Abstract
Coalescent methods in population genetics aim to detect biodiversity patterns, evolutionary mechanisms, and signatures of historical changes in effective population sizes with respect to the species fidelity. Restriction site-associated DNA sequencing (RADseq) was used to evaluate the population dynamics of invertebrate species within the same localities. New sequencing technologies, such as the ones employed by population genetics, could be used to improve the management and sustainability of marine and aquaculture resources. Sea cucumbers (Holothuria tubolosa) showed genetic differentiation patterns favoring limited gene flow between studied areas. Similar results for clams (Venus verrucosa) suggest local adaptation and low-dispersal abilities for sessile organisms. On the contrary, cuttlefish (Sepia officinalis) exhibited a panmictic pattern, resulting in a single genetic stock in the area. The larvae settlement duration may be responsible for such interspecies variations. Interspecies demographic modeling revealed different environmental pressures of historical events’ signatures with respect to the three invertebrates. Sea cucumbers favor a post-glacial bottleneck event followed by a more recent recovery, whereas cuttlefish favor an expansion before the late glacial maximum. Lastly, clams showed a constant effective population size in the area. The results of historical demographic changes in natural populations provide opportunities for critical evaluation and management in terms of the conservation of the species in the area.
1. Introduction
One of the fundamental questions in evolutionary biology concerns the environment’s potential to shape populations on a spatial scale, with respect to the correlation between site fidelity heterogeneity, selection pressures, and adaptation [1,2]. Environmental heterogeneity shows that species are dependent on climate alterations [3], fitness differences [4], and population dynamics [5]. These processes could be responsible for generating intraspecific diversity among populations [6], or even interspecific differentiation regarding the same geographic regions that differ climatically [7]. Ecological conditions lead to regional scales of local adaptation, implying a potential influence on marine species’ population dynamics [8,9]. Despite the absence of strong barriers in the marine realm, marine organisms show significant population structuring with respect to climatic variation, e.g., [10,11]. However, evolutionary forces, such as adaptation and genetic drift, make it difficult to understand the relevant evolutionary mechanisms that determine structure. On the other hand, spatial genetic structures are evaluated on the landscape level and on geographic distance, with respect to environmental attributes (e.g., dispersal limitation, climate differences) [12]. Apart from short-time forces (e.g., anthropogenic factors), species biogeography has been broadly shaped by large-scale climatic shifts (especially in the Quaternary period) see [13]. Effective population sizes [14], re-colonization events, and fragmented distributions have been largely affected by such demographic changes [15]. On the other hand, bottleneck events or an expansion-based demographic trajectory could lead to differentiated genetic diversity over time and space [16,17,18,19].
Although research into the genetic acclimation of several invertebrates, such as arthropoda, corals and limpets, urchins, and oysters, e.g., [20,21,22,23,24,25] is promising for ecological risk assessment, it is still quite limited. While much progress has been made in recent years, it is clear that continued work on population genetics and demographics is critical, since difficult sustainability and management decisions must be made. However, to the best of our knowledge, this study represents the first report concerning the eastern basin of the Mediterranean Sea, which examines genetic diversity between various Sepia officinalis (cuttlefish) (Linnaeus, 1758), Holothuria tubulosa (sea cucumber) (Gmelin, 1791), and Venus verrucosa (clam) (Linnaeus, 1758) populations that inhabit different marine areas in the North Aegean Sea. Nevertheless, studies within the North Aegean Sea show different patterns for a variety of marine organisms, revealing either high connectivity [26], limited dispersal abilities, or strong barriers to gene flow [27], showing contrasting patterns of gene flow and adaptive genetic variation. Regional heterogeneity aside, the latter may be due to species ecology (e.g., larval settlement) [7] or high selection pressures (e.g., pathogens) [4,28]. Thus, in order to determine the role of the environment in defining the genetic profile of the above three invertebrates, double digest RAD (ddRAD) sequencing was employed (as a promising approach for the detection of molecular adaptation) [29], for an in depth understanding of the evolutionary mechanisms that may explain the genetic diversity and adaptive variation of the three different invertebrates. We tested the hypothesis that geographic areas with respect to abiotic factors (such as temperature, salinity, and chlorophyll-a concentrations) are associated with different levels of genetic diversity among the three species. Additionally, we used coalescent methods, including Approximate Bayesian Computations (ABC), to identify the demographic history of each species in the study area, trying to determine the genomic signatures of historical changes in population size.
2. Materials and Methods
2.1. Animal and Tissue Collection
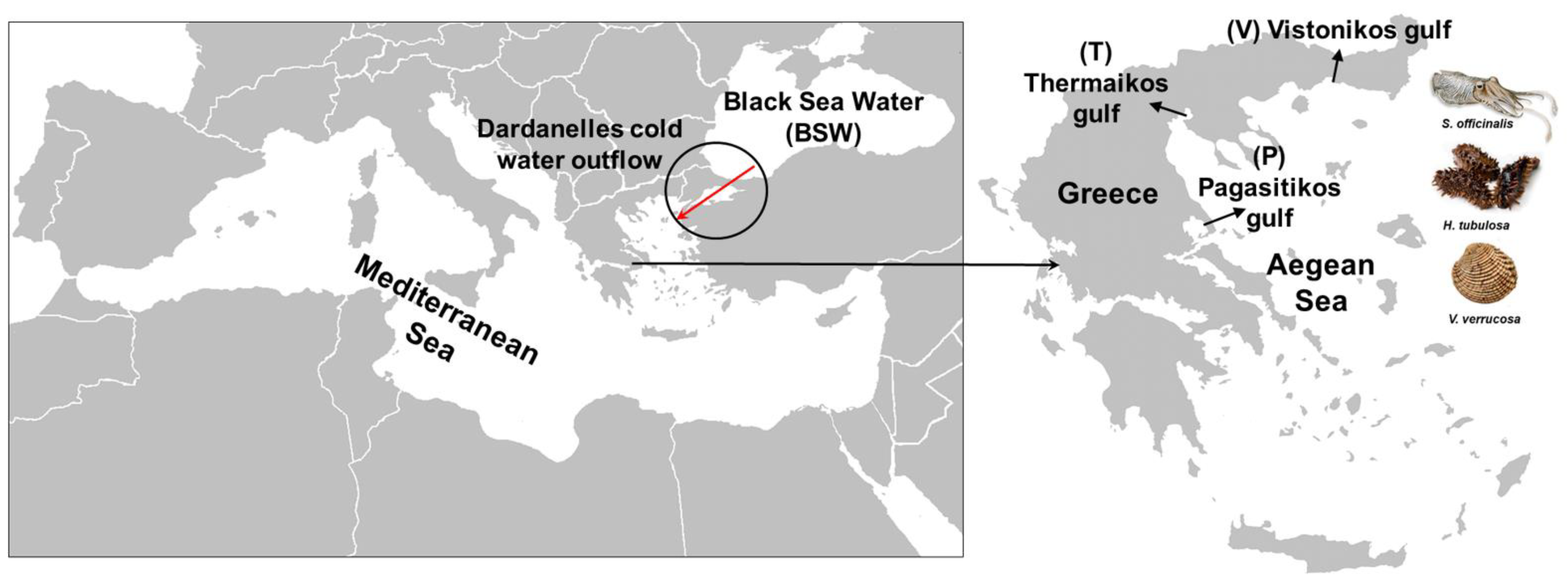
The method used for the collection of animals is explained in detail in Feidantsis et al. [30,31]. S. officinalis, H. tubulosa, and V. verrucosa were collected from the Thermaikos, Pagasitikos, and Vistonikos gulfs (Figure 1). Individuals of the examined species were sampled between 10 and 21 April 2018, 7 and 15 June 2018, 4 and 12 November 2018, and 6 and 11 January 2019. The examined species (6 individuals per gulf per species = 18 individuals per species) were collected by divers and local fishermen (0–10 m depth). Immediately after species sampling, individuals were dissected with high precision anatomic tools, and tissues were immediately placed in DNase- and RNase-free Eppendorf tubes, and thereafter frozen in liquid nitrogen. The frozen samples were transferred to the laboratory, where they were stored at −80 °C for genetic analysis.
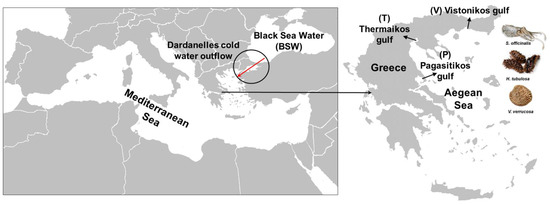
Figure 1.
Sampling sites of the three studied invertebrate species; P: Pagasitikos, T: Thermaikos, V: Vistonikos (red arrow indicates the Dardanelles cold water outflow).
2.2. Genomic Analysis
Muscle tissue was used for DNA extraction using the PureLink™ Genomic DNA Kit. A DNA library was constructed from 54 samples (6 individuals of each species from three locations) following the ddRADseq protocol described in Peterson et al. [29]. We chose a 6 bp cutter (HindIII) and a 4 bp cutter (MspI) based on in silico simulations with the R package SimRAD (Lepais and Weir, 2014) [32]. The fragment size selection window was 300–500 bp. Sequencing was paired end (2 × 151 bp) in one lane on an Illumina NextSeq_500. Reads were trimmed to 110 bp and demultiplexed using the process_radtags command of the software STACKS [33]. After quality control (samples with less than 1 million reads were rejected), 47 samples were retained (see Table 1). Paired reads were mapped against the Holothuria glaberina genome [34] for H. tubulosa samples, Mercenaria mercenaria for V. verrucosa samples [35] and Sepia pharaonis [36] for S. officinalis using BWA v. 0.7.12 [37]. Each resulting sam file was converted to bam format using SAMtools v. 1.3. [38].

Table 1.
Weir and Cockerham FST pairwise differences among the studied populations. (A) sea cucumber (H. tubulosa), (B) cuttlefish (S. officinalis), (C) clam (V. verrucosa).
The command SelectVariants was employed to filter out indels and non-biallelic single nucleotide polymorphisms (SNPs). Thereafter, the command VariantFiltration and the following settings: –filterExpression ‘QD < 2.0||FS > 60.0||MQ < 40.0||MQRankSum←12.5||ReadPosRankSum←8.0n, were employed in order for SNPs to be filtered based on mapping quality. The QUAL score (QD) was normalized by allele depth for a variant, and the Phred-scaled p-value (FS) used Fisher’s exact tests in order to detect strand bias. The MQRankSum command set the z-score from a Wilcoxon rank-sum test of Alt versus Ref read mapping qualities, and ReadPosRankSum did this for reading position bias. Loci were assembled using the GATK HaplotypeCaller [39].
VCF files were further filtered based on both genotype quality and depth of coverage greater than 5 using VCFTools [40], as well as to retain only SNPs genotyped in at least 80% of the individuals. Thereafter, SNPs with a depth of coverage twice the mean depth of coverage were discarded in order to remove potentially paralogous loci. All individuals with more than 20% missing data were removed and only variants with MAF greater than 0.05 were retained. Finally, SNPs showing significant departures from Hardy–Weinberg equilibrium with an alpha level of 0.05 were filtered out using the software PLINK v. 1.9 [41]. The latter was employed in order to prune out putatively linked loci using an r2 threshold of 0.5 as well.
Genetic diversity indices (Number of alleles, Number of effective alleles, HOBS, HEXP and FIS), estimates of heterozygosity, Structure, Principal Component Analysis (PCA) were performed using the program SAMBAR [42] through the R platform [43] following authors recommendations. Weir & Cockerham [44] FST estimates, along with associated significance values, were generated using the Stampp program.
The R package LEA, that enables ecological association studies from the R command line, was employed. The package can perform analyses of population structure and genome scans for adaptive alleles from large genomic data sets [45]. It derives advantages from R programming functionalities in order to adjust significance values for multiple testing issues and to visualize results. The significance of an environmental effect was measured by a z-score statistic computed at each locus using the latent factor models (lfmm) assuming there are K latent factors [46]. The faststructure software was employed for the optimal assignment of the analyzed individuals aiming to estimate the K value, reflecting the theoretically most likely number of populations [47]. This program provides correct null-hypothesis testing procedures and p-values for ecological association tests. Parameters used were set at 1 × 106 iterations, 25% burn in followed by 3 repetitions. The factors used were temperature, salinity, oxygen (Table S6 for mean values of the three abiotic factors). The results were visualized via Manhattan plots using the R package qqman [48]. Multiple comparisons on the GWAS plots were corrected using the Bonferroni type I error correction [49].
The amino acid changes, in each of the variants that LEA showed significant associations, were annotated and predicted using the SnpEff program [50]. This software uses an in-house build function and produces a required database for each of the reference genome. Then, the previously produced VCF file and the gff file of the reference genome was used for the downstream pipeline of the program. Accession numbers of reference genomes used in the present study were: Holothuria glaberrina: GCA_009936505.2, Sepia pharaonis: GCA_903632075.3 and Mercenaria mercenaria: GCA_014805675.2.
2.3. Demographic Analyses
The software DIYABC v2.0 [51] was used to analyze the SNP data of H. tubulosa, S. officinalis and V. verrucosa in order to infer demographic history based on the approximate Bayesian computation (ABC) approach.
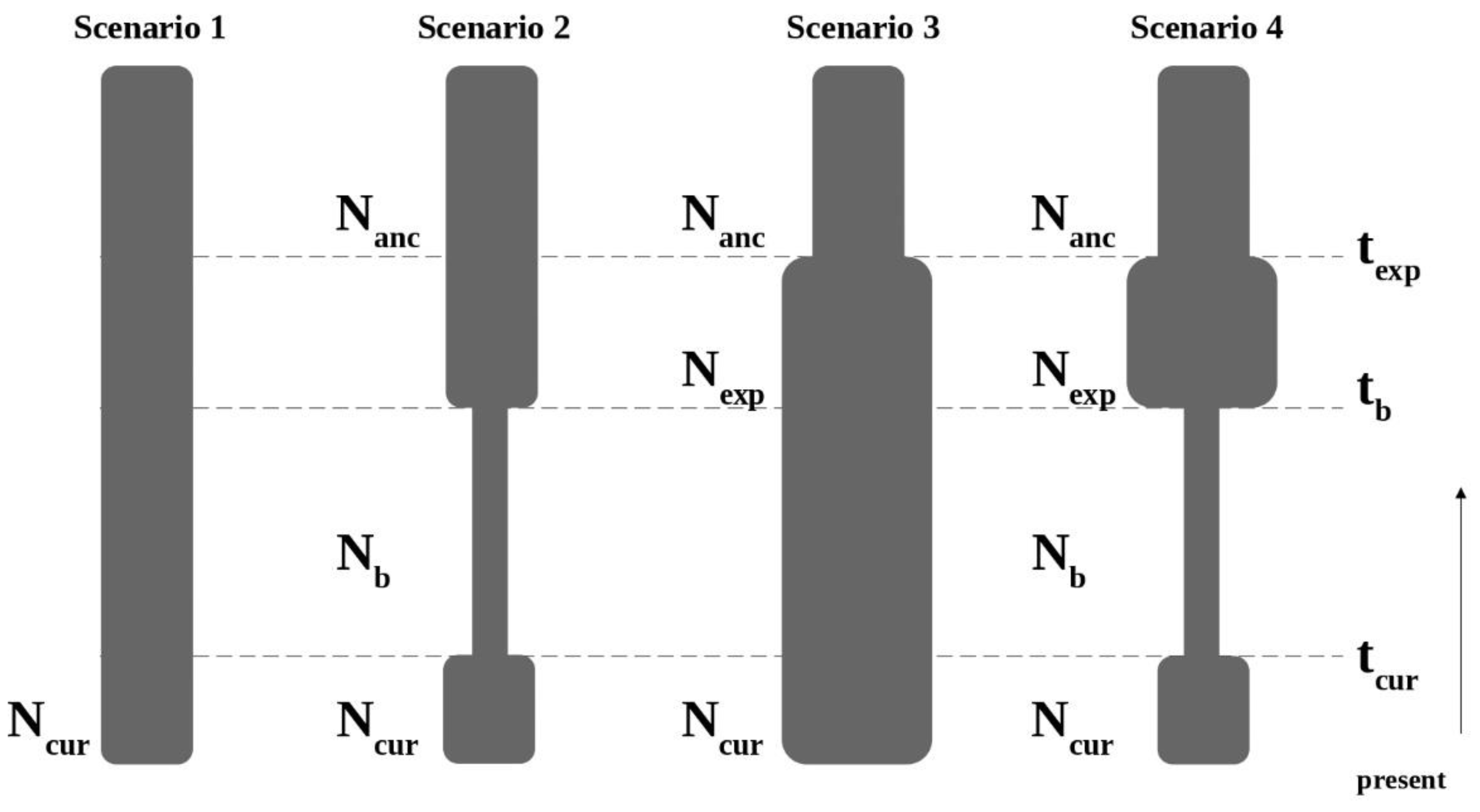
Four simple scenarios were tested for each species and each geographical area (Figure 2). The scenarios were as follows:
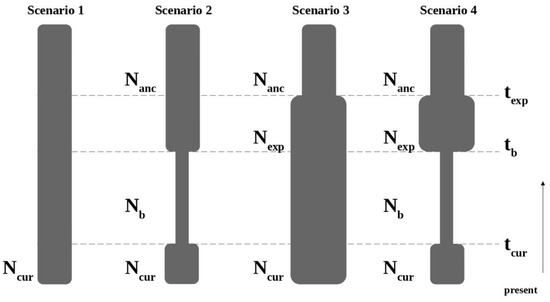
Figure 2.
Demographic scenarios tested with approximate Bayesian computation using DIYABC. Scenarios are defined by relative changes in effective population size (N) at given times (t) since present. Scenario 1: Constant population size model; Scenario 2: Bottleneck with population recovery model; Scenario 3: Population expansion model; Scenario 4: Bottleneck with expansion model.
Scenario 1: Constant population size model: The effective population size was constant at N1 from the past to the present.
Scenario 2: Bottleneck model: The effective population size experienced a bottleneck from an ancestral population (Nanc) to bottlenecked population (Nb) at tb followed by population size recovery from Nb to preset population (N1) at current time (tcur).
Scenario 3: Population expansion model: The effective population size increased from an ancestral population (Nanc) to present population (N1) at texp.
Scenario 4: Bottleneck and expansion model: The effective population size experienced an expansion from an ancestral population (Nanc) to expanded population (Nexp) at texp, followed by a bottleneck from Nexp to bottlenecked population (Nb) at tb, with a population size recovery from Nb to present population (N10) at current time (tcur).
3. Results
Not all the samples revealed useful information regarding the number of sequence reads. For sea cucumber and cuttlefish, the final panel of individuals consisted of 6, 5 and 5 for Pagasitikos, Thermaikos and Vistonikos, respectively, whereas for clam the final panel consisted of 5 individuals per geographical area (Supplementary Information Table S1 for the final samples per species and population).
3.1. H. tubulosa
3.1.1. SNPs Calling
An average of 5,180,850 sequence reads were generated from which we called 5,274,813 raw SNPs. Application of the filtering criteria described in the Materials and Methods resulted in a final dataset of 16,142 high quality filtered SNPs (Supplementary Table S1 for a downstream of applied filtering and remained SNPs).
3.1.2. Genetic Differentiation and Diversity
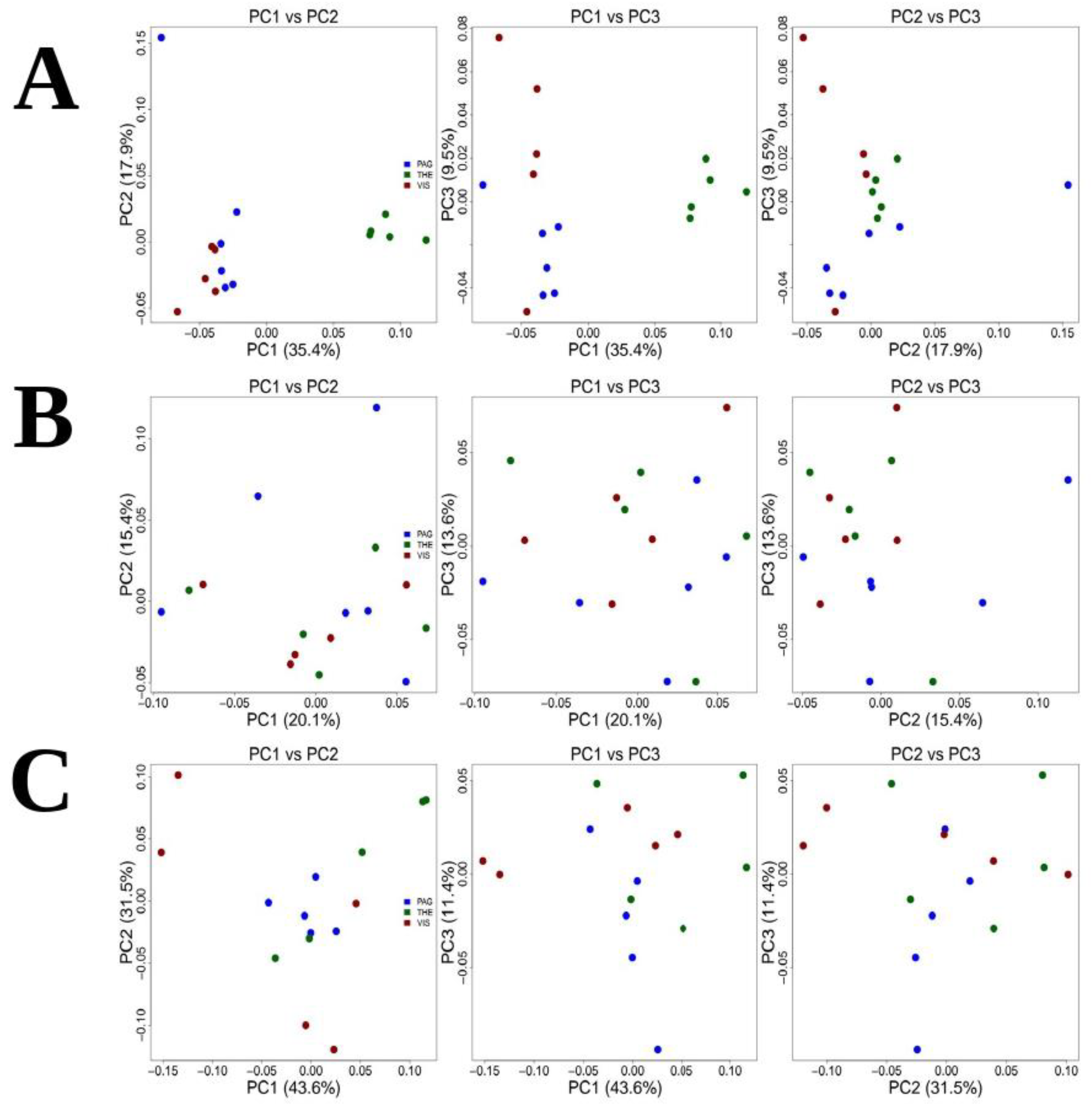
Genetic indices are shown in Table S7. To test for genetic differences, pairwise FST values were evaluated. These were highly significant for comparisons involving Pagasitikos and Thermaikos (FST = 0.110, p < 0.000001), and Thermaikos and Vistonikos (FST = 0.123, p < 0.000001). On the other hand, for the comparison between Pagasitikos and Vistonikos, a relatively low but significant differentiation was detected (FST = 0.013, p < 0.000001) (Table 1A). Principal Component Analysis (PCA) exhibited the presence of two genetic clusters showing that Thermaikos clearly separated apart from Pagasitikos and Vistonikos, while Pagasitikos population clustered together with Vistonikos population (Figure 3A). Structure analysis revealed similar patterns with PCA (Figure S6).
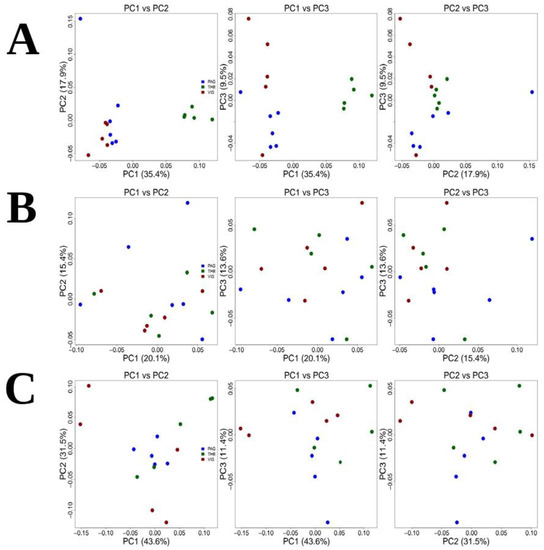
Figure 3.
Principal Component Analysis for (A) Holothuria tubulosa, (B) Sepia officinalis and (C) Venus verrucosa.
3.1.3. Genomics and Environmental Association Analysis
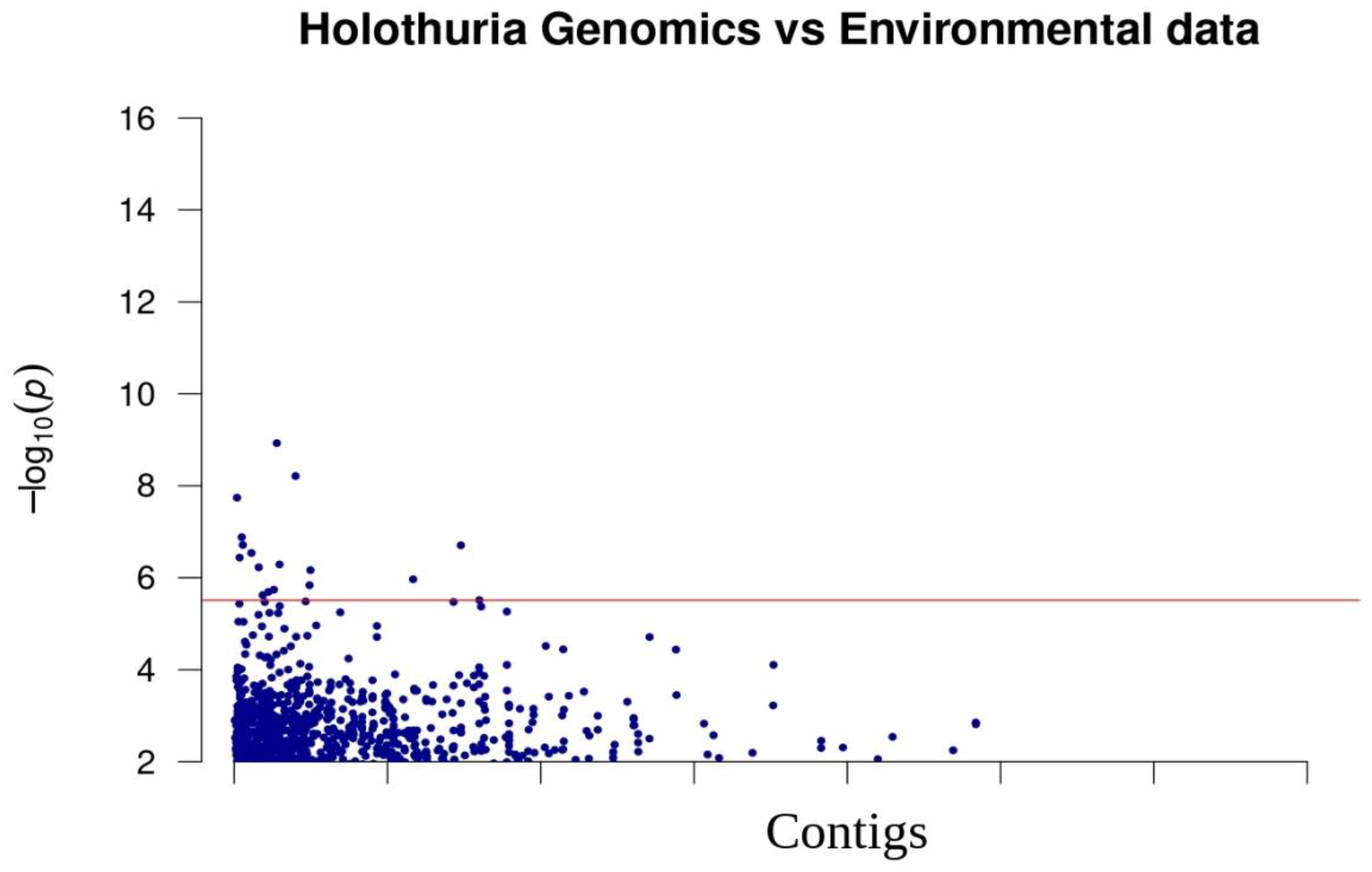
The R program LEA revealed that there is a highly significant (p < 3.09 × 10−6, after correction for multiple tests) correlation between 17 SNPs and oceanographic features (Figure 4). The snpEff program predicted that these outliers are located in intergenic regions.
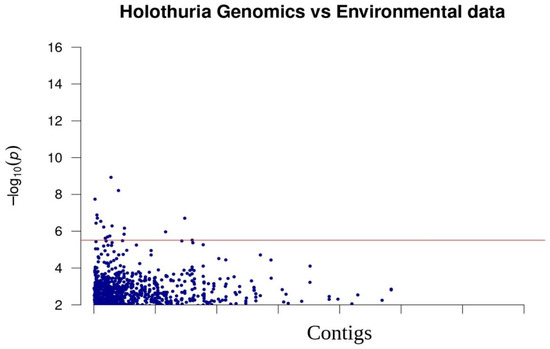
Figure 4.
Manhattan plot of the association between genomic SNPs and environmental factors (temperature, salinity, oxygen) for the sea cucumber. Blue line suggests the threshold of Type I error correction (3.09 × 10−6). Y axis is the negative log10 of p values and X axis is the SNP position.
3.1.4. Coalescent Demographic Analyses
Coalescent simulator showed that the most likely demographic scenario was model 3 (Supplementary Table S3—logistic and direct regression of the tested scenarios, Supplementary Figures S1–S3). Model 3 supports bottleneck with an evident population recovery for all three geographical areas.
In Pagasitikos, the bottleneck event occurred approximately 8.2 thousand generations ago (Table 2). Since sea cucumber reproduction occurs annually, its generation is 1 year [52]. This implies that the shrink of the population occurred 8.2 thousand years ago, after the Late Glacial Maximum (LGM), resulting in a decline of effective population size at 2.4 thousand individuals. A significant recovery of the effective population size to approximately 41 thousand individuals 4.9 thousands years ago.

Table 2.
Prior distribution settings and parameter estimates (mode and median) for the DIYABC analyses, using 4 × 106 data sets simulated under supported scenarios for the three invertebrate species (95% confidence intervals are shown for each of the parameters).
In Thermaikos and Vistonikos, the bottleneck event occurred 6.9 and 7.8 thousand years ago, respectively, also after the LGM. For these two regions the bottlenecked population was estimated at 5.9 thousand for Thermaikos and 3.4 thousand for Vistonikos, with a recovery population of 34 and 59 thousand for Thermaikos (4.28 thousand years ago) and Vistonikos (4.25 thousand years ago), respectively. The latter exhibited that the recovery in Vistonikos was higher compared to the other two regions (Table 2).
3.2. S. officinalis
3.2.1. SNPs Calling
An average of 3,209,914 sequence reads were generated from which we called 71,171 raw SNPs. Application of the filtering criteria described in the Materials and Methods resulted in a final dataset of 1083 high quality filtered SNPs (Supplementary Table S1 for a downstream of applied filtering and remained SNPs).
3.2.2. Genetic Differentiation and Diversity
Genetic indices are shown in Table S7. To test for genetic differences, pairwise FST values were evaluated. These were no significant for comparisons involving all geographical areas: Pagasitikos and Thermaikos (FST = 0.0003, p = 0.485), Thermaikos and Vistonikos (FST = 0.006, p = 0.184), Pagasitikos and Vistonikos (FST = −0.007, p = 0.92). (Table 1B).
3.2.3. Genomics and Environmental Association Analysis
The R program LEA did not reveal significant (p < 4.62 × 10−5, after correction for multiple tests) association between genomic data and oceanographic features.
3.2.4. Coalescent Demographic Analyses
Coalescent simulator showed that the most likely demographic scenario was model 3 (Supplementary Table S4—logistic and direct regression of the tested scenarios, Supplementary Figure S4) with evident expansion for all three geographical areas.
In Pagasitikos, the expansion event occurred approximately 535 thousand generations ago (Table 2). Since the generation time for cuttlefish is still unknown, we used the simplest assumption of 1 year/generation. Thus, this implies that the expansion of the population occurred 535 thousand years ago, before the Late Glacial Maximum (LGM), during the middle Pleistocene, resulting in a recent population effective size of 73 thousand individuals.
In Thermaikos and Vistonikos the expansion event occurred 314 and 397 thousand years ago, respectively, also before the LGM. For these two regions the expanded population was estimated at 767 thousand for Thermaikos and 784 thousand for Vistonikos (Table 2).
3.3. V. verrucosa
3.3.1. SNPs Calling
An average of 2,936,946 sequence reads were generated from which we called 799,035 raw SNPs. Application of the filtering criteria described in the Materials and Methods resulted in a final dataset of 3477 high quality filtered SNPs, with a main depth of coverage over 10×. (Supplementary Table S1 for a downstream of applied filtering and remained SNPs).
3.3.2. Genetic Differentiation and Diversity
Genetic indices are shown in Table S7. To test for genetic differences, pairwise FST values were evaluated. These were highly significant for comparisons involving all geographical areas: Pagasitikos and Thermaikos (FST = 0.047, p < 0.000001), Thermaikos and Vistonikos (FST = 0.099, p < 0.000001), Pagasitikos and Vistonikos (FST = −0.041, p < 0.000001) (Table 1C). Principal Component analysis (PCA) of the full dataset, showed a somewhat differentiation between the examined areas which reflects the FST pairwise differences (Figure 3C). Structure analysis revealed similar patterns with PCA (Figure S6).
3.3.3. Genomics and Environmental Association Analysis
The R program LEA did not reveal significant (p < 1.44 × 10−5, after correction for multiple tests) association between genomic data and oceanographic features.
3.3.4. Coalescent Demographic Analyses
Coalescent simulator showed that the most likely demographic scenario was model 1 (Supplementary Table S5—logistic and direct regression of the tested scenarios, Supplementary Figure S5) with constant population effective size for all three geographical areas. Population constant sizes were estimated at approximately 40, 38 and 59 thousand individuals for Pagasitikos, Thermaikos and Vistonikos, respectively.
In Figure S3 (Supplementary Information) are presented the posterior distributions of the parameters of the model of choice for each species.
4. Discussion
This study using RAD-sequencing investigates the population diversity and selection regarding the population history and adaptive importance of three non-model invertebrate species inhabiting the same geographical regions. Moreover, it assesses the utility of genotyping-by-sequencing approach for cross-species analyses that lack reference genomes regarding empirical data and demographic analyses. One of the striking outcomes of the present study was no shared demographic history across the three species, suggesting that environmental pressures may differentially act in species inhibiting the same geographical area, resulting in different evolutionary mechanisms for interspecies population dynamics in the study area. These results are of significant importance since population genetics could be used to improve the management and sustainability of marine and aquaculture resources [53]. To this end, several studies have highlighted the importance of molecular tools used in population genetics towards the design of management strategies and sustainability of species belonging to clams, cephalopods and holothuria. The latter is rather important, especially for aquatic species which belong to natural populations, as the ones examined in the present study [54,55,56]. As these species are not cultured (at least in the examined regions), these management strategies can serve as economic incentives to artisanal fishers. Moreover, due to the ongoing climate change and the threat that heat intensity by 2050 may lead to extinction 15–37% of the species, the necessity of research examining all levels of biological organizations in marine species for conservation and management strategies is highlighted [56,57,58].
4.1. Limitations of the Sampling Scheme
Despite the parameters’ optimization for the ddRAD analysis, a relative difference in the number of RAD loci and SNPs was found among the studied species. This might be due to technical issues (DNA quality and/or applied enzymes) or due to the choice of the cross-reference genomes for SNP calling. H. tubulosa individuals favored the higher number of reads which may explain the higher level of polymorphic SNPs that were identified compared to the other two species. Especially for the S. officinalis, which resulted the lowest number of SNPs, we acknowledge the fact that the chosen reference genome of Sepia pharaonis (which diverged approximately 48 mya) [59], might be the reason for less homologous loci and thus less SNPs to be called. On the other hand, the observed difference in genomic diversity among the three invertebrate species may be attributable to differences in demographic history. For instance, the sea cucumber population that was subjected in a bottleneck phenomenon in the past, is presently relatively smaller based on its effective size (34 to 59 thousands), compared to cuttlefish characterized by higher numbers of population effective size (767 to 784 thousands).
On the other hand, our study deals with a relatively small number of individuals, which may lead to ambiguous and/or misinterpretation of the outcomes. Indeed, past studies, using other molecular markers such as microsatellites, indicated that larger sample sizes were necessary (with a size of 25 to 30 individuals) for population genetic analyses [60]. However, recent studies using high-throughput sequencing argue that population genetics can be performed with small sample sizes [61,62]. Qu et al. (2020) [63], using resampling techniques in order to create simulated populations on their empirical data, showed that a sample size greater than four individuals (n ≥ 4) has little impact on estimates of genetic diversity using genome-wide high-throughput techniques. Moreover, many earlier studies comparing older markers and RAD-seq, demonstrate that the greater power availed by genome scan sampling reveals patterns that may otherwise have been missed [4,58], and therefore overcomes the relatively small sample size. However, other factors including dispersal ability, demographic history and overall population dynamics of the studied species should be considered for any effort to determine an ideal sampling scheme [62,64].
4.2. Population Structure and Genetic Differentiation
Overall, a different pattern of population structure was evident among the studied species, yielding somewhat mixed results. For instance, FST pairwise calculations revealed significant differentiation among all three geographical regions in sea cucumber and clam, whereas Principal Component Analysis (PCA) revealed fewer species-specific genetic clusters for the same species.
For sea cucumber, FST values revealed a clear genetic differentiation among all three geographical regions resulting in a high-significant genetic break in the North Aegean Sea, suggesting a limited low dispersal ability between the three regions. PCA results suggest two genetic clusters in the area. The first genetic cluster consists of Pagasitikos and Vistonida Gulf, and the second one consists of Thermaikos Gulf. The latter implies that nearly all Pagasitikos and Vistonida genotypes are related to each other, with however, high differentiated allele frequencies.
Interestingly, clam and cuttlefish populations showed a somehow similar pattern with respect to PCA plots. At this point, PCA did not reveal clear genetic clusters among populations, thus indicating a possible presence of subpopulations within all three studied areas for the two species. Despite the absence of association with respect to geographical areas, FST values document a very different story among the two species. Cuttlefish individuals showed no differentiation at all, with low pairwise values. Cuttlefish showed extensive migration strategies for reproduction and food acquisition [65,66] that may explain the genetic correlation which was observed between gulf populations examined herein. However, cuttlefish genetic diversity across the Mediterranean Sea was relatively high among the geographically distant populations, due to a low dispersion capability. Specifically, it seems that the species exhibits a different degree of genetic exchange among subpopulations, rather than a continuous population across the Mediterranean Sea [67,68,69]. Among the Mediterranean areas, the Sardinian coast exhibits the highest degree of diversity, independently of the known oceanographic factors (such as currents and fronts). The latter may be attributed to the fact that Sardinia represents a contact zone between the East and West populations [70]. These Sardinian samples included several haplotypes from the “Atlantic” lineage, several haplotypes shared with the Croatia-Italian samples and even an isolated, genetically unique sample [70]. This high degree of genetic diversification has also been demonstrated for Greece and Cyprus populations, emphasizing the fact of long-distance dispersal incapability. The latter concerning Greek marine areas, is attributed to the fact that Drábková et al. [70] have collected samples only from one area (Thermaikos gulf). However, we showed that no genetic diversity exists between North Aegean subpopulations. Therefore, dispersal with unrestricted gene flow and possible effect of some oceanographic factors is possible in this smaller subarea of the Aegean Sea.
On the other hand, clam showed a highly significant differentiation between the three examined areas, suggesting potential genetic breaks for the species in the North Aegean Sea. Although it is quite difficult to define such genetic breaks, the presence of this limited dispersal is similar to sea cucumber. It has been shown that climate change effects establish different genotypes among intertidal animals, due to different physical, or biotic conditions [29]. This pattern may explain clam’s adaptive physiological performance. In other bivalves, such as mytilids, intraspecies and intrapopulation genetic diversity (in nucleotide and haplotype diversity terms) has also been observed [71,72,73]. However, other bivalve species populations in the Aegean Sea, such as the horse mussel (Modiolus barbatus), revealed low levels of interpopulation genetic differentiation [74,75]. The latter is mostly attributed to the long duration (up to 6 months) of its pelagic larval stage [76]. To our knowledge no information on the larval duration of V. verrucosa exists.
The fact that our data detected fine-scale population differentiation within the North Aegean Sea for the sea cucumber and the clam, might also be a consequence of higher genetic resolution compared to cuttlefish, with respect to the number of the analysed SNPs. However, our results may explain this differentiation with respect to oceanographic features and how they affect plaktonic larval duration. Studies have shown that the time a pelagic larva spends in the open sea before settlement is strongly associated with water temperature [77]: the hotter the water, the shorter the metamorphosis is [78]. However, larval duration may not be the only evolutionary mechanism that sea cucumber and clam employ in order to display a higher level of genetic differentiation.
4.3. Genetic Diversity and Environment
Our results showed that only sea cucumber is significantly related to sea water physicochemical parameters, such as temperature and salinity, recorded in Pagasitikos, Thermaikos and Vistonikos gulfs. The different sampling areas exhibit a significantly different geomorphology (please refer to Feidantsis et al. [30,31] for detailed data). Specifically, Pagasitikos (a semi-enclosed with shallow depths) gulf is connected with the Central Aegean Sea through the Trikeri Channel, which is relatively narrow and deep [79]. On the other hand, Thermaikos gulf, which exhibits minimum tides and is located in the Northwest Aegean Sea, is connected to a complicated deltaic ecosystem, including 4 rivers (Aliakmonas, Axios, Gallikos and Loudias) which are discharged along the coastline. Since some major Greek ports are located in gulfs such as Thermaikos and Pagasitikos, these areas may suffer from degradation of ecosystems, which is largely seen in their water quality and habitat modifications [80]. Compared to the above and the rest of the Aegean Sea (where seabed topologies of various and different depths are observed), Vistonikos gulf exhibits lower sandy depths with few rocky areas, and therefore a greater uniformity [81,82]. The part of the North Aegean Sea, where Vistonikos is located, is largely influenced by the cold Black Sea outflow. This cold influx affects surface sea temperature, which seasonally remains lower compared to Thermaikos and Vistonikos [30,31,83,84,85].
4.4. Demographic History
Despite these differences in SNPs variability and population structure of the three species, we also found different signatures of historical population demography with bottleneck remaining evident for sea cucumber, expansion for cuttlefish and no change in Nexp favoring a constant size for the clam species. That pattern was seen across all geographic areas for each species. For sea cucumber this bottleneck event was followed by a recovery in contemporary genomes of the species in all three geographical regions. This overall species-specific demography suggests that oceanographic features may play a key-role to interspecies population dynamics.
Our finding of a demographic bottleneck in sea cucumber occurred approximately six to eight thousand years ago in the North Aegean Sea. This period occurred after the Last Glacial Maximum (LGM), when a rapid warming affected the sea levels and subsequently the oceanography of the species fidelity. The population effective size declined from an ancestral population of about 0.5 million down to a few thousands, thus signifying a massive loss of genetic diversity. However, the recovery of the populations that occurred about four thousand years ago, reflecting local adaptation and potential signatures of selection, was significant for the species. However, due to the fact that historical data are missing, the return of the genomic diversity to sea cucumber and these potential signatures of selection are limited in this study.
In contrast with sea cucumber, demographic models supported an ancestral expansion of the cuttlefish half million generations ago, although the generation time for cuttlefish is unknown. Even if we make the simplest assumption that the generation time is 1 year, the time of the expansion event occurred in the Middle Pleistocene. This result may be consistent with the absence of genetic differentiation associated with local habitat dependence and climate change. Possibly, the high dispersal of the species may be established by abiotic factors and evolutionary forces. Species during Pleistocene glaciations were re-distributed creating refugia [77], followed by post-glacial dispersal and expansion [86]. Additionally, the observed lack of spatial genetic differentiation could partially have been explained by rapid population growth [87].
Our results showed that clam did not go through any historical event, implying a constant population effective size in the area, with limited gene flow according to FST pairwise values. Therefore, different evolutionary mechanisms may have been recruited for the adaptation of the species. Although analysis showed that a clam’s history consists of several periods of constant size, one should bear in mind that its history may also consist of potential instantaneous changes of sizes between periods, suggesting a more complex framework of the demography expected in natural populations [88]. However, due to smaller sample size and lower number of loci, constant size scenarios may be supported with lower precision during the estimation of summary statistics [89].
The lack of similar demographic history in the North Aegean Sea could be explained by environmental differences or differences in the historical time frame of exploitation, recovery, and expansion between regions. In this system in the study area, the historical demography presented herein provided evidence for managing species. Despite the differences in the population dynamic of the three invertebrates, signatures of their different demographic history are apparent. Consistently, genetic diversity and historical signatures are correlated with species ecology. The different demography can be influenced by both abiotic and biotic factors, and changes in the environment may also be responsible for diversity patterns [90]. There are several examples of climate change effects among intertidal animals, in which larval/juvenile survival, or post-settlement selection leads to different genotypes, which become established in different physical or biotic conditions [91].
5. Conclusions
The data obtained in the present work clearly indicate the fact that economically important marine invertebrate species’ physiological performance is not solely imposed on phenotypic plasticity. Genetic diversity, as observed mostly in geographically different populations of sea cucumber and clam, seems to drive this species effectiveness in cellular responses’ integration, in comparison to the panmictic cuttlefish. The latter does not face intense environmental variations, as it is a benthopelagic species. Since climate change affects also microhabitats, local climate can be responsible for the abundance and even extinction of various species. Thus, further research examining all levels of biological organizations, from genes to organisms, is needed in order to obtain marine species conservation strategies and management plans, since according to the worst-case scenario, the heat intensity by 2050, will lead to extinction of several species. Results such as the ones presented herein and the existing literature highlight the fact that field studies examining population genetics and the physiological backgrounds of marine organisms are particularly important since they elucidate species adaptation capacity and their subsequent sustainable harvesting and population recovery.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/su142114380/s1, Table S1: Number of RAD-seq reads per individual for all species and populations. It is indicated the percentage of the reads that have been mapped to the reference genomes. Holothuria glaberina was used for Holothuria tubulosa, Mercenaria mercenaria for Venus verrucosa samples and Sepia pharaonis for Sepia officinalis; Table S2: Filtering steps that were carried out to generate the final high-quality SNP dataset starting from the raw SNPs output by the GATK pipeline. The number of retained SNPs after each step is reported. GQ: Genotype quality; DP: Genotype depth of coverage; IGR: Individual genotyping rate; maxDP: Depth of coverage (twice the mean depth of coverage of the raw dataset) greater than 416.24 for H. tubulosa, 176.62 for S. officinalis and 21.76 for V. verrucosa, were removed; MAF: Minimum Allele Frequency; hwe: Hardy–Weinberg equilibrium; ld r2: Linkage Disequilibrium pruning r2; Table S3: Confidence intervals for ABC analysis for Holothuria tubulosa populations for both direct and logistic regression of the scenarios comparison; Table S4: Confidence intervals for ABC analysis for Sepia officinalis populations for both direct and logistic regression of the scenarios comparison; Table S5: Confidence intervals for ABC analysis for Venus verrucosa populations for both direct and logistic regression of the scenarios comparison; Figure S1: Posterior (green line) and prior (red line) distribution plots of ABC analysis based on 4 × 106 simulated data sets of historical effective population sizes and time of historical events based on scenario 2 of H. tubulosa for Pagasitikos population; Nanc: Ancestral population, Nb: bottlenecked population, Ncur: recovered population, tb: time of bottleneck event, tcur: time of recovery; Figure S2: Posterior (green line) and prior (red line) distribution plots of ABC analysis based on 4 × 106 simulated data sets of historical effective population sizes and time of historical events based on scenario 2 of H. tubulosa for Thermaikos population; Nanc: Ancestral population, Nb: bottlenecked population, Ncur: recovered population, tb: time of bottleneck event, tcur: time of recovery; Figure S3: Posterior (green line) and prior (red line) distribution plots of ABC analysis based on 4 × 106 simulated data sets of historical effective population sizes and time of historical events based on scenario 2 of H. tubulosa for Vistonikos population; Nanc: Ancestral population, Nb: bottlenecked population, Ncur: recovered population, tb: time of bottleneck event, tcur: time of recovery; Figure S4: Posterior (green line) and prior (red line) distribution plots of ABC analysis based on 4 × 106 simulated data sets of historical effective population sizes and time of historical events based on scenario 3 of S. officinalis for Pagasitikos (A), Thermaikos (B) and Vistonikos (C) population; Nanc: Ancestral population, Nexp: expanded population, texp: time of expansion; Figure S5: Posterior (green line) and prior (red line) distribution plots of ABC analysis based on 4 × 106 simulated data sets of historical effective population sizes and time of historical events based on scenario 1 of V. verrucosa for Pagasitikos (A), Thermaikos (B) and Vistonikos (C) population; Ncur: constant population effective size; Table S6: Mean values of temperature, salinity and oxygen recorded during seasonal samplings; Table S7: Genetic indices per species and population. No: Number of alleles; Ea: Effective alleles; HOBS: observed heterozygosity; HEXP: expected heterozygosity, FIS: fixation inbreeding index; Figure S6: Structure plots and Delta-K against K for Structure run given the reduction of Ln from K = 2 for all populations made using HARVESTER. A: Holothuria; B: Sepia; C: Venus.
Author Contributions
K.F.: Conceptualization, Methodology, Investigation, Data curation, Visualization, Validation, Funding Acquisition, Project Administration, Writing Original Draft Preparation—Review and Editing; G.A.G.: Methodology, Investigation, Data curation, Formal analysis, Writing Original Draft Preparation—Review and Editing; A.E.: Conceptualization, Validation, Review and Editing; B.M.: Conceptualization; A.S.: Conceptualization; M.H.: Conceptualization; C.A.: Methodology; J.S.: Methodology; D.V.: Conceptualization, Visualization, Validation, Supervision, Funding Acquisition, Project Administration, Writing Original Draft Preparation—Review and Editing. All authors have read and agreed to the published version of the manuscript.
Funding
The present study was carried out under the University of Thessaly invitation: “Call for interest for postgraduate PhD research”, is implemented by the University of Thessaly and funded by the Stavros Niarchos Foundation (SNF).
Institutional Review Board Statement
Animals received proper care in compliance with the “Guidelines for the Care and Use of Laboratory Animals” published by US National Institutes of Health (NIH publication No 85–23, revised 1996) and the “Principles of laboratory animal care” published by the Greek Government (160/1991) based on EU regulations (86/609). The protocol as well as surgery and sacrifice were approved by the Committee on the Ethics of Animal Experiments of the Directorate of Veterinary Services of Prefecture of Thessaloniki under the license number EL 54 BIO 05.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the NCBI repository (accession number: PRJNA771498). https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA771498&o=acc_s%3Aa, accessed on 15 October 2021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dionne, M.; Miller, K.M.; Dodson, J.J.; Caron, F.; Bernatchez, L. Clinal variation in MHC diversity with temperature: Evidence for the role of host-pathogen interaction on local adaptation in Atlantic salmon. Evolution 2007, 61, 2154–2164. [Google Scholar] [CrossRef]
- Temunović, M.; Franjić, J.; Satovic, Z.; Grgurev, M.; Frascaria-Lacoste, N.; Fernández-Manjarrés, J. Environmental heterogeneity explains the genetic structure of continental and Mediterranean populations of Fraxinus angustifolia Vahl. PLoS ONE 2012, 7, e42764. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, L.J.; Lucas, M.C.; Winfield, I.J.; Hoelzel, A.R. Environmental factors associated with genetic and phenotypic divergence among sympatric populations of Arctic charr (Salvelinus alpinus). J. Evol. Biol. 2011, 24, 1906–1917. [Google Scholar] [CrossRef] [PubMed]
- Gkafas, G.A.; de Jong, M.; Exadactylos, A.; Raga, J.A.; Aznar, F.J.; Hoelzel, A.R. Sex-specific impact of inbreeding on pathogen load in the striped dolphin. Proc. R. Soc. B Biol. Sci. 2020, 287, 20200195. [Google Scholar] [CrossRef] [PubMed]
- Hoelzel, A.R.; Hey, J.; Dahlheim, M.E.; Nicholson, C.; Burkanov, V.; Black, N. Evolution of population structure in a highly social top predator, the killer whale. Mol. Biol. Evol. 2007, 24, 1407–1415. [Google Scholar] [CrossRef]
- Segura-García, I.; Rojo-Arreola, L.; Rocha-Olivares, A.; Heckel, G.; Gallo-Reynoso, J.P.; Hoelzel, R. Eco-Evolutionary Processes Generating Diversity Among Bottlenose Dolphin, Tursiops truncatus, Populations off Baja California, Mexico. Evol. Biol. 2018, 45, 223–236. [Google Scholar] [CrossRef]
- Konstantinidis, I.; Gkafas, G.A.; Karamitros, G.; Lolas, A.; Antoniadou, C.; Vafidis, D.; Exadactylos, A. Population structure of two benthic species with different larval stages in the eastern Mediterranean Sea. J. Environ. Prot. Ecol. 2017, 18, 930–939. [Google Scholar]
- Gkafas, G.A.; Exadactylos, A.; Rogan, E.; Raga, J.A.; Reid, R.; Hoelzel, A.R. Biogeography and temporal progression during the evolution of striped dolphin population structure in European waters. J. Biogeogr. 2017, 44, 2681–2691. [Google Scholar] [CrossRef]
- Gaither, M.R.; Gkafas, G.A.; De Jong, M.; Sarigol, F.; Neat, F.; Regnier, T.; Moore, D.; Gröcke, D.R.; Hall, N.; Liu, X.; et al. Genomics of habitat choice and adaptive evolution in a deep-sea fish. Nat. Ecol. Evol. 2018, 2, 680–687. [Google Scholar] [CrossRef]
- Crawford, D.L.; Oleksiak, M.F. Ecological population genomics in the marine environment. Brief. Funct. Genom. 2016, 15, 342–351. [Google Scholar] [CrossRef]
- Sarropoulou, X.; Tsaparis, D.; Tsagarakis, K.; Badouvas, N.; Tsigenopoulos, C.S. Different patterns of population structure and genetic diversity of three mesopelagic fishes in the Greek Seas. Mediterr. Mar. Sci. 2022, 23, 536–545. [Google Scholar] [CrossRef]
- Kozak, K.H.; Wiens, J.J. Does niche conservatism promote speciation? A case study in North American salamanders. Evolution 2006, 60, 2604–2621. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 183–195. [Google Scholar] [CrossRef]
- Bargelloni, L.; Alarcon, J.A.; Alvarez, M.C.; Penzo, E.; Magoulas, A.; Palma, J.; Patarnello, T. The Atlantic-Mediterranean transition: Discordant genetic patterns in two seabream species, Diplodus puntazzo (Cetti) and Diplodus sargus (L.). Mol. Phylogenet. Evol. 2005, 36, 523–535. [Google Scholar] [CrossRef] [PubMed]
- De Bruyn, M.; Hoelzel, A.R.; Carvalho, G.R.; Hofreiter, M. Faunal histories from Holocene ancient DNA. Trends Ecol. Evol. 2011, 26, 405–413. [Google Scholar] [CrossRef]
- Nei, M.; Maruyama, T.; Chakraborty, R. The bottleneck effect and genetic variability in populations. Evolution 1975, 29, 1–10. [Google Scholar] [CrossRef]
- Hansson, B.; Bensch, S.; Hasselquist, D.; Lillandt, B.G.; Wennerberg, L.; Von Schantz, T. Increase of genetic variation over time in a recently founded population of great reed warblers (Acrocephalus arundinaceus) revealed by mirosatellites and DNA fingerprinting. Mol. Ecol. 2000, 9, 1529–1538. [Google Scholar] [CrossRef]
- Excoffier, L.; Foll, M.; Petit, R.J. Genetic consequences of range expansion. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 481–501. [Google Scholar] [CrossRef]
- Hoban, S.M.; Gaggiotti, O.E.; Bertorelle, G. The number of markers and samples needed for detecting bottlenecks under realistic scenarios, with and without recovery: A simulation-based study. Mol. Ecol. 2013, 22, 3444–3450. [Google Scholar] [CrossRef]
- Schmidt, P.S.; Rand, D.M. Adaptive maintenance of genetic polymorphism in an intertidal barnacle: Habitat- and life-stage-specific survivorship of mpi genotypes. Evolution 2001, 55, 1336–1344. [Google Scholar] [CrossRef]
- Meyer, E.; Aglyamova, G.V.; Matz, M.V. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol. Ecol. 2011, 20, 3599–3616. [Google Scholar] [CrossRef] [PubMed]
- Rivière, G.; He, Y.; Tecchio, S.; Crowell, E.; Gras, M.; Sourdaine, P.; Guo, X.; Favrel, P. Dynamics of DNA methylomes underlie oyster development. PLoS Genet. 2017, 13, e1006807. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.S.; Thorne, M.A.; King, M.; Hipperson, H.; Hoffman, J.I.; Peck, L.S. Life in the intertidal: Cellular responses, methylation and epigenetics. Funct. Ecol. 2018, 32, 1982–1994. [Google Scholar] [CrossRef]
- Dixon, G.; Liao, Y.; Bay, L.K.; Matz, M.V. Role of gene body methylation in acclimatization and adaptation in a basal metazoan. Proc. Natl. Acad. Sci. USA 2018, 115, 13342–13346. [Google Scholar] [CrossRef]
- Strader, M.E.; Wong, J.M.; Kozal, L.C.; Leach, T.S.; Hofmann, G.E. Parental environments alter DNA methylation in offspring of the purple sea urchin, Strongylocentrotus purpuratus. J. Exp. Mar. Biol. Ecol. 2019, 517, 54–64. [Google Scholar] [CrossRef]
- Exadactylos, A.; Vafidis, D.; Tsigenopoulos, C.; Gkafas, G. High Connectivity of the White Seabream (Diplodus sargus, L. 1758) in the Aegean Sea, Eastern Mediterranean Basin. Animals 2019, 9, 979. [Google Scholar] [CrossRef]
- Seyhan-Ozturk, D.; Engin, S. Genetic diversity of marbled goby populations in the Anatolian coasts of the north-eastern Mediterranean. J. Mar. Biol. Assoc. UK 2021, 101, 419–429. [Google Scholar] [CrossRef]
- Sironi, M.; Cagliani, R.; Forni, D.; Clerici, M. Evolutionary insights into host–pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 2015, 16, 224–236. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Feidantsis, K.; Michaelidis, B.; Raitsos, D.Ε.; Vafidis, D. Seasonal cellular stress responses of commercially important invertebrates at different habitats of the North Aegean Sea. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2020, 250, 110778. [Google Scholar] [CrossRef]
- Feidantsis, K.; Michaelidis, B.; Raitsos, D.Ε.; Vafidis, D. Seasonal metabolic and oxidative stress responses of commercially important invertebrate species correlation with their habitat. Mar. Ecol. Prog. Ser. 2021, 658, 27–46. [Google Scholar] [CrossRef]
- Lepais, O.; Weir, J.T. SimRAD: An R package for simulation–based prediction of the number of loci expected in RADseq and similar genotyping by sequencing approaches. Mol. Ecol. Resour. 2014, 14, 1314–1321. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Medina-Feliciano, J.G.; Pirro, S.; García-Arrarás, J.E.; Mashanov, V.; Ryan, J.F. Draft Genome of the Sea Cucumber Holothuria glaberrima, a Model for the Study of Regeneration. Front. Mar. Sci. 2021, 8, 603410. [Google Scholar] [CrossRef]
- Song, H.; Guo, X.; Sun, L.; Wang, Q.; Han, F.; Wang, H.; Wray, G.A.; Davidson, P.; Wang, Q.; Hu, Z.; et al. The hard clam genome reveals massive expansion and diversification of inhibitors of apoptosis in Bivalvia. BMC Biol. 2021, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Li, R.; Zhao, Y.; Migaud, H.; Wang, C.; Bekaert, M. Pharaoh Cuttlefish, Sepia pharaonis, Genome Reveals Unique Reflectin Camouflage Gene Set. Front. Mar. Sci. 2021, 8, 639670. [Google Scholar] [CrossRef]
- Li, H. Exploring single–sample SNP and INDEL calling with whole–genome de novo assembly. Bioinformatics 2012, 28, 1838–1844. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Durbin, R. Genome Project Data Processing Subgroup Genome Project Data Processing Subgroup. 2009 The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Applebaum, S.W.; Heifetz, Y. Density-dependent physiological phase in insects. Annu. Rev. Entomol. 1999, 44, 317–341. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.J.; de Jong, J.F.; Hoelzel, A.R.; Janke, A. SambaR: An R package for fast, easy and reproducible population-genetic analyses of biallelic SNP datasets. Mol. Ecol. Resour. 2021, 21, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 15 November 2020).
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Frichot, E.; François, O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Frichot, E.; Schoville, S.D.; Bouchard, G.; François, O. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol. Biol. Evol. 2013, 30, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and Manhattan plots. Biorxiv 2014, 005165. Available online: https://www.biorxiv.org/content/early/2014/05/14/005165 (accessed on 10 December 2020).
- Simes, R.J. An improved Bonferroni procedure for multiple tests of significance. Biometrika 1986, 73, 751–754. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Cornuet, J.-M.; Pudlo, P.; Veyssier, J.; Dehne-Garcia, A.; Gautier, M.; Leblois, R.; Marin, J.-M.; Estoup, A. DIYABC v2.0: A software to make Approximate Bayesian Computation inferences about population history using Single Nucleotide Polymorphism, DNA sequence and microsatellite data. Bioinformatics 2014, 30, 1187–1189. [Google Scholar] [CrossRef]
- Dereli, H.; Çulha, S.T.; Çulha, M.; Özalp, B.H.; Tekinay, A.A. Reproduction and population structure of the sea cucumber Holothuria tubulosa in the Dardanelles Strait, Turkey. Mediterr. Mar. Sci. 2016, 17, 47–55. [Google Scholar] [CrossRef]
- Berkeley, S.A.; Hixon, M.A.; Larson, R.J.; Love, M.S. Fisheries sustainability via protection of age structure and spatial distribution of fish populations. Fisheries 2004, 29, 23–32. [Google Scholar] [CrossRef]
- McKeown, N.J.; Shaw, P.W. Microsatellite loci for studies of the common cuttlefish, Sepia officinalis. Conserv. Genet. Resour. 2014, 6, 701–703. [Google Scholar] [CrossRef][Green Version]
- Ravago-Gotanco, R.; Kim, K.M. Regional genetic structure of sandfish Holothuria (Metriatyla) scabra populations across the Philippine archipelago. Fish. Res. 2019, 209, 143–155. [Google Scholar] [CrossRef]
- Helmuth, B.; Broitman, B.R.; Yamane, L.; Gilman, S.E.; Mach, K.; Mislan, K.A.S.; Denny, M.W. Organismal climatology: Analyzing environmental variability at scales relevant to physiological stress. J. Exp. Biol. 2010, 213, 955–1003. [Google Scholar] [CrossRef] [PubMed]
- Osovitz, C.J.; Hofmann, G.E. Marine macrophysiology: Studying physiological variation across large spatial scales in marine systems. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 147, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, G.E.; Todgham, A.E. Living in the Now: Physiological Mechanisms to Tolerate a Rapidly Changing Environment. Annu. Rev. Physiol. 2010, 72, 127–145. [Google Scholar] [CrossRef]
- Pardo-Gandarillas, M.C.; Torres, F.I.; Fuchs, D.; Ibáñez, C.M. Updated molecular phylogeny of the squid family Ommastrephidae: Insights into the evolution of spawning strategies. Mol. Phylogenet. Evol. 2018, 120, 212–217. [Google Scholar] [CrossRef]
- Hale, M.L.; Burg, T.M.; Steeves, T.E. Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE 2012, 7, e45170. [Google Scholar] [CrossRef]
- Willing, E.M.; Dreyer, C.; Oosterhout, C.V. Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 2012, 7, e42649. [Google Scholar] [CrossRef]
- Nazareno, A.G.; Bemmels, J.B.; Dick, C.W.; Lohmann, L.G. Minimum sample sizes for population genomics: An empirical study from an Amazonian plant species. Mol. Ecol. Resour. 2017, 17, 1136–1147. [Google Scholar] [CrossRef]
- Qu, W.-M.; Liang, N.; Wu, Z.-K.; Zhao, Y.-G.; Chu, D. Minimum sample sizes for invasion genomics: Empirical investigation in an invasive whitefly. Ecol. Evol. 2020, 10, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Yang, S.; Shang, G.; Wei, W. Effects of predation risk on behavior, hormone levels, and reproductive success of plateau pikas. Ecosphere 2017, 8, e01643. [Google Scholar] [CrossRef]
- Pierce, G.J.; Valavanis, V.; Guerra, A.; Jereb, P.; Orsi-Relini, L.; Bellido-Millán, J.M.; Katara, I.; Piatkowski, U.; Pereira, J.; Balguerias, E.; et al. A review of cephalopod–environment interactions in European Seas. Hydrobiologia 2008, 612, 49–70. [Google Scholar] [CrossRef]
- Keller, S.; Valls, M.; Hidalgo, M.; Quetglas, A. Influence of environmental parameters on the life-history and population dynamics of cuttlefish Sepia officinalis in the western Mediterranean. Estuar. Coast. Shelf Sci. 2014, 145, 31–40. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Guerra, Á.; Sanjuan, A. Allozyme differentiation in the cuttlefish Sepia officinalis (Mollusca: Cephalopoda) from the NE Atlantic and Mediterranean. Heredity 1999, 83, 280–289. [Google Scholar] [CrossRef][Green Version]
- Pérez-Losada, M.; Guerra, A.; Carvalho, G.R.; Sanjuan, A.; Shaw, P.W. Extensive population subdivision of the cuttlefish Sepia officinalis (Mollusca: Cephalopoda) around the Iberian Peninsula indicated by microsatellite DNA variation. Heredity 2002, 89, 417–424. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Nolte, M.J.; Crandall, K.A.; Shaw, P.W. Testing hypotheses of population structuring in the Northeast Atlantic Ocean and Mediterranean Sea using the common cuttlefish Sepia officinalis. Mol. Ecol. 2007, 16, 2667–2679. [Google Scholar] [CrossRef]
- Drábková, M.; Jachníková, N.; Tyml, T.; Sehadová, H.; Ditrich, O.; Myšková, E.; Hypša, V.; Štefka, J. Population co-divergence in common cuttlefish (Sepia officinalis) and its dicyemid parasite in the Mediterranean Sea. Sci. Rep. 2019, 9, 14300. [Google Scholar] [CrossRef]
- Koehn, R.K.; Milkman, R.; Mitton, J.B. Population genetics of marine pelecypods. IV. Selection, migration and genetic differentiation in the blue mussel Mytilus edulis. Evolution 1976, 30, 2–32. [Google Scholar] [CrossRef]
- Koehn, R.K. Physiology and biochemistry of enzyme variation: The interface of ecology and population genetics. In Ecological Genetics: The Interface; Springer: New York, NY, USA, 1978; pp. 51–72. [Google Scholar]
- Giantsis, I.A.; Abatzopoulos, T.J.; Angelidis, P.; Apostolidis, A.P. Mitochondrial Control Region Variability in Mytilus galloprovincialis Populations from the Central-Eastern Mediterranean Sea. Int. J. Mol. Sci. 2014, 15, 11614–11625. [Google Scholar] [CrossRef]
- Giantsis, I.A.; Exadactylos, A.; Feidantsis, K.; Michaelidis, B. First insights towards the population genetic structure and the phylogeographic status of the horse mussel (Modiolus barbatus) from the eastern Mediterranean. J. Mar. Biol. Assoc. UK 2019, 99, 1111–1118. [Google Scholar] [CrossRef]
- De La Rosa-Velez, J.; Farfan, C.; Cervantes-Franco, M.A. Geographic pattern of genetic variation in Modiolus capax (Conrad, 1837) from the Gulf of California. Cienc. Mar. 2000, 26, 585–606. [Google Scholar] [CrossRef][Green Version]
- Halanych, K.M.; Vodoti, E.T.; Sundberg, P.; Dahlgren, T.G. Phylogeography of the horse mussel Modiolus modiolus. J. Mar. Biol. Assoc. UK 2013, 93, 1857–1869. [Google Scholar] [CrossRef]
- Vendrami, D.L.J.; De Noia, M.; Telesca, L.; Handal, W.; Charrier, G.; Boudry, P.; Eberhart-Philips, L.; Hoffman, J.I. RAD sequencing sheds new light on the genetic structure and local adaptation of European scallops and resolves their demographic histories. Sci. Rep. 2019, 9, 7455. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, A.R.; Barnes, D.A. Aspects of veliger larval growth and byssus drifting of the spat of Pecten maximus and Aequipecten (Chlamys) opercularis. ICES J. Mar. Sci. 1992, 49, 417–423. [Google Scholar] [CrossRef]
- Pancucci-Papadopoulou, M.A.; Zenetos, A.; Corsini-Foka, M.; Politou, C.-Y. Update of marine aliens in Hellenic waters. Mediterr. Mar. Sci. 2006, 6, 147–158. [Google Scholar] [CrossRef][Green Version]
- Dimiza, M.D.; Koukousioura, O.; Triantaphyllou, M.V.; Dermitzakis, M.D. Live and dead benthic foraminiferal assemblages from coastal environments of the Aegean Sea (Greece): Distribution and diversity. Rev. Micropaléontol. 2016, 59, 19–32. [Google Scholar] [CrossRef]
- Androulidakis, Y.S.; Krestenitis, Y.N.; Psarra, S. Coastal upwelling over the North Aegean Sea: Observations and simulations. Cont. Shelf Res. 2017, 149, 32–51. [Google Scholar] [CrossRef]
- Poulos, S.E.; Drakopoulos, P.G.; Collins, M.B. Seasonal variability in sea surface oceanographic conditions in the Aegean Sea (Eastern Mediterranean): An overview. J. Mar. Syst. 1997, 13, 225–244. [Google Scholar] [CrossRef]
- Zodiatis, G.; Alexandri, S.; Pavlakis, P.; Jonsson, L.; Kallos, G.; Demetropoulos, A.; Georgiou, G.; Theodorou, A.; Balopoulos, E. Tentative study of flow patterns in the North Aegean Sea using NOAA-AVHRR images and 2D model simulation. Ann. Geophys. 1997, 14, 1221–1231. [Google Scholar] [CrossRef]
- Kourafalou, V.H.; Barbopoulos, K. High resolution simulations on the North Aegean Sea seasonal circulation. Ann. Geophys. 2003, 21, 251–265. [Google Scholar] [CrossRef]
- Xue, D.X.; Wang, H.Y.; Zhang, T.; Liu, J.X. Population genetic structure and demographic history of Atrina pectinata based on mitochondrial DNA and microsatellite markers. PLoS ONE 2014, 9, e95436. [Google Scholar] [CrossRef] [PubMed]
- Cammen, K.M.; Schultz, T.F.; Don Bowen, W.; Hammill, M.O.; Puryear, W.B.; Runstadler, J.; Wenzel, F.W.; Wood, S.A.; Kinnison, M. Genomic signatures of population bottleneck and recovery in Northwest Atlantic pinnipeds. Ecol. Evol. 2018, 8, 6599–6614. [Google Scholar] [CrossRef] [PubMed]
- Hoban, S.; Kelley, J.L.; Lotterhos, K.E.; Antolin, M.F.; Bradburd, G.; Lowry, D.B.; Poss, M.L.; Reed, L.K.; Storfer, A.; Whitlock, M.C. Finding the genomic basis of local adaptation: Pitfalls, practical solutions, and future directions. Am. Nat. 2016, 188, 379–397. [Google Scholar] [CrossRef]
- Navascués, M.; Leblois, R.; Burgarella, C. Demographic inference through approximate-Bayesian-computation skyline plots. PeerJ 2017, 5, e3530. [Google Scholar] [CrossRef] [PubMed]
- Benton, M.J. The Red Queen and the Court Jester: Species diversity and the role of biotic and abiotic factors through time. Science 2009, 323, 728–732. [Google Scholar] [CrossRef]
- An, H.S.; Kim, E.M.; Park, J.Y. Isolation and characterization of microsatellite markers for the clam Ruditapes philippinarum and cross-species amplification with the clam Ruditapes variegate. Conserv. Genet. 2009, 10, 1821–1823. [Google Scholar] [CrossRef]
- Somero, G.N. The cellular stress response and temperature: Function, regulation, and evolution. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2020, 333, 379–397. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).