Genomic Characterization of the Three Balkan Livestock Guardian Dogs
Abstract
1. Introduction
2. Materials and Methods
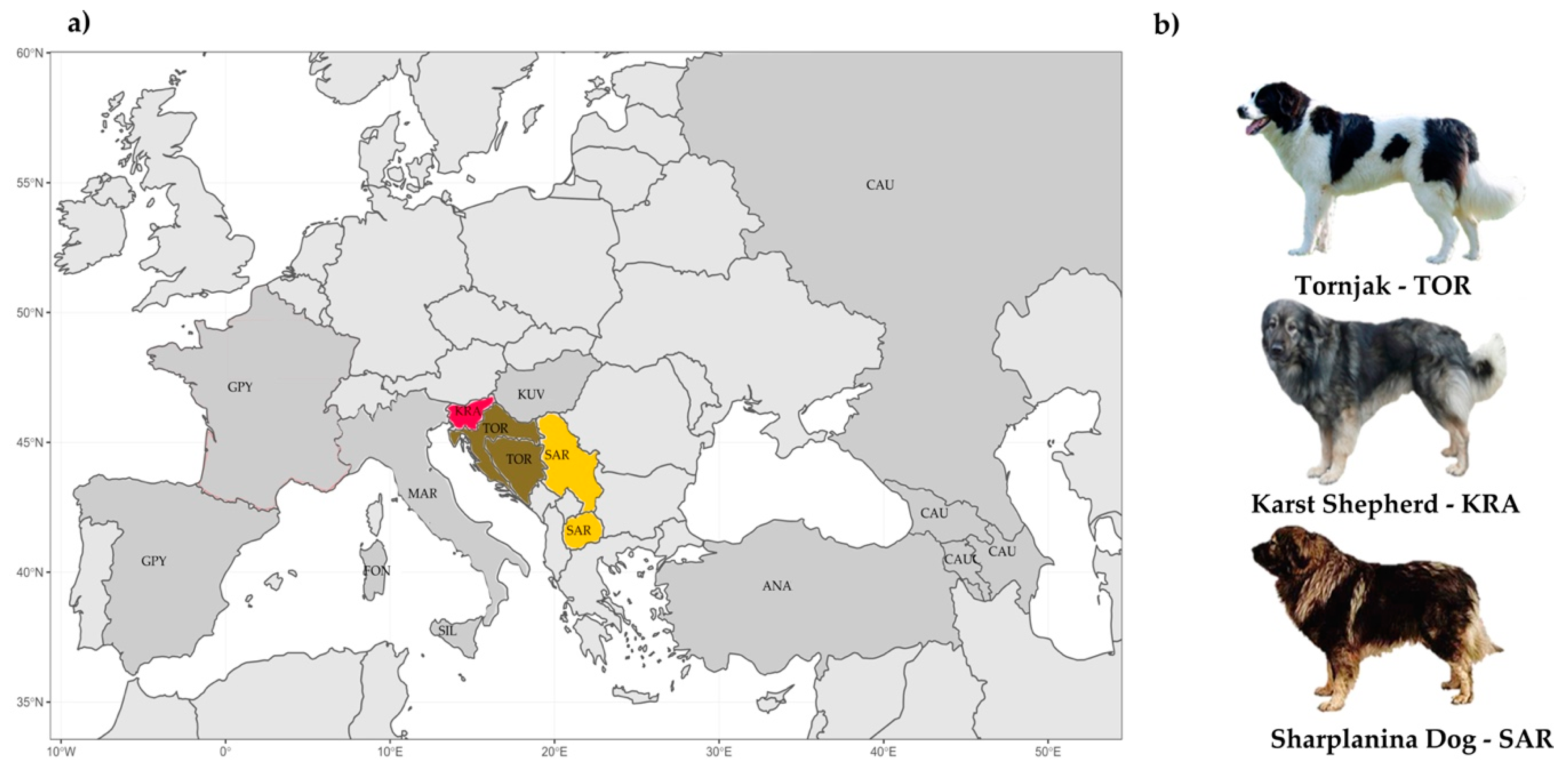
2.1. Sampling, DNA Extraction, Breed Origin, and Genomic Information
2.1.1. Sampling, DNA Extraction, and Genotyping
2.1.2. Publicly Available Genomic Information
2.2. Genomic Diversity, Inbreeding, and Molecular Coancestry
2.2.1. Genomic Diversity and Inbreeding
2.2.2. Runs of Homozygosity Inbreeding
2.2.3. Molecular Coancestry
2.3. Population Structure and Gene Flow
2.3.1. Discriminant Analysis of Principal Components
2.3.2. Phylogenetic Analyses and Gene Flow
2.3.3. Unsupervised Clustering with STRUCTURE Analyses
3. Results
3.1. Genetic Diversity, Inbreeding, and Molecular Coancestry
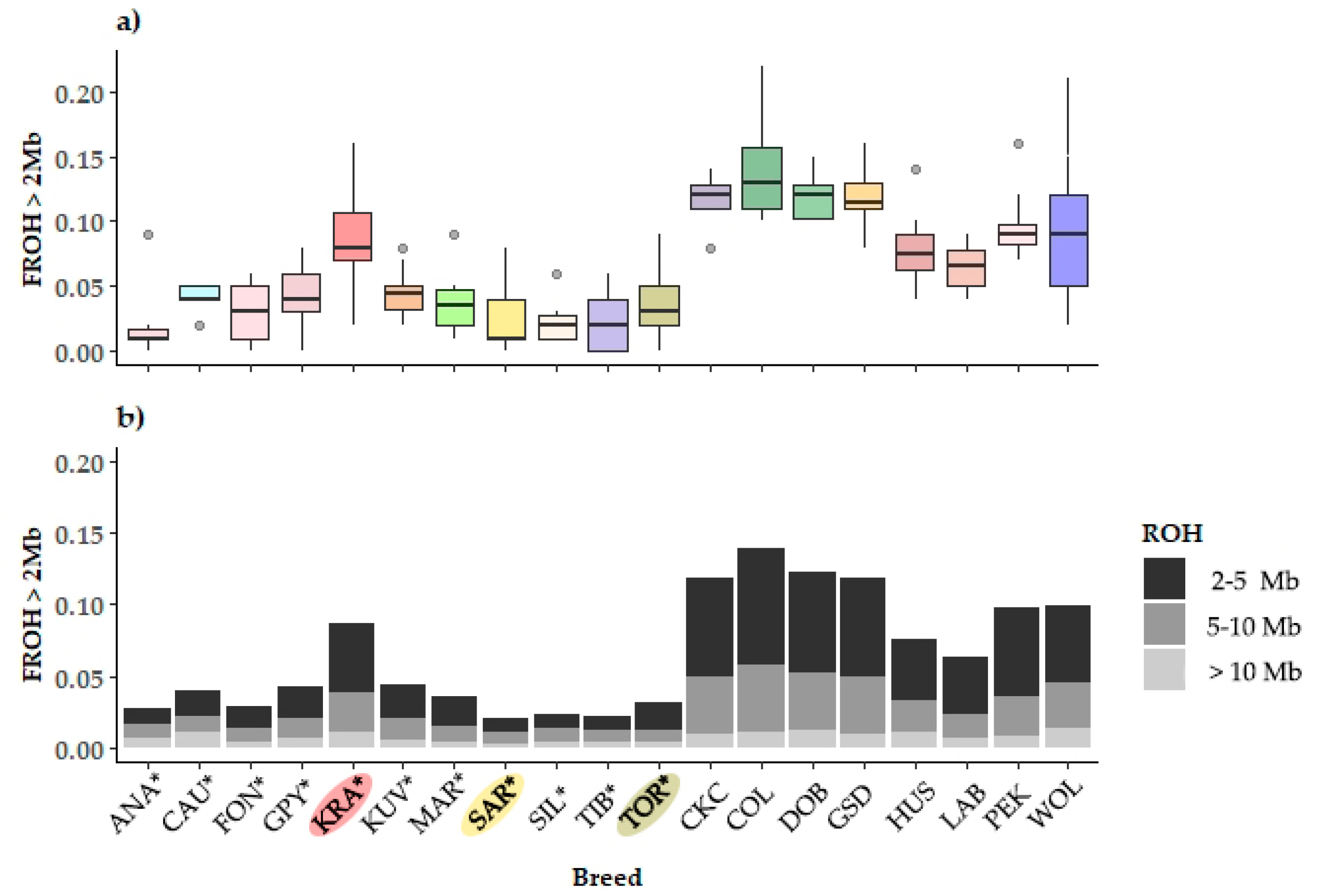
3.1.1. Genomic Diversity and Inbreeding
3.1.2. Runs of Homozygosity, Inbreeding, and Molecular Coancestry
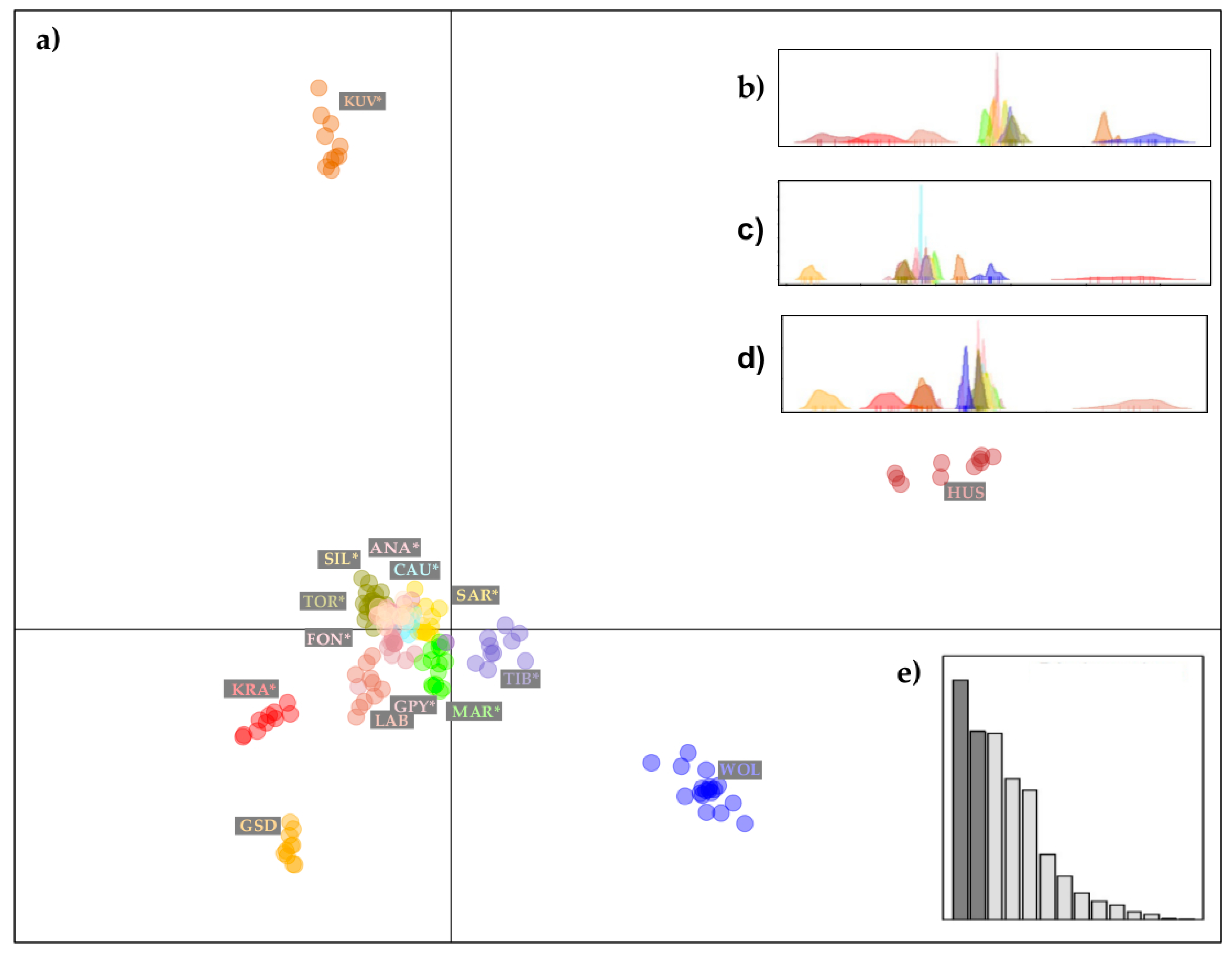
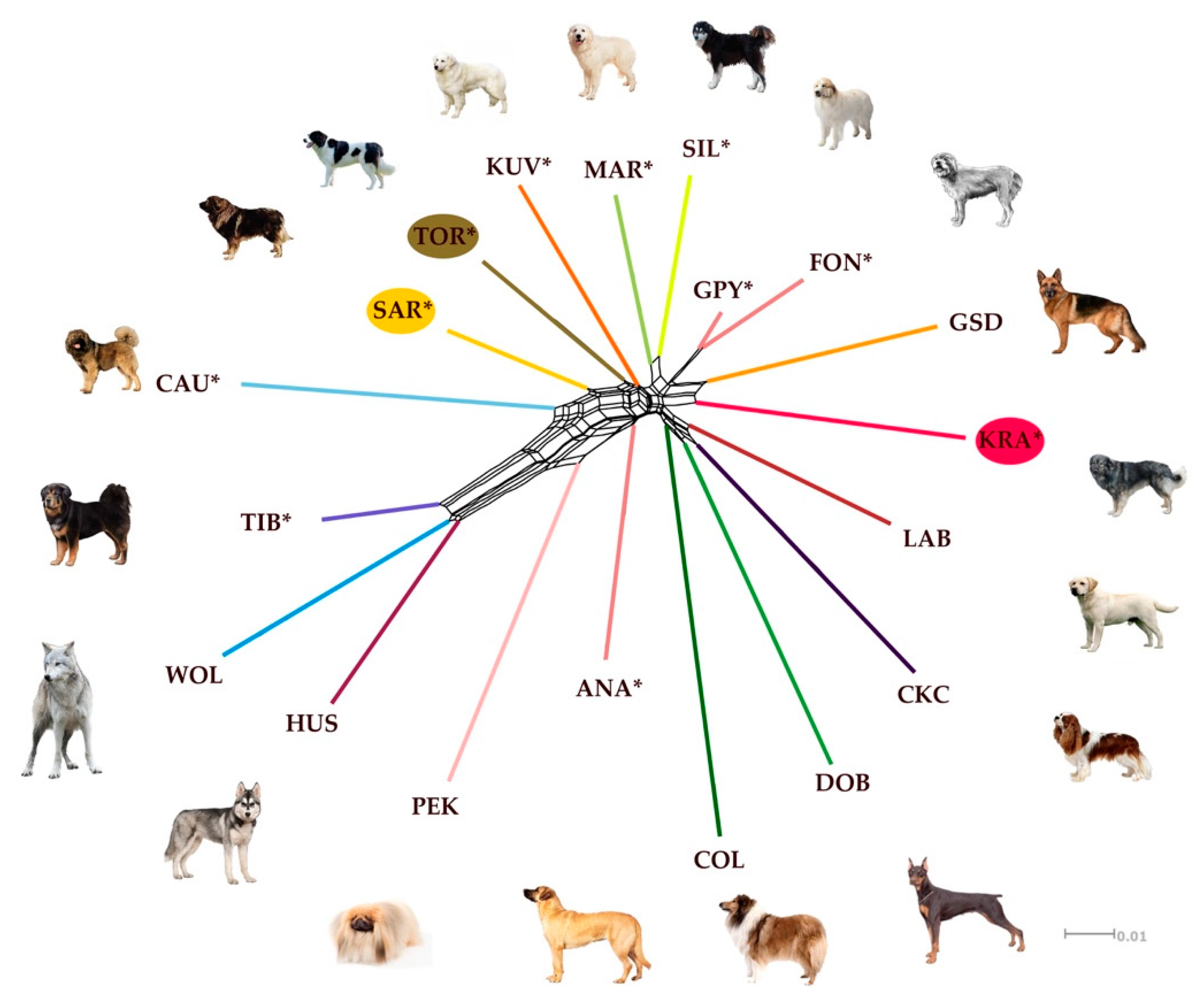
3.2. Population Structure and Gene Flow
Discriminant Analysis of Principal Components and Phylogenetic Analyses with Gene Flow
3.3. Population Structure and Admixture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yilmaz, O.; Coskun, F.; Ertugrul, M. Livestock damage by carnivores and use of livestock guardian dogs for its prevention in Europe—A review. J. Livest. Sci. 2015, 6, 23–35. [Google Scholar]
- Linnell, J.D.C.; Lescureux, N. Livestock Guarding Dogs: Cultural Heritage Icons with a New Relevance for Mitigating Conservation Conflicts; Norwegian Institute for Nature Research: Trondheim, Norway, 2015; p. 76. [Google Scholar]
- Frantz, L.A.F.; Mullin, V.E.; Pionnier-Capitan, M.; Lebrasseur, O.; Ollivier, M.; Perri, A.; Linderholm, A.; Mattiangeli, V.; Teasdale, M.D.; Dimopoulos, E.A.; et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science 2016, 352, 1228–1231. [Google Scholar] [CrossRef]
- Freedman, A.H.; Gronau, I.; Schweizer, R.M.; Ortega-Del Vecchyo, D.; Han, E.; Silva, P.M.; Galaverni, M.; Fan, Z.; Marx, P.; Lorente-Galdos, B.; et al. Genome Sequencing Highlights the Dynamic Early History of Dogs. PLoS Genet. 2014, 10, e1004016. [Google Scholar] [CrossRef]
- Gagliardi, R.; Silvia, L.; García, C.; Arruga, M.V. Microsatellite characterization of Cimarron Uruguayo dogs. Genet. Mol. Biol. 2010, 34, 165–168. [Google Scholar] [CrossRef]
- Irion, D.N. Analysis of Genetic Variation in 28 Dog Breed Populations With 100 Microsatellite Markers. J. Hered. 2003, 94, 81–87. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J.; Zody, M.C.; et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef]
- Parker, H.G.; Dreger, D.L.; Rimbault, M.; Davis, B.W.; Mullen, A.B.; Carpintero-Ramirez, G.; Ostrander, E.A. Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridization on Modern Dog Breed Development. Cell Rep. 2017, 19, 697–708. [Google Scholar] [CrossRef]
- Ceh, E.; Dovc, P. Population structure and genetic differentiation of livestock guard dog breeds from the Western Balkans. J. Anim. Breed. Genet. 2014, 131, 313–325. [Google Scholar] [CrossRef]
- Dimitrijević, V.; Savić, M.; Tarić, E.; Stanišić, L.; Stanimirović, Z.; Tabaković, A.; Aleksić, M.J. Genetic Characterization of the Yugoslavian Shepherd Dog—Sharplanina, a Livestock Guard Dog from the Western Balkans. Acta Vet. Brno 2020, 70, 329–345. [Google Scholar] [CrossRef]
- Bigi, D.; Marelli, S.P.; Randi, E.; Polli, M. Genetic characterization of four native Italian shepherd dog breeds and analysis of their relationship to cosmopolitan dog breeds using microsatellite markers. Animal 2015, 9, 1921–1928. [Google Scholar] [CrossRef]
- Bigi, D.; Marelli, S.P.; Liotta, L.; Frattini, S.; Talenti, A.; Pagnacco, G.; Polli, M.; Crepaldi, P. Investigating the population structure and genetic differentiation of livestock guard dog breeds. Animal 2018, 12, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- VonHoldt, B.M.; Pollinger, J.P.; Lohmueller, K.E.; Han, E.; Parker, H.G.; Quignon, P.; Degenhardt, J.D.; Boyko, A.R.; Earl, D.A.; Auton, A.; et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 2010, 464, 898–902. [Google Scholar] [CrossRef]
- Ostrander, E.A.; Wayne, R.K.; Freedman, A.H.; Davis, B.W. Demographic history, selection and functional diversity of the canine genome. Nat. Rev. Genet. 2017, 18, 705–720. [Google Scholar] [CrossRef] [PubMed]
- Bergström, A.; Frantz, L.; Schmidt, R.; Ersmark, E.; Lebrasseur, O.; Girdland-Flink, L.; Lin, A.T.; Storå, J.; Sjögren, K.-G.; Anthony, D.; et al. Origins and genetic legacy of prehistoric dogs. Science 2020, 370, 557–564. [Google Scholar] [CrossRef]
- Sinding, M.-H.S.; Gopalakrishnan, S.; Ramos-Madrigal, J.; de Manuel, M.; Pitulko, V.V.; Kuderna, L.; Feuerborn, T.R.; Frantz, L.A.F.; Vieira, F.G.; Niemann, J.; et al. Arctic-adapted dogs emerged at the Pleistocene–Holocene transition. Science 2020, 368, 1495–1499. [Google Scholar] [CrossRef]
- Janeš, M.; Zorc, M.; Cubric-Curik, V.; Curik, I.; Dovc, P. Population structure and genetic history of Tibetan Terriers. Genet. Sel. Evol. 2019, 51, 79. [Google Scholar] [CrossRef] [PubMed]
- Lampi, S.; Donner, J.; Anderson, H.; Pohjoismäki, J. Variation in breeding practices and geographic isolation drive subpopulation differentiation, contributing to the loss of genetic diversity within dog breed lineages. Canine Med. Genet. 2020, 7, 5. [Google Scholar] [CrossRef]
- Letko, A.; Minor, K.M.; Jagannathan, V.; Seefried, F.R.; Mickelson, J.R.; Oliehoek, P.; Drögemüller, C. Genomic diversity and population structure of the Leonberger dog breed. Genet. Sel. Evol. 2020, 52, 61. [Google Scholar] [CrossRef]
- Puente, J.M.A.; Barro, Á.L.P.; de la Haba Giraldo, M.R.; Bermejo, J.V.D.; González, F.J.N. Does Functionality Condition the Population Structure and Genetic Diversity of Endangered Dog Breeds under Island Territorial Isolation? Animals 2020, 10, 1893. [Google Scholar] [CrossRef]
- Talenti, A.; Dreger, D.L.; Frattini, S.; Polli, M.; Marelli, S.; Harris, A.C.; Liotta, L.; Cocco, R.; Hogan, A.N.; Bigi, D.; et al. Studies of modern Italian dog populations reveal multiple patterns for domestic breed evolution. Ecol. Evol. 2018, 8, 2911–2925. [Google Scholar] [CrossRef]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395. [Google Scholar] [CrossRef]
- Ferenčaković, M.; Sölkner, J.; Curik, I. Estimating autozygosity from high-throughput information: Effects of SNP density and genotyping errors. Genet. Sel. Evol. 2013, 45, 42. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of Homozygosity in European Populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 3319242776. [Google Scholar]
- Saura, M.; Fernández, A.; Rodríguez, M.C.; Toro, M.A.; Barragán, C.; Fernández, A.I.; Villanueva, B. Genome-Wide Estimates of Coancestry and Inbreeding in a Closed Herd of Ancient Iberian Pigs. PLoS ONE 2013, 8, e78314. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Collins, C. A tutorial for Discriminant Analysis of Principal Components (DAPC) using adegenet. Rvignette 2015. Available online: http://adegenet.r-forge.r-project.org/files/tutorial-dapc.pdf (accessed on 23 December 2020).
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Cogan, N.O.I.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2003, 4, 137–138. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F -statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Paraskevopoulou, C.; Theodoridis, A.; Johnson, M.; Ragkos, A.; Arguile, L.; Smith, L.; Vlachos, D.; Arsenos, G. Sustainability Assessment of Goat and Sheep Farms: A Comparison between European Countries. Sustainability 2020, 12, 3099. [Google Scholar] [CrossRef]
- Wiener, P.; Sánchez-Molano, E.; Clements, D.N.; Woolliams, J.A.; Haskell, M.J.; Blott, S.C. Genomic data illuminates demography, genetic structure and selection of a popular dog breed. BMC Genom. 2017, 18, 609. [Google Scholar] [CrossRef]
- Druml, T.; Curik, I.; Baumung, R.; Aberle, K.; Distl, O.; Sölkner, J. Individual-based assessment of population structure and admixture in Austrian, Croatian and German draught horses. Heredity 2007, 98, 114–122. [Google Scholar] [CrossRef]
- Dreger, D.L.; Davis, B.W.; Cocco, R.; Sechi, S.; Di Cerbo, A.; Parker, H.G.; Polli, M.; Marelli, S.P.; Crepaldi, P.; Ostrander, E.A. Commonalities in Development of Pure Breeds and Population Isolates Revealed in the Genome of the Sardinian Fonni’s Dog. Genetics 2016, 204, 737–755. [Google Scholar] [CrossRef]
- Sechi, S.; Polli, M.; Marelli, S.; Talenti, A.; Crepaldi, P.; Fiore, F.; Spissu, N.; Dreger, D.L.; Zedda, M.; Dimauro, C.; et al. Fonni’s dog: Morphological and genetic characteristics for a breed standard definition. Ital. J. Anim. Sci. 2017, 16, 22–30. [Google Scholar] [CrossRef]

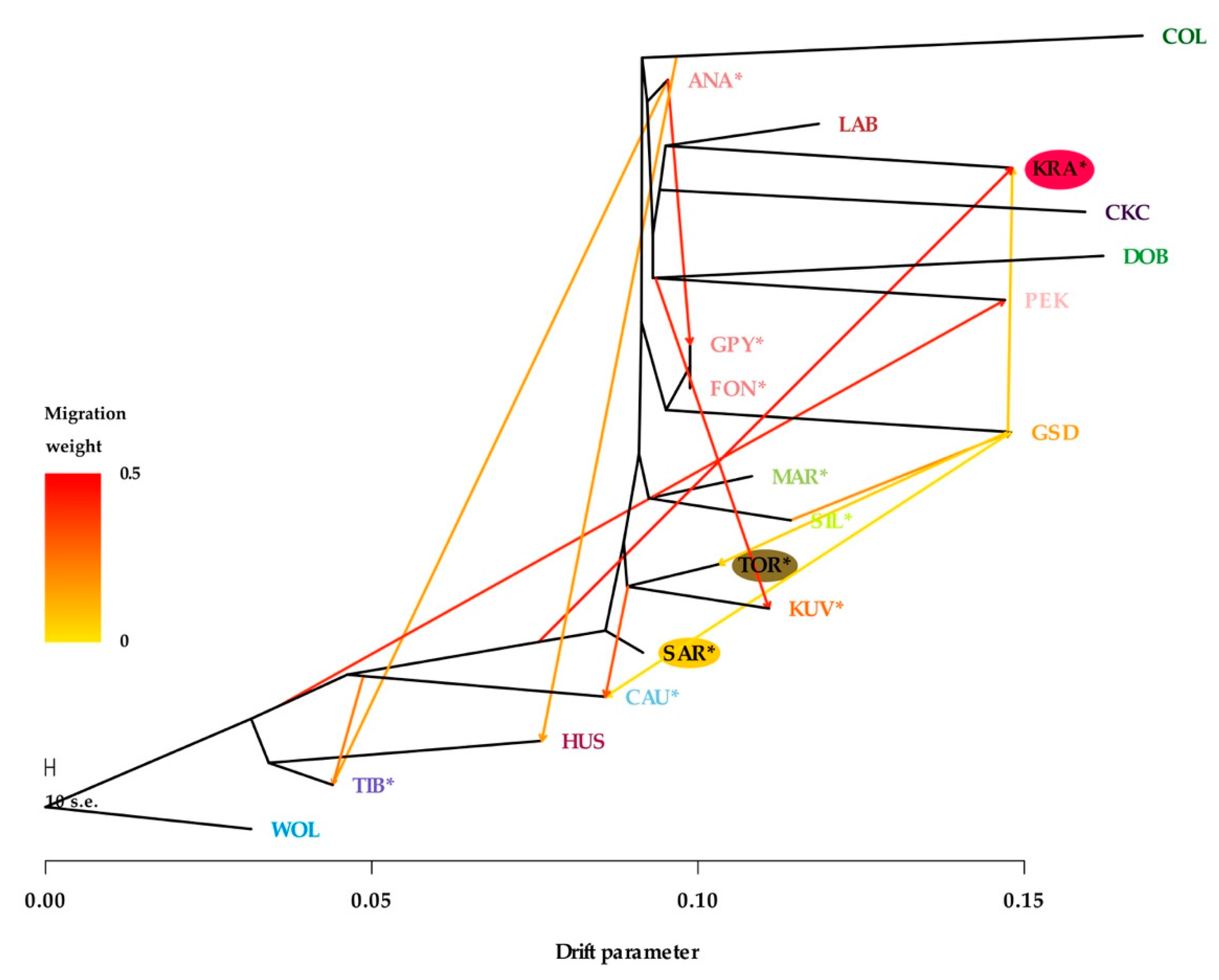
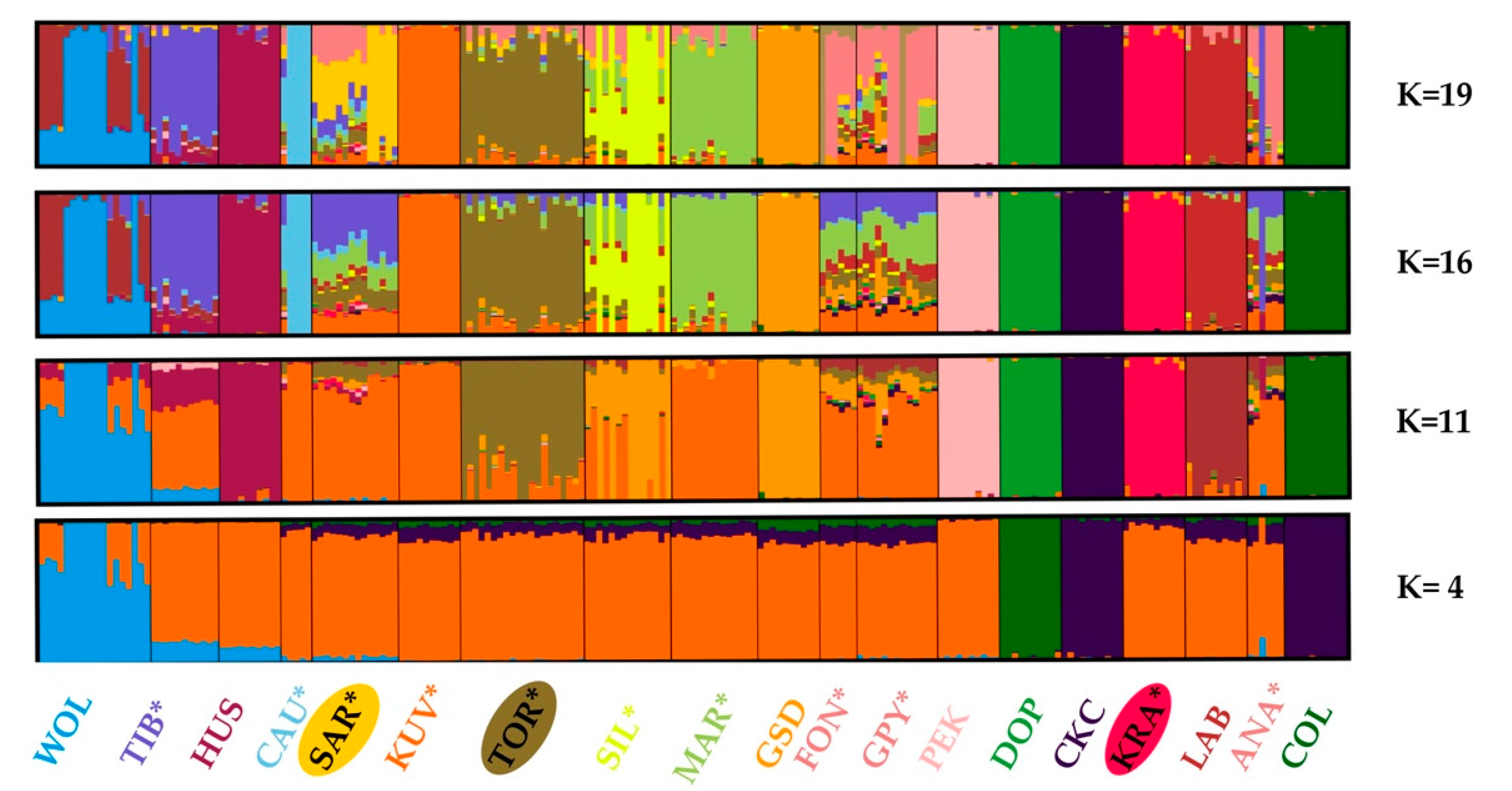
| Breed (Code) | Animals | Ho ± SE | He ± SE | FIS ± SE | FROH>2Mb ± SE |
|---|---|---|---|---|---|
| Livestock guarding dogs (LGD) | 123 | 0.290 ± 0.003 | 0.296 ± 0.003 | 0.015 ± 0.010 | 0.035 ± 0.003 |
| Anatolian Shepherd (ANA) | 6 | 0.294 ± 0.018 | 0.329 ± 0.000 | 0.105 ± 0.056 | 0.026 ± 0.013 |
| Caucasian Shepherd (CAU) | 5 | 0.278 ± 0.009 | 0.251 ± 0.000 | −0.107 ± 0.034 | 0.038 ± 0.004 |
| Fonni’s Dog (FON) | 6 | 0.300 ± 0.018 | 0.319 ± 0.000 | 0.073 ± 0.056 | 0.025 ± 0.010 |
| Great Pyrenees (GPY) | 13 | 0.277 ± 0.011 | 0.328 ± 0.000 | 0.157 ± 0.032 | 0.043 ± 0.007 |
| Karst Shepherd (KRA) | 10 | 0.241 ± 0.003 | 0.222 ± 0.000 | −0.081 ± 0.014 | 0.087 ± 0.012 |
| Kuvazs (KUV) | 10 | 0.279 ± 0.007 | 0.288 ± 0.000 | 0.033 ± 0.024 | 0.045 ± 0.006 |
| Maremmano-Abruzzese Sheepdog (MAR) | 14 | 0.291 ± 0.006 | 0.296 ± 0.000 | 0.016 ± 0.022 | 0.036 ± 0.005 |
| Sharplanina Dog (SAR) | 14 | 0.315 ± 0.005 | 0.315 ± 0.000 | 0.000 ± 0.017 | 0.020 ± 0.007 |
| Pastore della Sila (SIL) | 14 | 0.302 ± 0.007 | 0.284 ± 0.000 | −0.064 ± 0.025 | 0.022 ± 0.004 |
| Tibetan Mastiff (TIB) | 11 | 0.289 ± 0.005 | 0.300 ± 0.000 | 0.036 ± 0.017 | 0.018 ± 0.005 |
| Tornjak (TOR) | 20 | 0.301 ± 0.003 | 0.301 ± 0.000 | 0.000 ± 0.010 | 0.033 ± 0.005 |
| Popular dog breeds (PDB) | 70 | 0.208 ± 0.004 | 0.220 ± 0.004 | 0.052 ± 0.010 | 0.105 ± 0.005 |
| Cavalier King Charles Spaniel (CKC) | 10 | 0.192 ± 0.004 | 0.199 ± 0.000 | 0.034 ± 0.019 | 0.119 ± 0.005 |
| Rough Collie (COL) | 10 | 0.173 ± 0.005 | 0.181 ± 0.000 | 0.044 ± 0.028 | 0.140 ± 0.012 |
| Doberman Pinscher (DOB) | 10 | 0.187 ± 0.004 | 0.193 ± 0.000 | 0.032 ± 0.023 | 0.120 ± 0.006 |
| German Shepherd Dog (GSD) | 10 | 0.208 ± 0.005 | 0.221 ± 0.000 | 0.058 ± 0.023 | 0.118 ± 0.007 |
| Husky (HUS) | 10 | 0.023 ± 0.007 | 0.237 ± 0.001 | 0.061 ± 0.030 | 0.077 ± 0.009 |
| Labrador Retriever (LAB) | 10 | 0.256 ± 0.006 | 0.277 ± 0.000 | 0.074 ± 0.020 | 0.065 ± 0.006 |
| Pekingese (PEK) | 10 | 0.215 ± 0.009 | 0.229 ± 0.000 | 0.061 ± 0.038 | 0.097 ± 0.008 |
| Gray wolf (WOL) | 18 | 0.177 ± 0.011 | 0.235 ± 0.000 | 0.247 ± 0.046 | 0.093 ± 0.014 |
| Breed | KRA* | SAR* | TOR* | ANA* | CAU* | COL | FON* | GPY* | GSD | KUV* | MAR | SIL* | TIB* | CKC | DOB | HUS | LAB | PEK | MFST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KRA* | 0.27 | ||||||||||||||||||
| SAR* | 0.18 | 0.13 | |||||||||||||||||
| TOR* | 0.20 | 0.07 | 0.15 | ||||||||||||||||
| ANA* | 0.19 | 0.04 | 0.06 | 0.12 | |||||||||||||||
| CAU* | 0.30 | 0.13 | 0.16 | 0.13 | 0.23 | ||||||||||||||
| COL | 0.40 | 0.25 | 0.26 | 0.26 | 0.38 | 0.33 | |||||||||||||
| FON* | 0.20 | 0.05 | 0.07 | 0.03 | 0.15 | 0.27 | 0.14 | ||||||||||||
| GPY | 0.17 | 0.04 | 0.06 | 0.00 | 0.12 | 0.23 | 0.00 | 0.11 | |||||||||||
| GSD | 0.32 | 0.19 | 0.19 | 0.19 | 0.30 | 0.40 | 0.18 | 0.16 | 0.27 | ||||||||||
| KUV* | 0.23 | 0.09 | 0.11 | 0.08 | 0.19 | 0.30 | 0.09 | 0.07 | 0.23 | 0.17 | |||||||||
| MAR* | 0.21 | 0.08 | 0.10 | 0.07 | 0.18 | 0.27 | 0.08 | 0.06 | 0.22 | 0.12 | 0.16 | ||||||||
| SIL* | 0.23 | 0.10 | 0.12 | 0.09 | 0.20 | 0.29 | 0.10 | 0.08 | 0.21 | 0.14 | 0.11 | 0.18 | |||||||
| TIB* | 0.23 | 0.09 | 0.12 | 0.07 | 0.17 | 0.30 | 0.11 | 0.09 | 0.25 | 0.14 | 0.14 | 0.16 | 0.17 | ||||||
| CKC | 0.37 | 0.23 | 0.24 | 0.22 | 0.35 | 0.43 | 0.24 | 0.20 | 0.37 | 0.27 | 0.25 | 0.27 | 0.28 | 0.30 | |||||
| DOB | 0.38 | 0.23 | 0.24 | 0.24 | 0.36 | 0.44 | 0.25 | 0.21 | 0.37 | 0.28 | 0.26 | 0.27 | 0.29 | 0.41 | 0.31 | ||||
| HUS | 0.33 | 0.18 | 0.21 | 0.18 | 0.28 | 0.39 | 0.20 | 0.18 | 0.34 | 0.23 | 0.22 | 0.24 | 0.16 | 0.37 | 0.38 | 0.26 | |||
| LAB | 0.24 | 0.11 | 0.13 | 0.09 | 0.21 | 0.31 | 0.10 | 0.08 | 0.25 | 0.15 | 0.13 | 0.15 | 0.16 | 0.27 | 0.29 | 0.25 | 0.19 | ||
| PEK | 0.33 | 0.19 | 0.20 | 0.18 | 0.29 | 0.40 | 0.20 | 0.17 | 0.34 | 0.23 | 0.22 | 0.24 | 0.21 | 0.36 | 0.38 | 0.31 | 0.25 | 0.26 | |
| WOL | 0.35 | 0.20 | 0.23 | 0.19 | 0.29 | 0.41 | 0.22 | 0.20 | 0.36 | 0.25 | 0.24 | 0.26 | 0.18 | 0.39 | 0.40 | 0.27 | 0.27 | 0.32 | 0.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janeš, M.; Zorc, M.; Ferenčaković, M.; Curik, I.; Dovč, P.; Cubric-Curik, V. Genomic Characterization of the Three Balkan Livestock Guardian Dogs. Sustainability 2021, 13, 2289. https://doi.org/10.3390/su13042289
Janeš M, Zorc M, Ferenčaković M, Curik I, Dovč P, Cubric-Curik V. Genomic Characterization of the Three Balkan Livestock Guardian Dogs. Sustainability. 2021; 13(4):2289. https://doi.org/10.3390/su13042289
Chicago/Turabian StyleJaneš, Mateja, Minja Zorc, Maja Ferenčaković, Ino Curik, Peter Dovč, and Vlatka Cubric-Curik. 2021. "Genomic Characterization of the Three Balkan Livestock Guardian Dogs" Sustainability 13, no. 4: 2289. https://doi.org/10.3390/su13042289
APA StyleJaneš, M., Zorc, M., Ferenčaković, M., Curik, I., Dovč, P., & Cubric-Curik, V. (2021). Genomic Characterization of the Three Balkan Livestock Guardian Dogs. Sustainability, 13(4), 2289. https://doi.org/10.3390/su13042289










