Beta-Lactam Susceptibility Profiles of Bacteria Isolated from the Ozama River in Santo Domingo, Dominican Republic
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Physico-Chemical Tests
2.2. Bacteria Isolation
2.3. Bruker BioTyper Bacterial Identification
2.4. Antibiotic Susceptibility Test
2.5. Data Analysis
2.6. Genomic DNA Extraction from Isolates
2.7. Genome Sequencing, Assembly, and Analysis
3. Results
3.1. Physical, Chemical, and Biological Parameters of Water
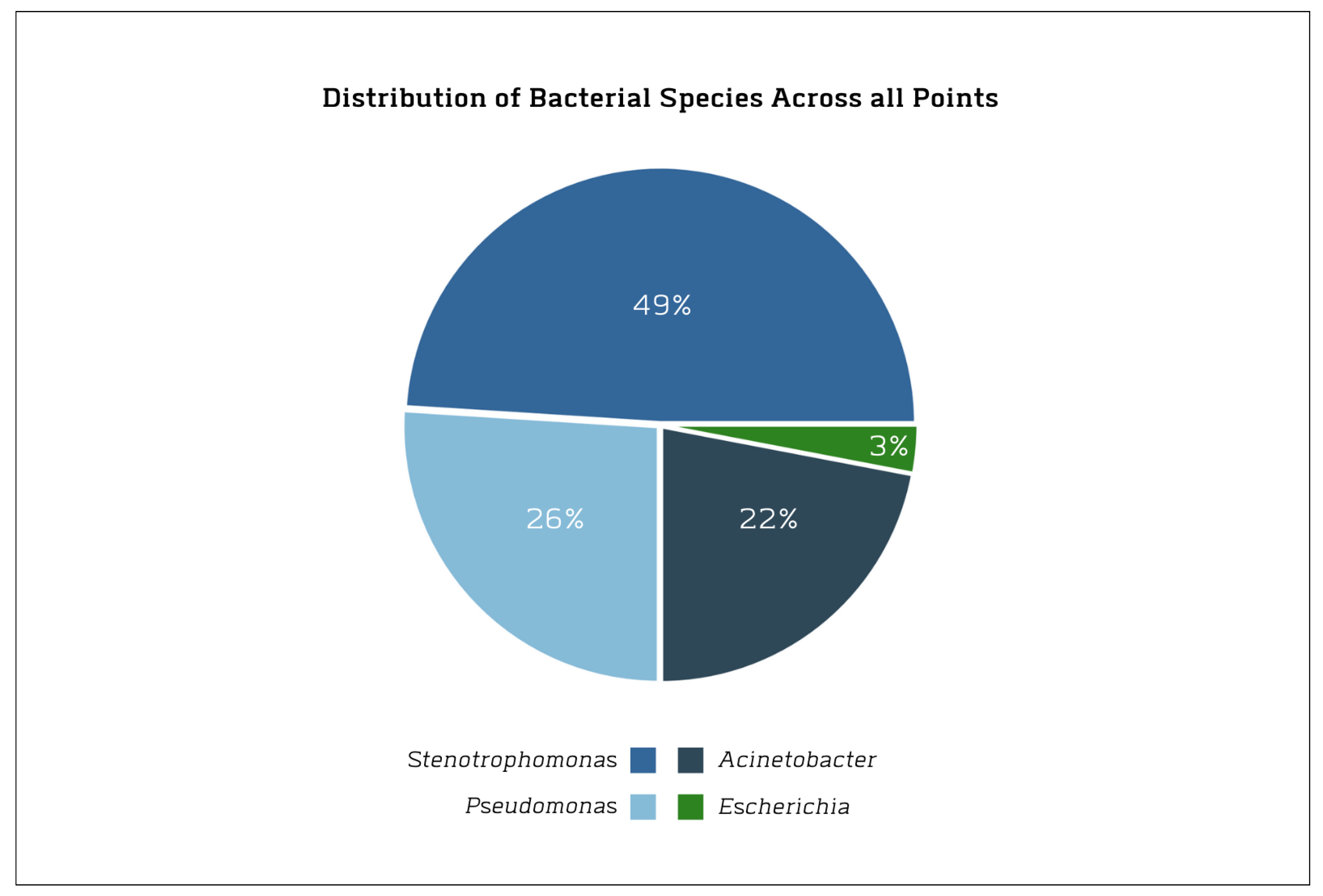
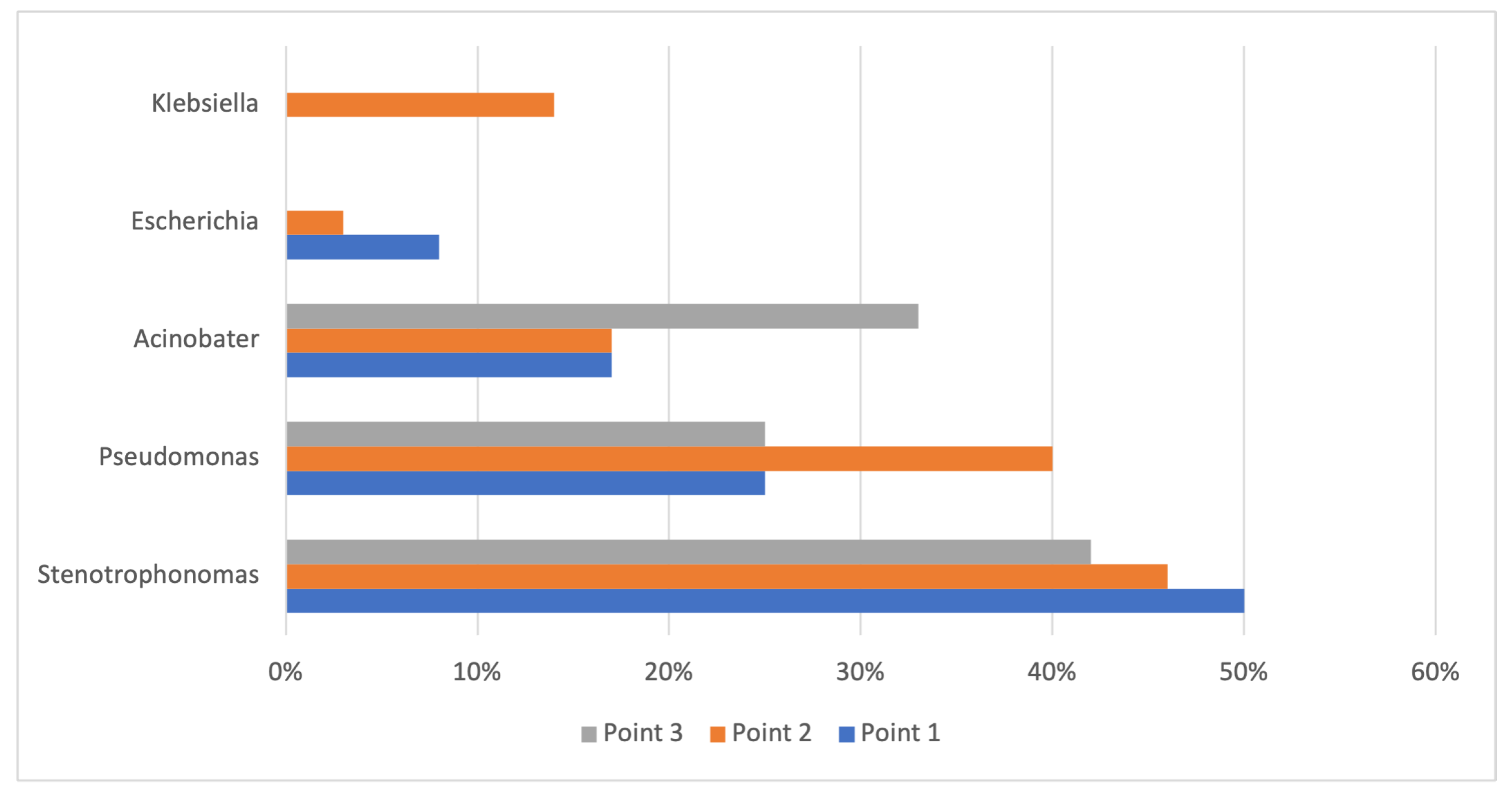
3.2. Composition and Distribution of the Isolates
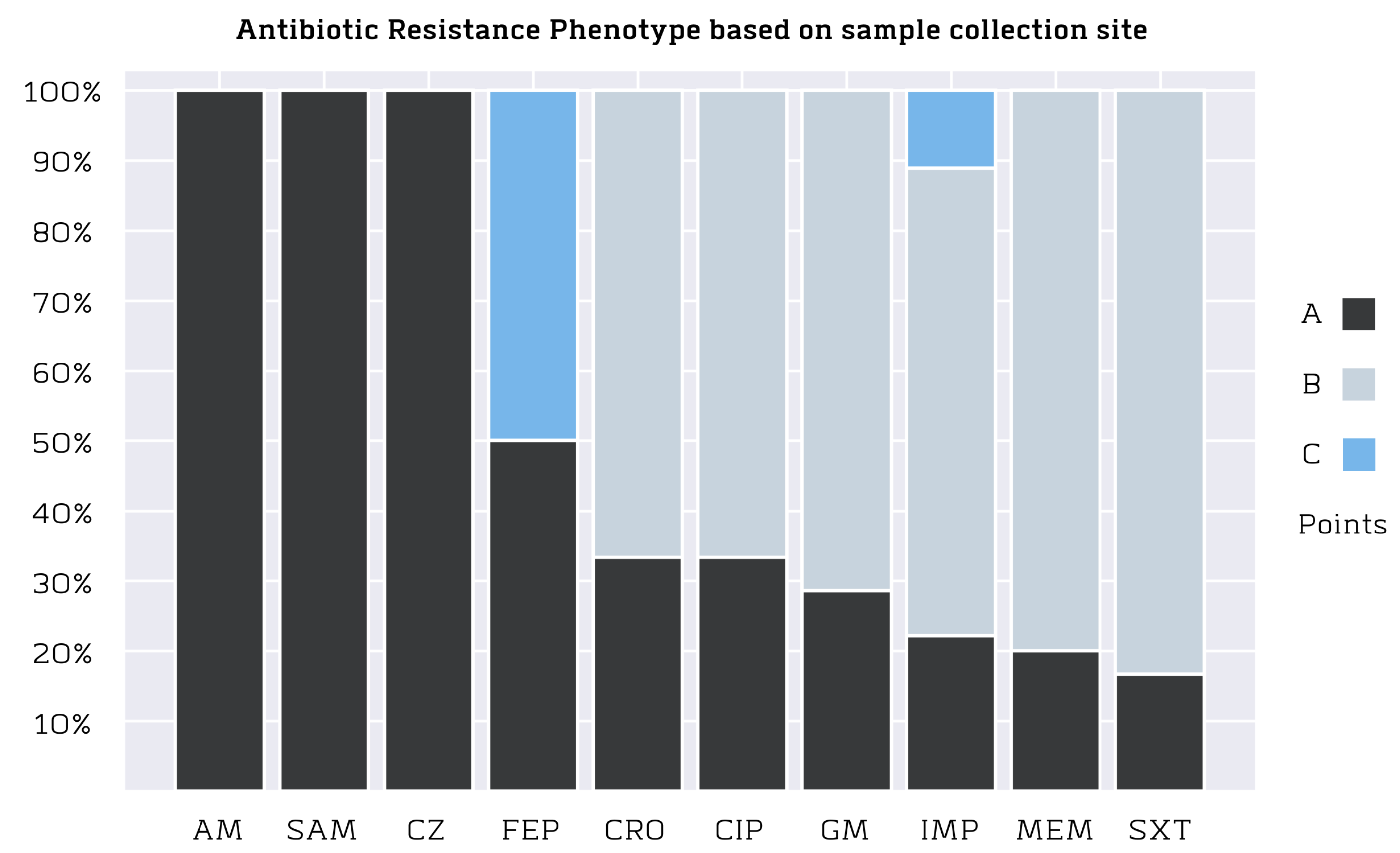
3.3. Antibiotic Resistance
3.4. Analysis of Multi-Resistance Genomes
3.4.1. Escherichia coli
3.4.2. Pseudomonas putida
3.4.3. Acinetobacter pittii
3.4.4. Klebsiella pneumoniae
3.4.5. Acinetobacter baumanii
4. Discussion
4.1. Physico-Chemical and Microbiological Analyses
4.2. Genomic and Resistome Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARB | Antibiotic-resistant bacteria |
| ARG | Antibiotic-resistant genes |
| ChromoAgar | Chromogenic culture |
| MIC | Minimum inhibitory concentration |
| RGI | Resistance Genes Identifier |
| COD | Chemical oxygen demand |
| BOD5 | Biological oxygen demands |
| TP | Total phosphorous |
| TN | Total Nitrogen |
| DO | Dissolved Oxygen |
| CTX | Cefotaxime |
| IMP | Imipenem |
References
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Biyela, P.; Lin, J.; Bezuidenhout, C. The role of aquatic ecosystems as reservoirs of antibiotic resistant bacteria and antibiotic resistance genes. Water Sci. Technol. 2004, 50, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef]
- Servais, P.; Passerat, J. Antimicrobial resistance of fecal bacteria in waters of the Seine river watershed (France). Sci. Total Environ. 2009, 408, 365–372. [Google Scholar] [CrossRef]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Calderón, V.V.; Bonnelly, R.; Del Rosario, C.; Duarte, A.; Baraúna, R.; Ramos, R.T.; Perdomo, O.P.; Rodriguez de Francisco, L.E.; Franco, E.F. Distribution of Beta-Lactamase Producing Gram-Negative Bacterial Isolates in Isabela River of Santo Domingo, Dominican Republic. Front. Microbiol. 2021, 11, 2973. [Google Scholar] [CrossRef]
- Freitas, D.Y.; Araújo, S.; Folador, A.R.; Ramos, R.T.; Azevedo, J.S.; Tacão, M.; Silva, A.; Henriques, I.; Baraúna, R.A. Extended spectrum beta-lactamase-producing gram-negative bacteria recovered from an Amazonian lake near the city of Belém, Brazil. Front. Microbiol. 2019, 10, 364. [Google Scholar] [CrossRef]
- Reddy, B.; Dubey, S.K. River Ganges water as reservoir of microbes with antibiotic and metal ion resistance genes: High throughput metagenomic approach. Environ. Pollut. 2019, 246, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Tacão, M.; Correia, A.; Henriques, I. Resistance to broad-spectrum antibiotics in aquatic systems: Anthropogenic activities modulate the dissemination of bla CTX-M-like genes. Appl. Environ. Microbiol. 2012, 78, 4134–4140. [Google Scholar] [CrossRef]
- World Health Organization. Overcoming Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2000. [Google Scholar]
- Stoll, C.; Sidhu, J.; Tiehm, A.; Toze, S. Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environ. Sci. Technol. 2012, 46, 9716–9726. [Google Scholar] [CrossRef]
- Yazdan, M.M.; Kumar, R.; Leung, S.W. The Environmental and Health Impacts of Steroids and Hormones in Wastewater Effluent, as Well as Existing Removal Technologies: A Review. Ecologies 2022, 3, 206–224. [Google Scholar] [CrossRef]
- Jiao, Y.N.; Chen, H.; Gao, R.X.; Zhu, Y.G.; Rensing, C. Organic compounds stimulate horizontal transfer of antibiotic resistance genes in mixed wastewater treatment systems. Chemosphere 2017, 184, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.F.; Zhang, L.; Balfour, A.J.; Garside, R.; Gaze, W.H. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ. Int. 2015, 82, 92–100. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Hsu, B.M.; Ji, W.T.; Chen, J.S.; Hsu, T.K.; Ji, D.D.; Tseng, S.F.; Chiu, Y.C.; Kao, P.M.; Huang, Y.L. Antibiotic resistance pattern and gene expression of non-typhoid Salmonella in riversheds. Environ. Sci. Pollut. Res. 2015, 22, 7843–7850. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Ministerio de Medio Ambiente y Recursos Naturales de la Republica Dominicana (MMARN). Ozama Basin Biophysical Description; MMARN: Santo Domingo, Dominican Republic, 2015. [Google Scholar]
- Oficina Nacional de Estadística de la República Dominicana. Total, Estimaciones y Proyecciones de la Poblacion. 2020. Available online: https://www.one.gob.do/datos-y-estadisticas/temas/estadisticas-demograficas/estimaciones-y-proyecciones-demograficas/ (accessed on 9 June 2022).
- Arroyo Rodríguez, S.C. Regeneración Urbana en los Márgenes del Rio Ozama en la Ciudad de Santo Domingo; Universidad Polictecnica de Valencia: Valencia, Spain, 2020. [Google Scholar]
- Chantada, A. Medio Ambiente, crisis y desarrollo: Reflexión en torno a los ríos Ozama e Isabela. Rev. Estud. Soc. 1991, 24, 12–17. [Google Scholar]
- Gutiérrez, W. Recopilacion Documental de Informaciones Relacionadas con la Cuenca, Calidad de Sus Aguas, el Saneamiento y Rehabilitacion del rio Ozama; Technical Report; Coalicion Rio: Santo Domingo, Dominican Republic, 2014. [Google Scholar]
- de la Rosa, A. En ríos Ozama e Isabela Descargan 54 cañadas y 241 Empresas del Gran Santo Domingo. 2018. Available online: https://www.diariolibre.com/actualidad/ciudad/en-rios-ozama-e-isabela-descargan-54-canadas-y-241-empresas-del-gran-santo-domingo-BO10697789 (accessed on 10 February 2022).
- Martínez, E.; Tió, R.C.; Tatis, L.R.; de León, P.; Salcedo, L. Calidad del agua en la república dominicana. Calid. Agua Las Am. 2019, 560, 559–560. [Google Scholar]
- Espinal, G.; Mendoza Gómez, C.L.; Contreras Pérez, J.B.; Vásquez, J. Contaminación Química y Bacteriológica de los Ríos Ozama e Isabela. Cienc. Soc. 1993, 18, 31–39. [Google Scholar] [CrossRef]
- Butler, M.S.; Gigante, V.; Sati, H.; Paulin, S.; Al-Sulaiman, L.; Rex, J.H.; Fernandes, P.; Arias, C.A.; Paul, M.; Thwaites, G.E.; et al. Analysis of the clinical pipeline of treatments for drug-resistant bacterial infections: Despite progress, more action is needed. Antimicrob. Agents Chemother. 2022, 66, e01991-21. [Google Scholar] [CrossRef] [PubMed]
- Association, A.P.H. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Denver, CO, USA, 2005. [Google Scholar]
- Strejcek, M.; Smrhova, T.; Junkova, P.; Uhlik, O. Whole-Cell MALDI-TOF MS Versus 16S rRNA Gene Analysis for Identification and Dereplication of Recurrent Bacterial Isolates. Front. Microbiol. 2018, 9, 1294. [Google Scholar] [CrossRef]
- Suzuki, Y.; Niina, K.; Matsuwaki, T.; Nukazawa, K.; Iguchi, A. Bacterial flora analysis of coliforms in sewage, river water, and ground water using MALDI-TOF mass spectrometry. J. Environ. Sci. Health Part A 2018, 53, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Melsted, P.; Halldórsson, B.V. KmerStream: Streaming algorithms for k-mer abundance estimation. Bioinformatics 2014, 30, 3541–3547. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.; François, P.; Farinelli, L.; Østerås, M.; Schrenzel, J. De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Res. 2008, 18, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 1–15. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, 000206. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). In Horizontal Gene Transfer; Springer: New York, NY, USA, 2020; pp. 285–294. [Google Scholar]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder-distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Kleinheinz, K.A.; Joensen, K.G.; Larsen, M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 2014, 4, e27943. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Johansson, M.H.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Ministério de Médio Ambiente de la Republica Dominicana. Norma Ambiental de Calidad de Aguas Superficiales y Costeras; Ministério de Médio Ambiente: Santo Domingo, Dominican Republic; p. 12.
- Castro, Y.A.; Agblevor, F.A. Biomethanation of invasive water hyacinth from eutrophic waters as a post weed management practice in the Dominican Republic: A developing country. Environ. Sci. Pollut. Res. 2020, 27, 14138–14149. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Pujols, N.S. Estudio de la calidad de las aguas superficiales y caracterizacion de la cuenca del Rio Cabon, afluente del Rio Ozama, ubicado en Santo Domingo Norte Republica Dominicana 2017. Ph.D. Thesis, Universidad Nacional Pedro Henriquez Urena, Santo Domingo, Dominican Republic, 2018. [Google Scholar]
- Ash, R.J.; Mauck, B.; Morgan, M. Antibiotic resistance of gram-negative bacteria in rivers, United States. Emerg. Infect. Dis. 2002, 8, 713. [Google Scholar] [CrossRef] [PubMed]
- Williams, L. Rid the rivers of rubbish [Plastics Pollution]. Eng. Technol. 2020, 15, 64–67. [Google Scholar] [CrossRef]
- Munthe, C.; Malmqvist, E.; Rönnerstrand, B. Non-prescription acquisition of antibiotics: Prevalence, motives, pathways and explanatory factors in the Swedish population. PLoS ONE 2022, 17, e0273117. [Google Scholar] [CrossRef]
- Beyene, K.A.; Sheridan, J.; Aspden, T. Prescription medication sharing: A systematic review of the literature. Am. J. Public Health 2014, 104, e15–e26. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Global Surveillance and Monitoring System for Substandard and Falsified Medical Products; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Haque, M.; Sartelli, M.; McKimm, J.; Bakar, M.A. Health care-associated infections—An overview. Infect. Drug Resist. 2018, 11, 2321. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed]

| Point | Parameter | Mean | Standard Deviation | Permitted Value | Units |
|---|---|---|---|---|---|
| Temperature | 33.3 | 2.49 | NA | C | |
| pH | 6.75 | 0.24 | 6.5–9.0 | None | |
| BOD | 17.04 | 6.88 | 5.0 | mg/L | |
| COD | 30.66 | 12.36 | 300.0 | mg/L | |
| A | TP | 1.0 | 0 | 0.025 | mg/L |
| TN | 2.4 | 1.2 | 30.0 | mg/L | |
| fDO | 6.98 | 0.19 | >5 | mg/L | |
| Turbidity | 2.39 | 0.024 | Und | NFU | |
| Coliforms | 7700 | 711.8 | 1000 | CFU/100 mL | |
| Water Class | NORDOM B | ||||
| Temperature | 31.67 | 0.94 | NA | C | |
| pH | 8.14 | 0.87 | 6.5–9.0 | None | |
| BOD | 67.81 | 3.87 | 5.0 | mg/L | |
| COD | 125.0 | 4.32 | 300.0 | mg/L | |
| B | TP | 1.0 | 0 | 0.025 | mg/L |
| TN | 2.7 | 0 | 30.0 | mg/L | |
| DO | 6.79 | 0.074 | 5 | mg/L | |
| Turbidity | 6.69 | 0.84 | Und | NFU | |
| Coliforms | 7166 | 1027.4 | 1000 | CFU/100 mL | |
| Water Class | NORDOM B | ||||
| Temperature | 29.3 | 0.47 | NA | C | |
| pH | 7.52 | 0.021 | 6.5–8.5 | None | |
| BOD | 163.0 | 53.78 | 2.0 | mg/L | |
| COD | 163.0 | 53.78 | 150.0 | mg/L | |
| C | TP | 1.0 | 0 | 0.025 | mg/L |
| TN | 1.2 | 0 | 20.0 | mg/L | |
| DO | 6.79 | 0.074 | 6.4 | mg/L | |
| Turbidity | 1.07 | 0.24 | Und | NFU | |
| Coliforms | 2100 | 535.41 | 1000 | CFU/100 mL | |
| Water Class | NORDOM A |
| ID | Species | Resistance Phenotype |
|---|---|---|
| DC1 | Acinetobacter baumannii | No resistance |
| DC2 | Escherichia coli | AM, CZ, FEP, CRO, SXT |
| DC5 | Acinetobacter baumannii | No resistance |
| DC8 | Escherichia coli | AM, SAM, CZ, FEP, CRO, CIP, SXT |
| DC9 | Acinetobacter baumannii | No resistance |
| DC10 | Acinetobacter pittii | No resistance |
| EC1 | Klebsiella pneumoniae | SAM, CZ, FEP, CRO, CIP, GM, SXT |
| EC4 | Klebsiella pneumoniae | SAM, CZ, FEP, CRO, CIP, GM, SXT |
| EC6 | Escherichia coli | SAM, CZ, FEP, CRO, CIP, GM, SXT |
| EC7 | Klebsiella pneumoniae | SAM, CZ, FEP, CRO, CIP, GM, SXT |
| EC11 | Acinetobacter baumannii | No resistance |
| EC13 | Acinetobacter baumannii | No resistance |
| EC14 | Acinetobacter baumannii | FEP, IPM |
| EC15 | Acinetobacter baumannii | MEM |
| EC16 | Acinetobacter baumannii | No resistance |
| EC16.2 | Klebsiella pneumoniae | SAM, FEP, CRO |
| FC1 | Acinetobacter pittii | No resistance |
| FC4 | Acinetobacter baumannii | No resistance |
| FC5 | Acinetobacter baumannii | No resistance |
| FC6 | Acinetobacter baumannii | No resistance |
| FC7 | Acinetobacter baumannii | FEP, MEM |
| FC8 | Acinetobacter baumannii | No resistance |
| FC12 | Acinetobacter baumannii | No resistance |
| Sample ID | Species | Genome Size (pb) | GC (%) | CDS | N50 | Plasmids |
|---|---|---|---|---|---|---|
| DC8 | Escherichia coli | 4,710,092 | 50.7 | 4664 | 148,992 | IncY, IncFIB |
| DC2 | Escherichia coli | 4,657,304 | 51.1 | 4637 | 56,323 | IncY |
| DC10 | Acinetobacter pitii | 3,907,124 | 38.6 | 3705 | 245,032 | - |
| EC4 | Klebsiella pneumoniae | 5,527,705 | 57.2 | 5457 | 201,357 | IncFBI(K), IncFII(K), Col440II |
| EC7 | Klebsiella pneumoniae | 5,424,764 | 57.3 | 5346 | 320,811 | IncFBI(K), IncFII(K0), Col440II |
| FC5 | Acinetobacter baumanii | 3,809,530 | 38.9 | 3622 | 324,095 | - |
| FC7 | Acinetobacter baumanii | 3,841,875 | 38.9 | 3660 | 323,883 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonnelly, R.; Queiroz Cavalcante, A.L.; Calderon, V.V.; Baraúna, R.A.; Jucá Ramos, R.T.; Rodríguez-Rodríguez, Y.; De Francisco, L.E.R.; Maroto Martín, L.O.; Perdomo, O.P.; Franco De Los Santos, E.F. Beta-Lactam Susceptibility Profiles of Bacteria Isolated from the Ozama River in Santo Domingo, Dominican Republic. Sustainability 2023, 15, 5109. https://doi.org/10.3390/su15065109
Bonnelly R, Queiroz Cavalcante AL, Calderon VV, Baraúna RA, Jucá Ramos RT, Rodríguez-Rodríguez Y, De Francisco LER, Maroto Martín LO, Perdomo OP, Franco De Los Santos EF. Beta-Lactam Susceptibility Profiles of Bacteria Isolated from the Ozama River in Santo Domingo, Dominican Republic. Sustainability. 2023; 15(6):5109. https://doi.org/10.3390/su15065109
Chicago/Turabian StyleBonnelly, Roberto, Ana Lidia Queiroz Cavalcante, Victor V. Calderon, Rafael Azevedo Baraúna, Rommel Thiago Jucá Ramos, Yaset Rodríguez-Rodríguez, Luis Enrique Rodríguez De Francisco, Luis Orlando Maroto Martín, Omar Paino Perdomo, and Edian Franklin Franco De Los Santos. 2023. "Beta-Lactam Susceptibility Profiles of Bacteria Isolated from the Ozama River in Santo Domingo, Dominican Republic" Sustainability 15, no. 6: 5109. https://doi.org/10.3390/su15065109
APA StyleBonnelly, R., Queiroz Cavalcante, A. L., Calderon, V. V., Baraúna, R. A., Jucá Ramos, R. T., Rodríguez-Rodríguez, Y., De Francisco, L. E. R., Maroto Martín, L. O., Perdomo, O. P., & Franco De Los Santos, E. F. (2023). Beta-Lactam Susceptibility Profiles of Bacteria Isolated from the Ozama River in Santo Domingo, Dominican Republic. Sustainability, 15(6), 5109. https://doi.org/10.3390/su15065109







