Assessment of Antifungal/Anti-Oomycete Activity of Frass Derived from Black Soldier Fly Larvae to Control Plant Pathogens in Horticulture: Involvement of Bacillus velezensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Pathogenic Fungi and Oomycetes
2.2. Gainesville Diet
2.3. Frass
2.4. Extracts of Frass/Gainesville Diet
2.5. Effect of Extracts on Plant Pathogen Mycelial Growth (Dual Culture Overlay Assay)
2.6. Isolation of Bacteria and Fungi from Gainesville Diet and Frass Extracts
2.7. Screening Isolates for Antagonistic Activity against Plant Pathogens (Dual Culture on Agar)
2.8. Identification of Isolates GV1-11 and FGV15-6
2.8.1. DNA Extraction and Whole Genome Sequencing
2.8.2. Genomic Sequence Analysis
2.9. Effect of Bacillus Velezensis on Plant Pathogen Mycelial Growth
3. Results
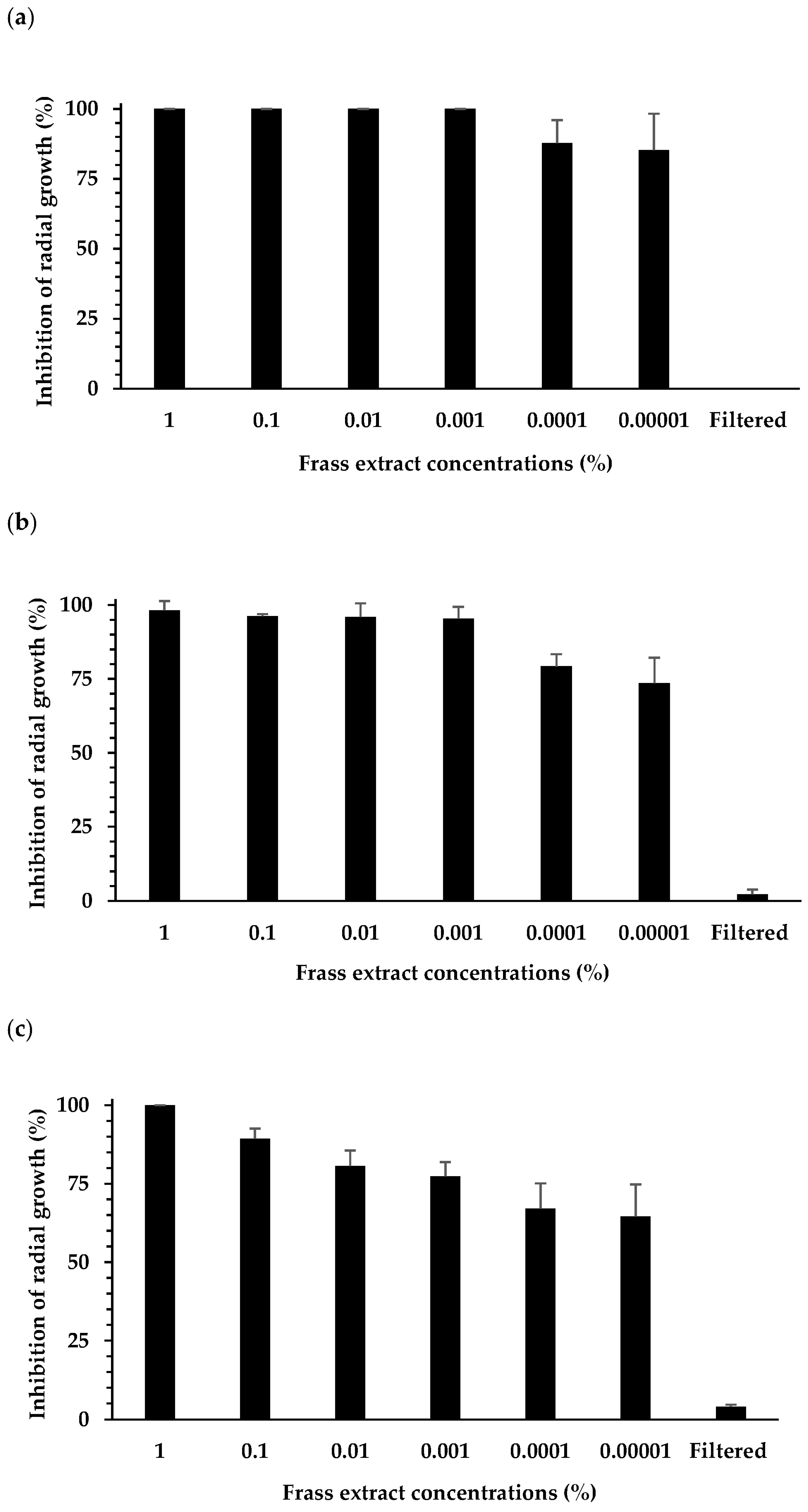
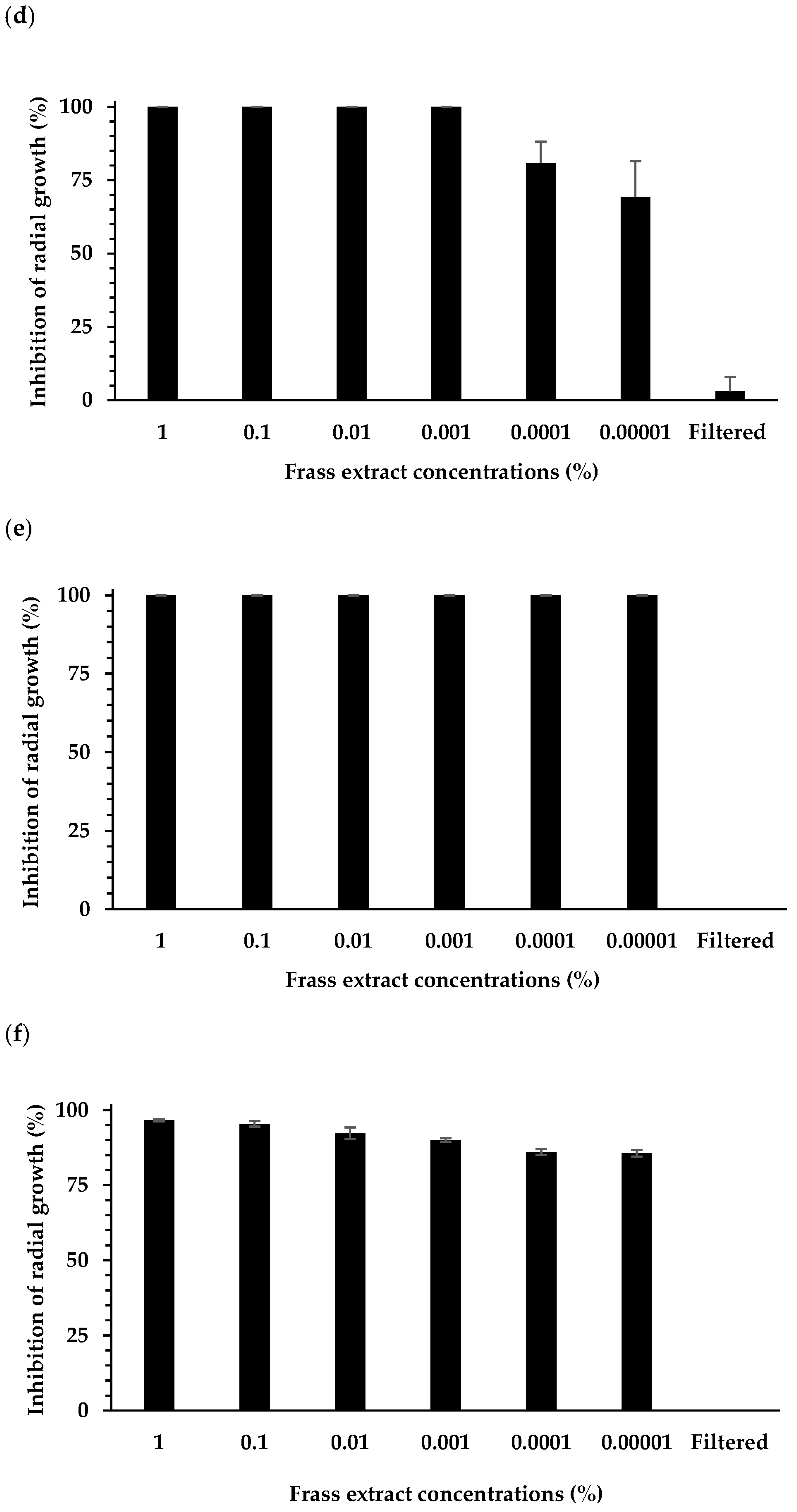
3.1. Effect of Extracts on Plant Pathogen Mycelial Growth
3.2. Screening Isolates for Antagonistic Activity against Plant Pathogens (Dual Culture on Agar)
3.3. Identification of Isolates
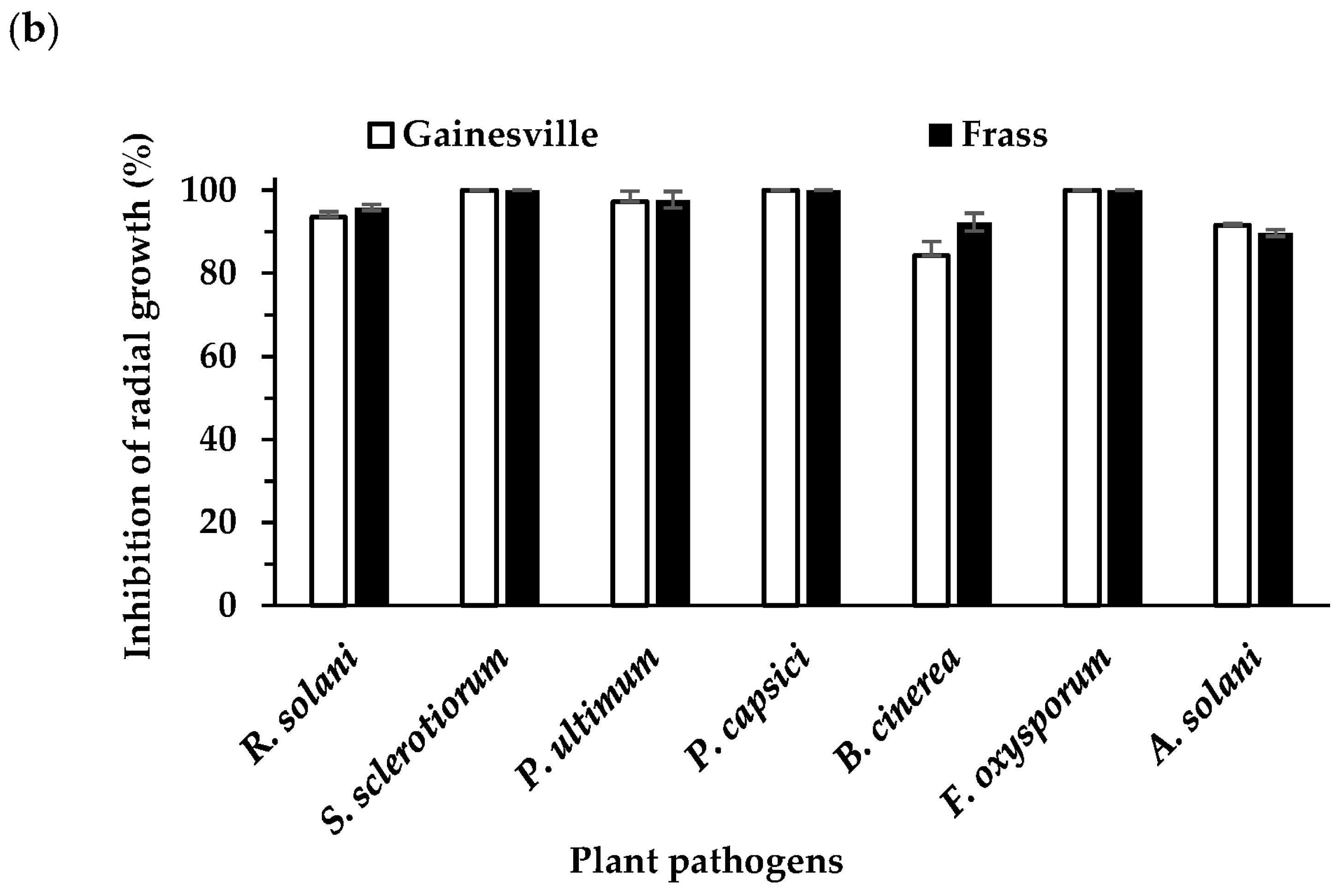
3.4. Effect of B. velezensis on Plant Pathogen Mycelial Growth (Dual Culture Overlay Assay)
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cullere, M.; Tasoniero, G.; Giaccone, V.; Miotti-Scapin, R.; Claeys, E.; De Smet, S.; Dalle Zotte, A. Black soldier fly as dietary protein source for broiler quails: Apparent digestibility, excreta microbial load, feed choice, performance, carcass and meat traits. Animal 2016, 10, 1923–1930. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, T.; Bosch, G. Insects: A protein-rich feed ingredient in pig and poultry diets. Anim. Front. 2015, 5, 45–50. [Google Scholar]
- Diener, S.; Zurbrügg, C.; Tockner, K. Conversion of organic material by black soldier fly larvae: Establishing optimal feeding rates. Waste Manag. Res. 2009, 27, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.X.; Tomberlin, J.K.; Vanlaerhoven, S. Ability of black soldier fly (Diptera:Stratiomyidae) larvae to recycle food waste. Environ. Entomol. 2015, 44, 406–410. [Google Scholar] [CrossRef]
- Ravi, H.K.; Degrou, A.; Costil, J.; Trespeuch, C.; Chemat, F.; Vian, M.A. Larvae mediated valorization of industrial, agriculture and food wastes: Biorefinery concept through bioconversion, processes, procedures, and products. Processes 2020, 8, 857. [Google Scholar] [CrossRef]
- Schmitt, E.; de Vries, W. Potential benefits of using Hermetia illucens frass as a soil amendment on food production and for environmental impact reduction. Curr. Opin. Green Sustain. Chem. 2020, 25, 100335. [Google Scholar] [CrossRef]
- Arabzadeh, G.; Delisle-Houde, M.; Tweddell, R.J.; Deschamps, M.-H.; Dorais, M.; Lebeuf, Y.; Derome, N.; Vandenberg, G. Diet composition influences growth performance, bioconversion of black soldier fly larvae: Agronomic value and in vitro biofungicidal activity of derived frass. Agronomy 2022, 12, 1765. [Google Scholar] [CrossRef]
- Beesigamukama, D.; Mochoge, B.; Korir, N.K.; Fiaboe, K.K.M.; Nakimbugwe, D.; Khamis, F.M.; Subramanian, S.; Dubois, T.; Musyoka, M.W.; Ekesi, S.; et al. Exploring black soldier fly frass as novel fertilizer for improved growth, yield, and nitrogen use efficiency of maize under field conditions. Front. Plant Sci. 2020, 11, 574592. [Google Scholar] [CrossRef]
- Beesigamukama, D.; Subramanian, S.; Tanga, C.M. Nutrient quality and maturity status of frass fertilizer from nine edible insects. Sci. Rep. 2022, 12, 7182. [Google Scholar] [CrossRef]
- Ding, M.-Q.; Yang, S.-S.; Ding, J.; Zhang, Z.-R.; Zhao, Y.-L.; Dai, W.; Sun, H.-J.; Zhao, L.; Xing, D.; Ren, N.; et al. Gut microbiome associating with carbon and nitrogen metabolism during biodegradation of polyethene in Tenebrio larvae with crop residues as co-diets. Environ. Sci. Technol. 2023, 57, 3031–3041. [Google Scholar] [CrossRef]
- Bruno, D.; Bonelli, M.; De Filippis, F.; Di Lelio, I.; Tettamanti, G.; Casartelli, M.; Ercolini, D.; Caccia, S. The intestinal microbiota of Hermetia illucens larvae is affected by diet and shows a diverse composition in the different midgut regions. Appl. Environ. Microbiol. 2019, 85, e01864-18. [Google Scholar] [CrossRef]
- Jiang, C.-L.; Jin, W.-Z.; Tao, X.-H.; Zhang, Q.; Zhu, J.; Feng, S.-Y.; Xu, X.-H.; Li, H.-Y.; Wang, Z.-H.; Zhang, Z.-J. Black soldier fly larvae (Hermetia illucens) strengthen the metabolic function of food waste biodegradation by gut microbiome. Microb. Biotechnol. 2019, 12, 528–543. [Google Scholar] [CrossRef]
- Penhallegon, R. Nitrogen-Phosphorus-Potassium Values of Organic Fertilizers; Oregon State University, Extension Service: Corvallis, OR, USA, 2003. [Google Scholar]
- Temple, W.D.; Radley, R.; Baker-French, J.; Richardson, F. Use of Enterra Natural Fertilizer (Black Soldier Fly Larvae Digestate) as a Soil Amendment; Enterra Feed Corporation: Vancouver, BC, Canada, 2013. [Google Scholar]
- Quilliam, R.S.; Nuku-Adeku, C.; Maquart, P.; Little, D.; Newton, R.; Murray, F. Integrating insect frass biofertilisers into sustainable peri-urban agro-food systems. J. Insects Food Feed 2020, 6, 315–322. [Google Scholar] [CrossRef]
- Hogsette, J.A. New diets for production of house flies and stable flies (Diptera: Muscidae) in the laboratory. J. Econ. Entomol. 1992, 85, 2291–2294. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Kreft, Ł.; Botzki, A.; Coppens, F.; Vandepoele, K.; Van Bel, M. PhyD3: A phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 2017, 33, 2946–2947. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, C.A.; Kim, S.-J.; Kwon, S.-W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and “Bacillus oryzicola” are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [PubMed]
- Shaviv, A. Advances in controlled-release fertilizers. Adv. Agron. 2001, 71, 1–49. [Google Scholar]
- Beesigamukama, D.; Mochoge, B.; Korir, N.K.; Fiaboe, K.K.M.; Nakimbugwe, D.; Khamis, F.M.; Subramanian, S.; Wangu, M.M.; Dubois, T.; Ekesi, S.; et al. Low-cost technology for recycling agro-industrial waste into nutrient-rich organic fertilizer using black soldier fly. Waste Manag. 2021, 119, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Klammsteiner, T.; Turan, V.; Fernández-Delgado Juárez, M.; Oberegger, S.; Insam, H. Suitability of black soldier fly frass as soil amendment and implication for organic waste hygienization. Agronomy 2020, 10, 1578. [Google Scholar] [CrossRef]
- Setti, L.; Francia, E.; Pulvirenti, A.; Gigliano, S.; Zaccardelli, M.; Pane, C.; Caradonia, F.; Bortolini, S.; Maistrello, L.; Ronga, D. Use of black soldier fly (Hermetia illucens (L.), Diptera: Stratiomyidae) larvae processing residue in peat-based growing media. Waste Manag. 2019, 95, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hidalgo, M.; Pascual, J.; de la Cruz, M.; Martín, J.; Kath, G.S.; Sigmund, J.M.; Masurekar, P.; Vicente, F.; Genilloud, O.; Bills, G.F. Prescreening bacterial colonies for bioactive molecules with Janus plates, a SBS standard double-faced microbial culturing system. Antonie Van Leeuwenhoek 2012, 102, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Blom, J.; Klenk, H.-P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis Form an “Operational Group B. amyloliquefaciens” within the B. subtilis Species Complex. Front. Microbiol. 2017, 8, 22. [Google Scholar]
- Chowdhury, S.P.; Dietel, K.; Rändler, M.; Schmid, M.; Junge, H.; Borriss, R.; Hartmann, A.; Grosch, R. Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS ONE 2013, 8, e68818. [Google Scholar] [CrossRef]
- Elanchezhiyan, K.; Keerthana, U.; Nagendran, K.; Prabhukarthikeyan, S.R.; Prabakar, K.; Raguchander, T.; Karthikeyan, G. Multifaceted benefits of Bacillus amyloliquefaciens strain FBZ24 in the management of wilt disease in tomato caused by Fusarium oxysporum f. sp. lycopersici. Physiol. Mol. Plant Pathol. 2018, 103, 92–101. [Google Scholar] [CrossRef]
- Schmiedeknecht, G.; Bochow, H.; Junge, H. Use of Bacillus subtilis as biocontrol agent. II. Biological control of potato diseases. J. Plant Dis. Prot. 1998, 105, 376–386. [Google Scholar]
- Sylla, J.; Alsanius, B.W.; Krüger, E.; Reineke, A.; Strohmeier, S.; Wohanka, W. Leaf microbiota of strawberries as affected by biological control agents. Phytopathology 2013, 103, 1001–1011. [Google Scholar] [CrossRef]
- Talboys, P.J.; Owen, D.W.; Healey, J.R.; Withers, P.J.; Jones, D.L. Auxin secretion by Bacillus amyloliquefaciens FZB42 both stimulates root exudation and limits phosphorus uptake in Triticum aestivum. BMC Plant Biol. 2014, 14, 51. [Google Scholar] [CrossRef]
- Yao, A.V.; Bochow, H.; Karimov, S.; Boturov, U.; Sanginboy, S.; Sharipov, A.K. Effect of FZB 24® Bacillus subtilis as a biofertilizer on cotton yields in field tests. Arch. Phytopathol. Plant Prot. 2006, 39, 323–328. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Schneider, K.; Vater, J.; Süssmuth, R.; Piel, J.; Borriss, R. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 2009, 140, 27–37. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Bouchard-Rochette, M.; Machrafi, Y.; Cossus, L.; Nguyen, T.T.A.; Antoun, H.; Droit, A.; Tweddell, R.J. Bacillus pumilus PTB180 and Bacillus subtilis PTB185: Production of lipopeptides, antifungal activity, and biocontrol ability against Botrytis cinerea. Biol. Control 2022, 170, 104925. [Google Scholar] [CrossRef]
- Arrebola, E.; Jacobs, R.; Korsten, L. Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. J. Appl. Microbiol. 2010, 108, 386–395. [Google Scholar] [CrossRef]
- DeFilippi, S.; Groulx, E.; Megalla, M.; Mohamed, R.; Avis, T.J. Fungal competitors affect production of antimicrobial lipopeptides in Bacillus subtilis strain B9-5. J. Chem. Ecol. 2018, 44, 374–383. [Google Scholar] [CrossRef]
- Gao, L.; Han, J.; Liu, H.; Qu, X.; Lu, Z.; Bie, X. Plipastatin and surfactin coproduction by Bacillus subtilis pB2-L and their effects on microorganisms. Antonie Van Leeuwenhoek 2017, 110, 1007–1018. [Google Scholar] [CrossRef]
- Gong, Q.; Zhang, C.; Lu, F.; Zhao, H.; Bie, X.; Lu, Z. Identification of bacillomycin D from Bacillus subtilis fmbJ and its inhibition effects against Aspergillus flavus. Food Control 2014, 36, 8–14. [Google Scholar] [CrossRef]
- Klammsteiner, T.; Walter, A.; Bogataj, T.; Heussler, C.D.; Stres, B.; Steiner, F.M.; Schlick-Steiner, B.C.; Arthofer, W.; Insam, H. The core gut microbiome of black soldier fly (Hermetia illucens) larvae raised on low-bioburden diets. Front. Microbiol. 2020, 11, 993. [Google Scholar] [CrossRef] [PubMed]




| GV1-11 | FGV15-6 | |||||
|---|---|---|---|---|---|---|
| dDDH | C.I. | G+C Content Difference | dDDH | C.I. | G+C Content Difference | |
| Bacillus Strain | (d4, in %) | (d4, in %) | (in %) | (d4, in %) | (d4, in %) | (in %) |
| B. amyloliquefaciens subsp. plantarum FZB42 | 88.2 | [85.7–90.3] | 0.63 | 87.8 | [85.3–89.9] | 0.59 |
| B. velezensis NRRL B-41580 | 84.4 | [81.7–86.8] | 0.47 | 84.1 | [81.4–86.6] | 0.43 |
| B. methylotrophicus KACC 13105 | 83.3 | [80.5–85.8] | 0.59 | 83.0 | [80.2–85.5] | 0.55 |
| B. vanillea XY18 | 56.5 | [53.8–59.2] | 0.48 | 57.2 | [54.5–60.0] | 0.44 |
| B. siamensis KCTC 13613 | 56.2 | [53.4–58.9] | 0.49 | 56.9 | [54.1–59.6] | 0.45 |
| B. amyloliquefaciens DSM 7 | 55.5 | [52.7–58.2] | 0.24 | 56.1 | [53.4–58.8] | 0.20 |
| B. nakamurai NRRL B-41091 | 30.7 | [28.3–33.2] | 0.58 | 31.7 | [29.3–34.2] | 0.62 |
| B. spizizenii TU-B-10 | 20.7 | [18.5–23.1] | 2.02 | 22.3 | [20.0–24.7] | 2.06 |
| B. axarquiensis NRRL B-41617 | 20.6 | [18.4–23.0] | 2.05 | 22.1 | [19.9–24.6] | 2.09 |
| B. subtilis NCIB 3610 | 20.6 | [18.4–23.1] | 2.45 | |||
| B. rugosus SPB7 | 20.5 | [18.3–22.9] | 2.72 | 22.0 | [19.7–24.4] | 2.76 |
| B. vallismortis DV1-F-3 | 20.4 | [18.1–22.8] | 2.08 | |||
| B. mojavensis KCTC 3706 | 20.3 | [18.0–22.7] | 2.18 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arabzadeh, G.; Delisle-Houde, M.; Vandenberg, G.W.; Derome, N.; Deschamps, M.-H.; Dorais, M.; Vincent, A.T.; Tweddell, R.J. Assessment of Antifungal/Anti-Oomycete Activity of Frass Derived from Black Soldier Fly Larvae to Control Plant Pathogens in Horticulture: Involvement of Bacillus velezensis. Sustainability 2023, 15, 10957. https://doi.org/10.3390/su151410957
Arabzadeh G, Delisle-Houde M, Vandenberg GW, Derome N, Deschamps M-H, Dorais M, Vincent AT, Tweddell RJ. Assessment of Antifungal/Anti-Oomycete Activity of Frass Derived from Black Soldier Fly Larvae to Control Plant Pathogens in Horticulture: Involvement of Bacillus velezensis. Sustainability. 2023; 15(14):10957. https://doi.org/10.3390/su151410957
Chicago/Turabian StyleArabzadeh, Ghazaleh, Maxime Delisle-Houde, Grant W. Vandenberg, Nicolas Derome, Marie-Hélène Deschamps, Martine Dorais, Antony T. Vincent, and Russell J. Tweddell. 2023. "Assessment of Antifungal/Anti-Oomycete Activity of Frass Derived from Black Soldier Fly Larvae to Control Plant Pathogens in Horticulture: Involvement of Bacillus velezensis" Sustainability 15, no. 14: 10957. https://doi.org/10.3390/su151410957
APA StyleArabzadeh, G., Delisle-Houde, M., Vandenberg, G. W., Derome, N., Deschamps, M.-H., Dorais, M., Vincent, A. T., & Tweddell, R. J. (2023). Assessment of Antifungal/Anti-Oomycete Activity of Frass Derived from Black Soldier Fly Larvae to Control Plant Pathogens in Horticulture: Involvement of Bacillus velezensis. Sustainability, 15(14), 10957. https://doi.org/10.3390/su151410957









