, D.; Ren, N.Q.; Lee, D.J. Isolation of Fe(III)-reducing fermentative bacterium Bacteroides sp. W7 in the anode suspension of a microbial electrolysis cell (MEC). Int. J. Hydrog. Energy 2010, 35, 3178–3182. [Google Scholar] [CrossRef]
- White, A.G.; Watts, G.S.; Lu, Z.Q.; Meza-Montenegro, M.M.; Lutz, E.A.; Harber, P.; Burgess, J.L. Environmental Arsenic Exposure and Microbiota in Induced Sputum. Int. J. Environ. Res. Public Health 2014, 11, 2299–2313. [Google Scholar] [CrossRef] [PubMed]
- Jabari, L.; Gannoun, H.; Cayol, J.L.; Hedi, A.; Sakamoto, M.; Falsen, E.; Ohkuma, M.; Hamdi, M.; Fauque, G.; Ollivier, B.; et al. Macellibacteroides fermentans gen. nov., sp. nov., a member of the family Porphyromonadaceae isolated from an upflow anaerobic filter treating abattoir wastewaters. Int. J. Syst. Evol. Microbiol. 2012, 62 (Pt 10), 2522–2527. [Google Scholar] [CrossRef]
- Setsuko, H.; Marcus, T.; Eri, H.; Hideyuki, T.; Koji, M.; Shinichi, T.; Shin, H.; Satoshi, H. Aquabacterium pictum sp. nov., the first aerobic bacteriochlorophyll a-containing fresh water bacterium in the genus Aquabacterium of the class Betaproteobacteria. Int. J. Microbiol. 2020, 70, 596–603. [Google Scholar]
- Chen, Z.; Zhong, X.; Zheng, M.; Liu, W.S.; Fei, Y.; Ding, K.; Li, Y.; Liu, Y.; Chao, Y.; Tang, Y.T.; et al. Indicator species drive the key ecological functions of microbiota in a river impacted by acid mine drainage generated by rare earth elements mining in South China. Environ. Microbiol. 2022, 24, 919–937. [Google Scholar] [CrossRef]
- Niu, Z.S.; Pan, H.; Guo, X.P.; Lu, D.P.; Feng, J.N.; Chen, Y.R.; Tou, F.Y.; Liu, M.; Yang, Y. Sulphate-reducing bacteria (SRB) in the Yangtze Estuary sediments: Abundance, distribution and implications for the bioavailibility of metals. Sci. Total Environ. 2018, 634, 296–304. [Google Scholar] [CrossRef]
- He, S.C.; Jiang, D.Y.; Hong, M.H.; Liu, Z.H. Hazard-free treatment and resource utilisation of electrolytic manganese residue: A review. J. Clean. Prod. 2021, 306, 127224. [Google Scholar] [CrossRef]
- Shu, J.; Wu, H.; Chen, M.; Peng, H.; Li, B.; Liu, R.; Liu, Z.; Wang, B.; Huang, T.; Hu, Z. Fractional removal of manganese and ammonia nitrogen from electrolytic metal manganese residue leachate using carbonate and struvite precipitation. Water Res. 2019, 153, 229–238. [Google Scholar] [CrossRef]
- Pereira, I.; Ramos, A.R.; Grein, F.; Marques, M.C.; Silva, S.; Venceslau, S.S. A Comparative Genomic Analysis of Energy Metabolism in Sulfate Reducing Bacteria and Archaea. Front. Microbiol. 2011, 2, 69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, X.; Li, Y.; Wang, G.; Wang, Z.; Wen, J. Microbial community and metabolic pathway succession driven by changed nutrient inputs in tailings: Effects of different nutrients on tailing remediation. Sci. Rep. 2017, 7, 474. [Google Scholar] [CrossRef]
- Chiang, Y.L.; Hsieh, Y.C.; Fang, J.Y.; Liu, E.H.; Huang, Y.C.; Chuankhayan, P.; Jeyakanthan, J.; Liu, M.Y.; Chan, S.I.; Chen, C.J. Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit. J. Bacteriol. 2009, 191, 7597–7608. [Google Scholar] [CrossRef] [PubMed]
- Raquel, R.A. The Membrane QmoABC Complex Interacts Directly with the Dissimilatory Adenosine 5′-Phosphosulfate Reductase in Sulfate Reducing Bacteria. Front. Microbiol. 2012, 3, 137. [Google Scholar]
- Berlicki, L. Inhibitors of Glutamine Synthetase and their Potential Application in Medicine. Mini Rev. Med. Chem. 2008, 8, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Leustek, T.; Martin, M.N.; Bick, J.A.; Davies, J.P. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Biol. 2000, 51, 141–165. [Google Scholar]
- Mehmood, M.A.; Fu, Y.P.; Zhao, H.Z.; Cheng, J.S.; Xie, J.T.; Jiang, D.H. Enrichment of bacteria involved in the nitrogen cycle and plant growth promotion in soil by sclerotia of rice sheath blight fungus. Stress Biol. 2022, 2, 32. [Google Scholar] [CrossRef]
- Cloutier, M.; Alcaide, T.; Duiker, S.; Bruns, M.A. Tillage intensity and plant rhizosphere selection shape bacterial-archaeal assemblage diversity and nitrogen cycling genes. Soil. Tillage Res. 2023, 225, 105525. [Google Scholar] [CrossRef]






| Sample | Pb (mg/L) | Fe (mg/L) | Mn (mg/L) | As (mg/L) | NH4+-N (mg/L) | NO2− (mg/L) | Eh (mV) | pH |
|---|---|---|---|---|---|---|---|---|
| EMR leachate | 0.23 | 7.15 | 4566.67 | 0.01 | 5245 | 1.805 | 241.88 | 6.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, G.; Lv, Y.; Yan, X.; Liu, X.; Zhu, X.; Zhang, M. Changes in Microbial and Metabolic Pathways of Solidifying Manganese and Removing Nitrogen from Electrolytic Manganese Residue by the Sulfate-Reducing Bacteria. Sustainability 2023, 15, 5215. https://doi.org/10.3390/su15065215
Ma G, Lv Y, Yan X, Liu X, Zhu X, Zhang M. Changes in Microbial and Metabolic Pathways of Solidifying Manganese and Removing Nitrogen from Electrolytic Manganese Residue by the Sulfate-Reducing Bacteria. Sustainability. 2023; 15(6):5215. https://doi.org/10.3390/su15065215
Chicago/Turabian StyleMa, Guoying, Ying Lv, Xiao Yan, Xingyu Liu, Xuezhe Zhu, and Mingjiang Zhang. 2023. "Changes in Microbial and Metabolic Pathways of Solidifying Manganese and Removing Nitrogen from Electrolytic Manganese Residue by the Sulfate-Reducing Bacteria" Sustainability 15, no. 6: 5215. https://doi.org/10.3390/su15065215
APA StyleMa, G., Lv, Y., Yan, X., Liu, X., Zhu, X., & Zhang, M. (2023). Changes in Microbial and Metabolic Pathways of Solidifying Manganese and Removing Nitrogen from Electrolytic Manganese Residue by the Sulfate-Reducing Bacteria. Sustainability, 15(6), 5215. https://doi.org/10.3390/su15065215






