1. Introduction
Today, the oceans are facing increasing pressures, many of them of anthropogenic origin, such as pollution, global warming, overexploitation or illegal activities, which generate biodiversity losses and deteriorate marine ecosystems, acting at different geographical and temporal scales. These pressures affect the capacity of aquatic ecosystems to maintain a healthy, safe and resilient state and jeopardize the provision of food of high nutritional quality and biodiversity [
1,
2]. For the maintenance of seafood provision for future generations, marine resources must be sustainably managed. Fecundity is one of the population parameters considered critical in estimating the reproductive potential of a fish stock [
3] and is thus of interest to fishery scientists as both a critical parameter of stock assessment [
4] and as a basic aspect of population dynamics [
5]. The importance of determining accurate fecundity estimates has led to many research efforts to provide simpler, faster and lower-cost methods for fisheries science [
6].
For practical purposes, fecundity is the number of mature oocytes that a fish can spawn, and it can vary from thousands to millions of eggs. Nowadays, the stereological method [
7,
8] is the most accurate technique to estimate fish fecundity by using histological images of fish ovaries [
9]. Stereology is a tridimensional interpretation of bidimensional sections of the 3D structure [
8], allowing the estimation of the number of particles (in our case, cells) and the volume that they occupy within the structure. In order to automate this process, it will be necessary to recognize the mature cells (oocytes) in the image and classify them into categories [
10]. Cell recognition is an image segmentation process, which is a relevant topic in computer vision [
11]. The segmentation of cells in routinely stained histological images is a challenging problem due to the high variability in images, caused by a number of factors, including differences in slide preparation (dye concentration, evenness of the cut, presence of foreign artifacts, damage of tissue sample, etc.) and the image acquisition system (presence of digital noise, specific features of slice scanner, different lighting conditions or variations in microscope focus throughout the image). Furthermore, biological heterogeneity among specimens (cell type or development state) and differences in the type of tissue under observation influence outcomes. A successful image segmentation approach will have to overcome these variations in a robust way in order to maintain high quality and accuracy in all situations.
Image segmentation divides an image into non-overlapping regions. Classically, the segmentation methods are categorized into thresholding, edge-based, region-based, morphological segmentation or watershed and hybrid strategies [
11]. Edge-based segmentation finds boundaries between regions based on local discontinuities in image properties (brightness, texture, color or numerical measures over local image patches). Region-based segmentation constructs regions directly based on similarities in image properties. Thresholding segmentation is accomplished via thresholds based on the distribution of pixels’ properties. Thresholding is the simplest and fastest method, but it is not suitable for complex images. Note that the results of edge-based and region-based methods may not be exactly the same. On the other hand, it is easy to construct regions from their borders and conversely to detect borders of existing regions.
The segmentation approaches can also be categorized by the families of algorithms into active contours [
12], graph cuts [
13], edge detectors [
14], clustering, other hybrid methods and deep learning (DL). The first ones are unsupervised, while the last one is normally used in a supervised manner. The active contour model family transforms image segmentation to an energy minimization problem, where the energy functional specifies the segmentation criterion and the unknown variables describe the contours of different regions. The active contour is a parametric model, with an explicit representation (called snakes) or an implicit representation (called level set algorithms). The level set methods can be categorized into edge-based and region-based, according to the image property embedded in the energy functional. The Chan–Vese model is the most representative region-based model [
15]. The graph cut models represent the image as a weighted graph, and the segmentation problem is translated to find a minimum cut in the constructed velocity graph via a maximum flow computation. Clustering algorithms can cluster any image pixels that can be represented by numeric attributes of those pixels. The most popular clustering algorithms can broadly be classified into graph-based like normalized cuts [
16] or Felzenszwalb–Huttenlocher segmentation algorithms [
17] and non-graph-based like k-means, meanshift [
18] or expectation maximization (EM).
The DL methods are usually complex and require the support of powerful computing resources [
19]. They have been used to segment different organs in radiological medical images [
20], among others. In the field of medical microscopic images, Jiang et al. [
21] reviewed the applications of DL in citological studies, principally to classify and detect cells or nuclei in citology images. Shujian Deng et al. reviewed the applications of DL for digital pathology image analysis [
22], including classification, detection and semantic segmentation. Due to the computational requirements of DL methods, they are applied to image patches or downsampled images. The most popular DL techniques for image segmentation are U-Net versions [
23], DeepLab [
24] and SegNet [
25].
The aim of this work is to design and evaluate algorithms to segment cells in histological images of fish gonads requiring low computational time and resources in order to (1) be executed on general-purpose computers in order to be used in all institutions around the world, without any specific computing equipment, in order to manage its marine resources, e.g., to be included in the STERapp software [
26]; and (2) be used in interactive systems, because the problem is too complex to provide totally automatic recognition and it may be necessary to rely on expert supervision before image quantification.
In our previous work [
27], we statistically evaluated different state-of-the-art, publicly available segmentation techniques to recognize cells in histological images of two fish species, although cell segmentation is still an open issue due to the complexity of these images. In the current paper, we propose a new segmentation approach, called MSCF (multi-scale Canny filter), to recognize cells based on the Canny filter, and we perform an extensive statistical evaluation using five fish species.
Section 2 describes the datasets used in the experimental work.
Section 3 describes the proposed MSCF segmentation algorithm and the measures used to report the performance. Finally, in
Section 4, we present and discuss the results obtained, and
Section 5 summarizes the main conclusions achieved.
4. Results
The MSCF algorithm is applied to the grey-level images of all datasets using the following parameters: (1) the Gaussian spread of the filter to smooth the images is some combination of the set
with
; and (2) the rate for selecting the two thresholds to execute the hysteresis process on the gradient image is some combination of the following pairs—
. In order to test the effect of varying both parameters, we performed experiments using one, two or three
values.
Table 2 shows the highest value
using one, two or three values of
for each fish species. The precision and recall and the best configuration for the rates to threshold are also presented. The
score gives a trade-off between the capability of the algorithm to detect pixels that belong to cells without detecting pixels outside the cells. The highest
ranges from 70.14% for
pouting to 80.33% for the
Four-spot-megrim species (see column
in the table). The best configuration, number of
s used and values of the thresholds depend on the fish species considered, as can be seen in
Table 2. Nevertheless, the differences in performance (
value) are small for different numbers of
s. The highest
is normally achieved using more than one value of
, except for the
pouting fish species. In order to identify the best configuration globally, the Friedman rank [
29] over all configurations and species is shown in
Table 3. The first position is for an MSCF that needs three scales
and one pair of thresholds
; in the second and third positions, the MSCF algorithm uses two scales
and also one pair of thresholds; and the first configuration using only one scale, as in the classical Canny filter, is ranked in 12th position. Thus, the best performance cannot be achieved using the classical Canny filter.
We compare the best configuration for the MSCF algorithm with other state-of-the-art segmentation techniques mentioned in
Section 1 with publicly available code. Specifically, we use the following approaches: k-means clustering (implemented by
kmeans function in the OpenCV library (
http://opencv.org (accessed on 12 July 20233))), mean shift (Authors’ code:
https://github.com/xylin/EDISON (accessed on 12 July 2023)) [
18], Chan–Vese (
http://dx.doi.org/10.5201/ipol.2012.g-cv (accessed on 12 July 2023)) [
15,
30], region merging (Authors’ code:
http://cs.brown.edu/~pff/segment/ (accessed on 12 July 2023)) [
17] and Govocitos [
10] segmentation methods. The configuration used for each mentioned technique is detailed in
Section S1 of the Supplementary Materials. Deep learning approaches are not considered in this comparison because they are normally used as supervised segmentation techniques and they would require large amounts of computational resources for the size of the images in our datasets. Moreover, these are high-resolution images, but DL should be applied on image patches or downsampled images. None of the options are acceptable in our case, because (1) image patches should be large enough to include a small number of whole cells, and therefore the patches would be too large for a DL setting; and (2) image downsampling to the size required by DL networks may significantly reduce the image information required to perform cell recognition.
The output of the MSCF algorithm is a set of contours
associated with the outline of the cells. The output of the remaining algorithms is a binary image
with cells and background. In the last case, the set of contours
is extracted from
using the Suzuki algorithm [
31]. The experts provide a minimum diameter for the matured cells,
, which depends on the fish species and the spatial resolution at which the images are acquired. In our case,
pixels for all fish species. At the same time, the cells are always rounded, so we assume that their roundness is lower than a value
. We consider
, slightly above 1, which is the circle roundness. Finally, in all algorithms, the contours
are filtered by size and roundness.
Table 4 shows the performance (classification of pixels) of all algorithms for the different fish species using the same minimum diameter,
pixels, which is the parameter used by the fishery experts in their daily work to distinguish immature from matured cells. Comparing the segmentation algorithms tested, the highest
value is provided by the MSCF algorithm for all fish species (
% for
pouting, 72.04% for
European pilchard, 80.33% for
Four-spot megrim and 71.16% for
Roughhead grenadier), except for the
hake species, where the best performance is provided by clustering (
%). The differences from the best algorithm to the poorest one are normally less than 10 points for all fish species: from 67.67% for the Chan–Vese method to 79.86% for clustering and the
hake species; from 46.6% for the Chan–Vese method to 70.14% for MSCF and
pouting; from 65.97% for the Chan–Vese method to 72.04 for MSCF and the
European pilchard species; from 65.09% for the Chan–Vese method to 80.33% for MSCF and the
Four-spot megrim species; and from 43.55% for the Chan–Vese method to 71.16 for MSCF and the
Roughhead grenadier species. Thus, the Chan–Vese method is the worst segmentation algorithm for this problem.
Due to the properties of our problem, the best algorithm from the pixel classification point of view would not be the best option. For example, imagine that the segmentation algorithm detects correctly the majority of pixels of a cell; thus, from the pixel point of view, its performance is good. However, if these correctly detected pixels are distributed into various regions, its performance would be rather poor in order to count and measure the cells, which is our goal in estimating fish fecundity.
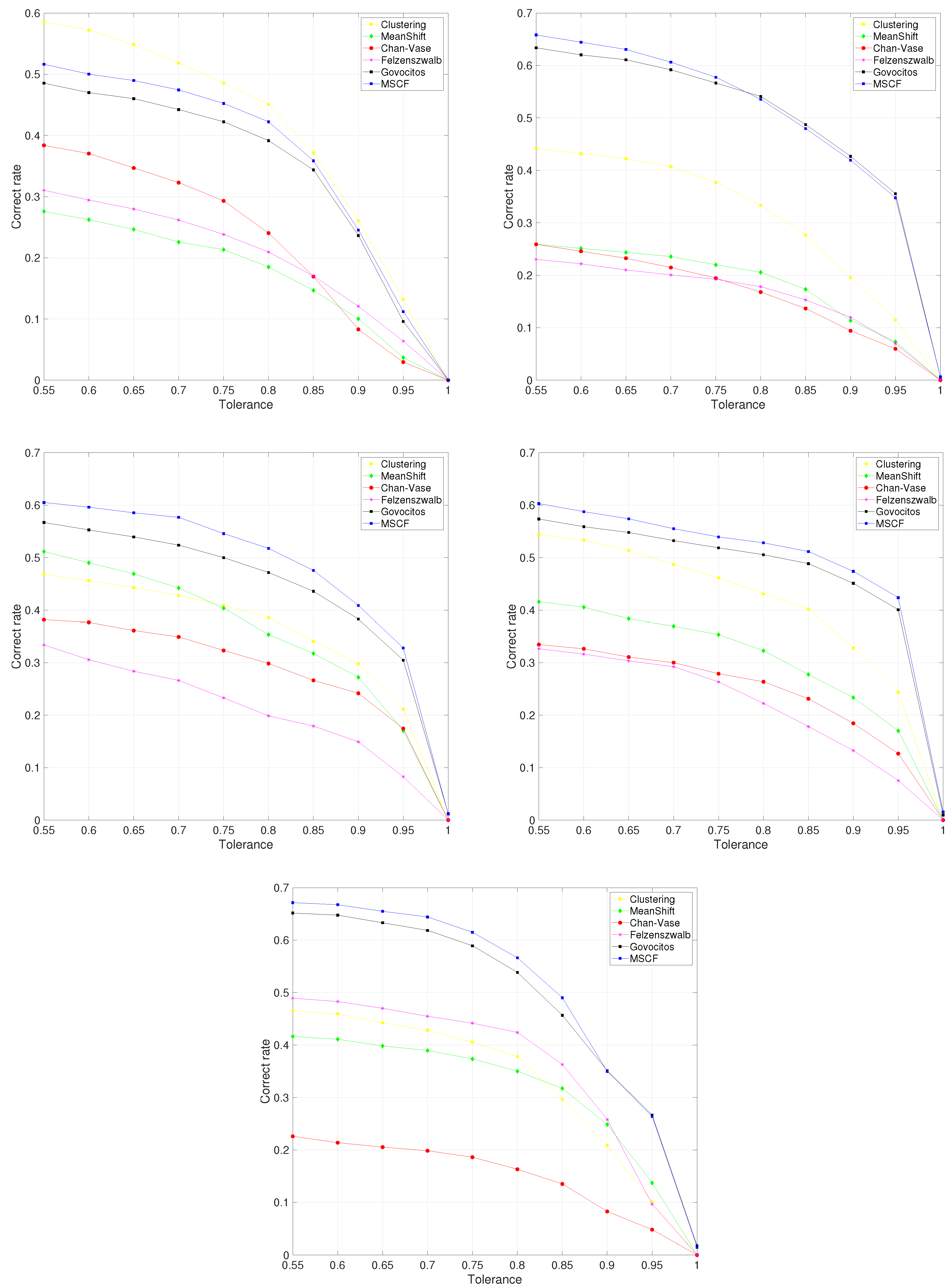
Figure 2 shows the variation in correct cell recognition for different values of tolerance for all the fish species. The MSCF algorithm achieved the best results, followed by the Govocitos algorithm, with the exception of the
European hake fish species, in which clustering provided the best results. The correct rate decreases as a stricter overlap between the computer-recognized and expert-annotated cell is required, i.e., for higher values of tolerance
T, being practically zero when we demand a perfect cell overlap. Experts believe that an overlap of 70% could be acceptable for practical purposes. For this value, the average percentage of cells correctly recognized is 51.83% for the
hake fish species and clustering segmentation algorithm, and 60.62%, 57.67%, 55.55% and 64.41% for the MSCF algorithm and the
pouting,
European pilchard,
Four-spot and
Roughhead grenadier species, respectively. The average performance for the different fish species ranges from 51.83% for
European hake to 64.41% for
Roughhead grenadier fish species.
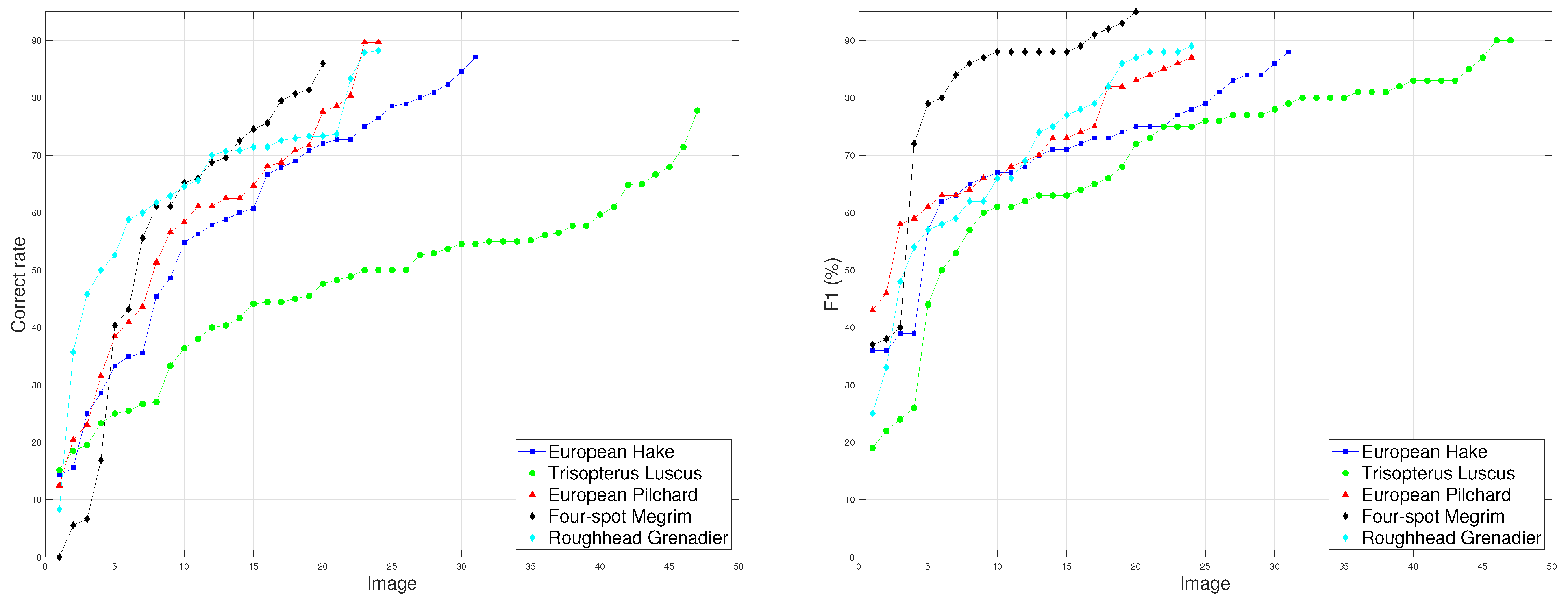
Figure 3 shows the performance of the MSCF algorithm using the best global configuration (
and
) for each image of all fish species. The left panel shows the correct cell recognition rate for a tolerance of
T = 0.7 and the right panel presents the
value. Among each fish species, there is great variability in performance in terms of the correct cell recognition rate and
value. This behavior is similar for all fish species. It is important to emphasize that these datasets were built to test the robustness of the STERapp software, and the images present high variability. The absolute performance at a pixel level (
value) is always higher than at the region level (correct cell recognition rate), which confirms the hypothesis that the pixels are correctly labeled as cells or background but the connectivity among pixels could not be correctly identified. Thus, we can conclude that the MSCF algorithm can be automatically run to recognize cells for some images but, for other images, the recognition of cells could be practically performed manually by the fishery experts. Thus, in order to provide a fully automatic tool to estimate fecundity, it will be necessary to adopt software like STERapp that combines automatic processing with an intuitive graphical interface to review the recognition results before image quantification [
27].
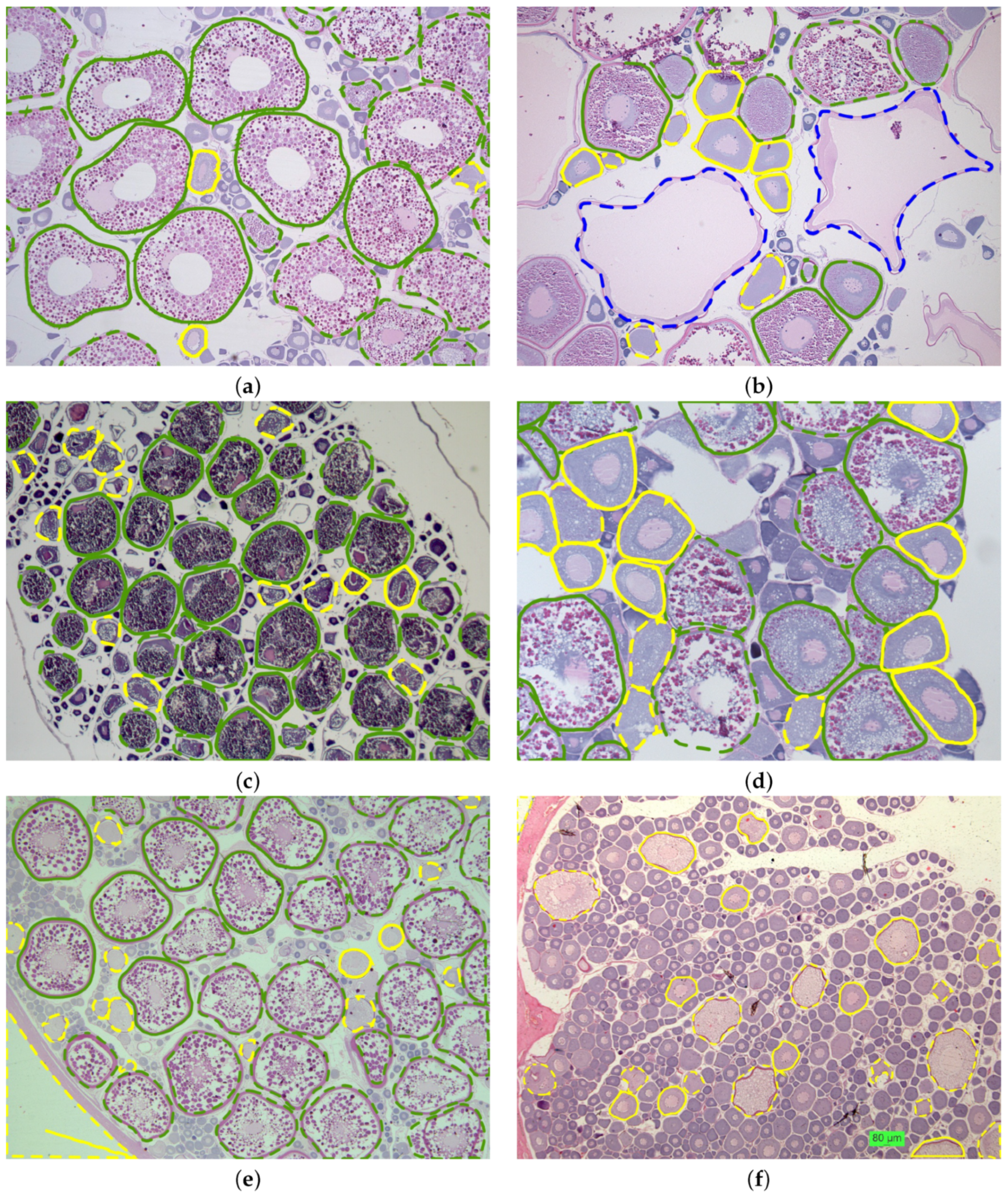
Figure 4 shows visual examples of the performance of the MSCF algorithm for the different fish species, where the cell contour annotated by the expert and the cell outline recognized by the algorithm are overlapped with the image in green and blue, respectively. In all cases, we use the minimum diameter
pixels. As can be seen visually in the images, some false positive cells are due to the detection of cells whose sizes are between matured and immature. In some cases, they were detected by the algorithm, but the experts considered them immature. This fact led us to perform additional experiments using a greater
value, providing slightly higher performance results. Thus, in the current version of STERapp [
27], the process of cell recognition can be achieved using two steps: firstly using a larger diameter and, if it is necessary, using a second diameter to add the undetected cells.
To determine which is the best algorithm globally,
Table 5 shows the Friedman ranking of the algorithms over all datasets for correct cell recognition. The MSCF algorithm achieved the first position, very near to 1. This means that it achieved the best performance in almost all the experiments. The second position, with a ranking of 2.2 (i.e., the second-best performance in almost all the experiments), was achieved by Govocitos, followed by clustering, meanshift, Felzenszwalb and Chan–Vese in the last position.
All the experiments were performed on a computer equipped with 32 GB of RAM and a 3.10 GHz processor under Linux Kubuntu 20.04. The average elapsed time of the segmentation algorithms per image, using the configuration that provided the best performance for each fish species, was 4.61 s for clustering, 2.97 s for Felzenszwalb, 3.83 s for Govocitos, 2.69 s for MSCF, 133.31 s for meanshift and 165.04 s for the Chan–Vese algorithm. The MSCF algorithm was the fastest one, followed by Felzenszwalb, Govocitos and clustering. In fact, the time needed by the first four algorithms was only a few seconds, which makes them suitable for image processing in interactive applications, while the last two require more than one minute and they are too slow for real-time applications.