Recent Advances in Understanding Mechanisms of Plant Tolerance and Response to Aluminum Toxicity
Abstract
1. Introduction
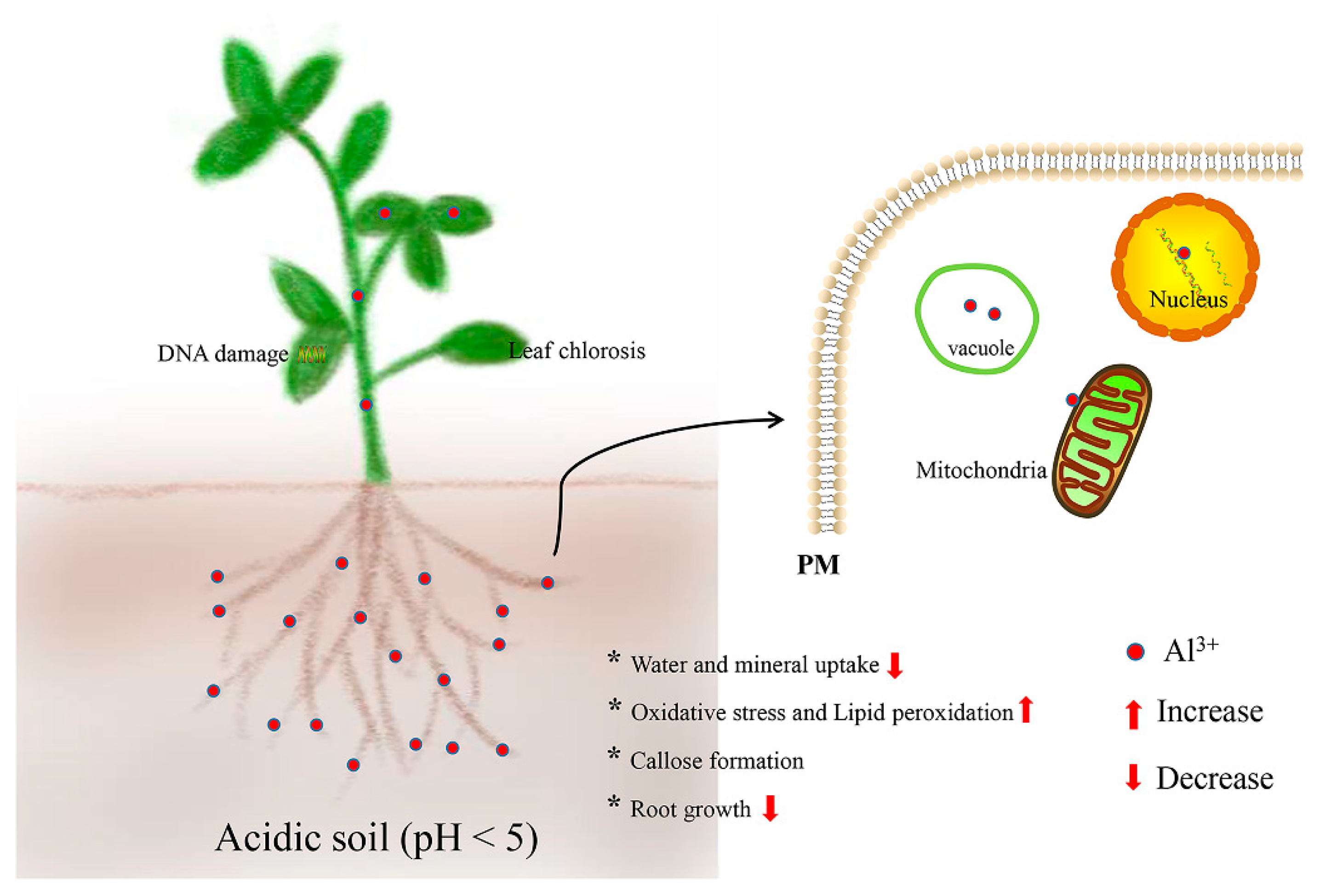
2. Effects of Aluminum (Al) Stress in Plants
3. Al Tolerance Mechanisms in Plants
3.1. External Exclusion Mechanisms
3.2. Internal Tolerance Mechanisms
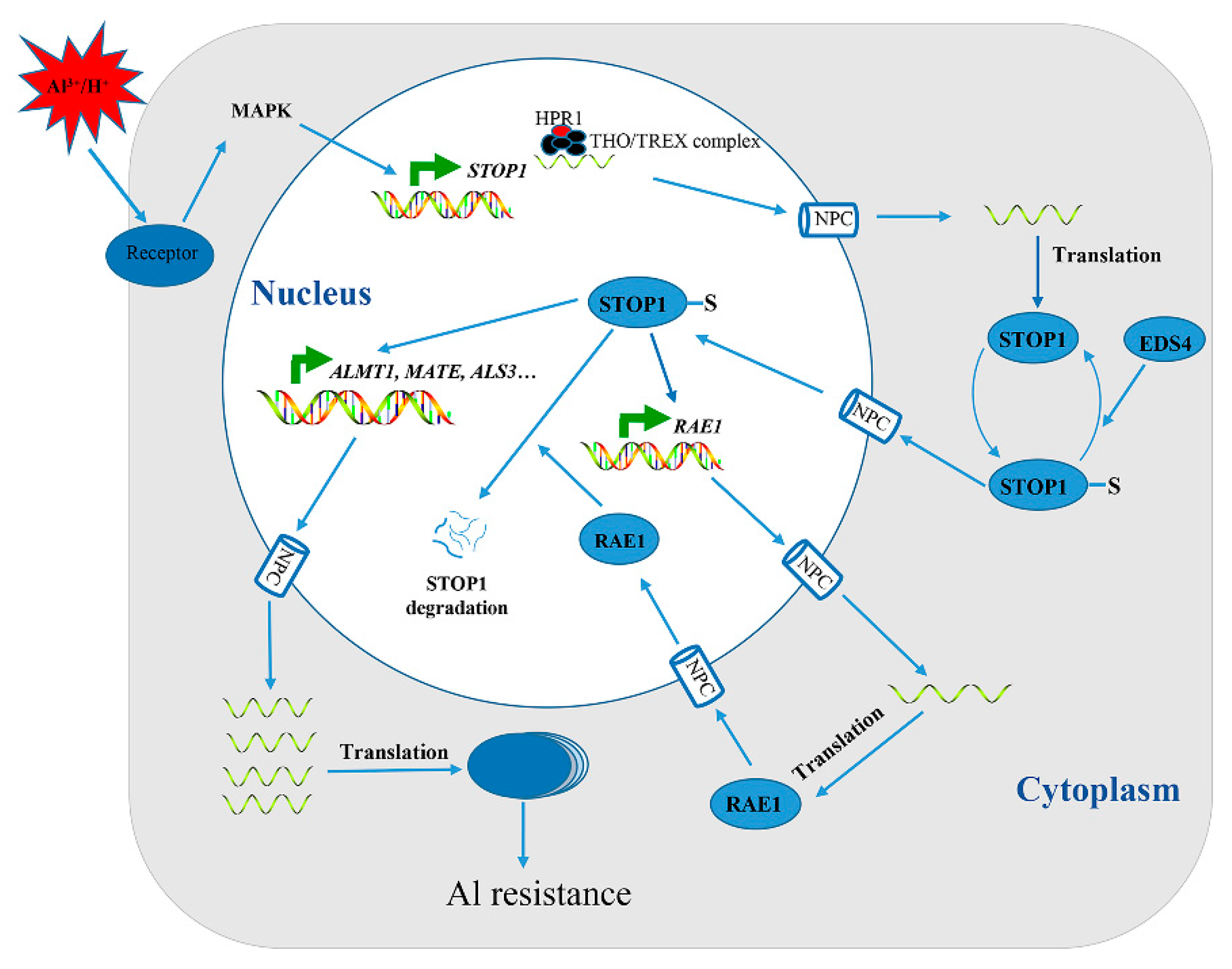
4. Transcription Factors Are Involved in Adaptation to Al Stress
5. Plant Hormones Involved in Adaptation to Al Stress
6. Small Molecules to Alleviate Al Toxicity
7. Plant Growth-Promoting Rhizobacteria (PGPRs) Alleviate Al Toxicity
8. Transgenic Approaches Manipulating Al-Tolerant Genes
9. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zheng, S.J. Crop production on acidic soils: Overcoming aluminium toxicity and phosphorus deficiency. Ann. Bot. 2010, 106, 183–184. [Google Scholar] [CrossRef]
- Wei, Y.; Jiang, C.; Han, R.; Xie, Y.; Liu, L.; Yu, Y. Plasma membrane proteomic analysis by TMT-PRM provides insight into mechanisms of aluminum resistance in tamba black soybean roots tips. PeerJ 2020, 8, e9312. [Google Scholar] [CrossRef]
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant Adaptation to Acid Soils: The Molecular Basis for Crop Aluminum Resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef]
- Eekhout, T.; Larsen, P.; De Veylder, L. Modification of DNA Checkpoints to Confer Aluminum Tolerance. Trends Plant Sci. 2017, 22, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.T.; Qi, Y.P.; Jiang, H.X.; Chen, L.S. Roles of organic acid anion secretion in aluminium tolerance of higher plants. BioMed Res. Int. 2013, 2013, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Liu, L.; Li, X.; Han, R.; Wei, Y.; Yu, Y. Insights into aluminum-tolerance pathways in Stylosanthes as revealed by RNA-Seq analysis. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Snowden, K.C.; Cardner, R.C. Five genes induced by aluminum in wheat (Triticum aestivum L.) roots. Plant Physiol. 1993, 103, 855–861. [Google Scholar] [CrossRef]
- Regisfer, P.G.; Richards, K.D.; Snowden, K.C.; Cardner, R.C. wali6 and wali7: Genes induced by aluminum in wheat (Triticum aestivum L.) roots. Plant Physiol. 1994, 105, 1455–1456. [Google Scholar]
- Muhammad, N.; Zvobgo, G.; Zhang, G.P. A review: The beneficial effects and possible mechanisms of aluminum on plant growth in acidic soil. J. Integr. Agric. 2019, 18, 1518–1528. [Google Scholar] [CrossRef]
- Wang, L.; Fan, X.W.; Pan, J.L.; Huang, Z.B.; Li, Y.Z. Physiological characterization of maize tolerance to low dose of aluminum, highlighted by promoted leaf growth. Planta 2015, 242, 1391–1403. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Eticha, D.; Stass, A.; Horst, W.J. Cell-wall pectin and its degree of methylation in the maize root-apex: Significance for genotypic differences in aluminium resistance. Plant Cell Environ. 2005, 28, 1410–1420. [Google Scholar] [CrossRef]
- Singh, S.; Tripathi, D.K.; Singh, S.; Sharma, S.; Dubey, N.K.; Chauhan, D.K.; Vaculík, M. Toxicity of aluminium on various levels of plant cells and organism: A review. Environ. Exp. Bot. 2017, 137, 177–193. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, W.; Cao, Y.; Cai, Y.; Lyi, S.M.; Wu, W.; Kang, Y.; Liang, C.; Liu, J. An exclusion mechanism is epistatic to an internal detoxification mechanism in aluminum resistance in Arabidopsis. BMC Plant Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Riaz, M.; Yan, L.; Wu, X.; Hussain, S.; Aziz, O.; Jiang, C. Mechanisms of organic acids and boron induced tolerance of aluminum toxicity: A review. Ecotoxicol. Environ. Saf. 2018, 165, 25–35. [Google Scholar] [CrossRef]
- Ma, J.F.; Furukawa, J. Recent progress in the research of external Al detoxification in higher plants: A minireview. J. Inorg. Biochem. 2003, 97, 46–51. [Google Scholar] [CrossRef]
- Lyu, B.; Guo, X.; Gao, D.; Kou, M.; Yu, Y.; Ma, J.; Chen, S.; Wang, H.; Zhang, Y.; Bao, X. Auxin metabolic network regulates the plant response to metalloids stress. J. Hazard. Mater. 2020, 405, 124250. [Google Scholar]
- Rahman, M.A.; Lee, S.H.; Ji, H.C.; Kabir, A.H.; Jones, C.S.; Lee, K.W. Importance of mineral nutrition for mitigating aluminum toxicity in plants on acidic soils: Current status and opportunities. Int. J. Mol. Sci. 2018, 19, 3073. [Google Scholar] [CrossRef]
- Zhou, J.; Li, Z.; Zhou, T.; Xin, Z.; Wu, L.; Luo, Y.; Christie, P. Aluminum toxicity decreases the phytoextraction capability by cadmium/zinc hyperaccumulator Sedum plumbizincicola in acid soils. Sci. Total Environ. 2020, 711, 134591. [Google Scholar] [CrossRef]
- He, H.; Li, Y.; He, L.F. Aluminum toxicity and tolerance in Solanaceae plants. S. Afr. J. Bot. 2019, 123, 23–29. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Naamala, J.; Dakora, F.D. Nature and mechanisms of aluminium toxicity, tolerance and amelioration in symbiotic legumes and rhizobia. Biol. Fertil. Soils 2018, 54, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Kopittke, P.M. Role of phytohormones in aluminium rhizotoxicity. Plant Cell Environ. 2016, 39, 2319–2328. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, M.; Liu, X.; Mao, Q.; Shi, C.; Kochian, L.V.; Liao, H. Aluminium is essential for root growth and development of tea plants (Camellia sinensis). J. Integr. Plant Biol. 2020, 62, 984–997. [Google Scholar] [CrossRef]
- Liu, Y.; Tao, J.; Cao, J.; Zeng, Y.; Li, X.; Ma, J.; Huang, Z.; Jiang, M.; Sun, L. The beneficial effects of aluminum on the plant growth in Camellia japonica. J. Soil Sci. Plant Nutr. 2020, 20, 1799–1809. [Google Scholar] [CrossRef]
- Sade, H.; Meriga, B.; Surapu, V.; Gadi, J.; Sunita, M.S.L.; Suravajhala, P.; Kavi Kishor, P.B. Toxicity and tolerance of aluminum in plants: Tailoring plants to suit to acid soils. BioMetals 2016, 29, 187–210. [Google Scholar] [CrossRef]
- Yang, J.l.; Fan, W.; Zheng, S.j. jian Mechanisms and regulation of aluminum-induced secretion of organic acid anions from plant roots. J. Zhejiang Univ. Sci. B 2019, 20, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Wang, N.; Dai, J.; Wang, T.; Kochian, L.V.; Liu, J.; Zuo, Y. AhFRDL1-mediated citrate secretion contributes to adaptation to iron deficiency and aluminum stress in peanuts. J. Exp. Bot. 2019, 70, 2873–2886. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, Z.; How, J.; Xu, H.; Chen, L.; Li, K. Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol. Biol. 2017, 95, 157–168. [Google Scholar] [CrossRef]
- Ezaki, B.; Higashi, A.; Nanba, N.; Nishiuchi, T. An S-adenosyl methionine synthetase (SAMS) gene from Andropogon virginicus L. confers aluminum stress tolerance and facilitates epigenetic gene regulation in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, X.X.; Wang, J.; Qi, C.; Wang, X.; Wang, G.; Li, M.; Li, X.; Guo, Y.D. BoALMT1, an al-induced malate transporter in cabbage, enhances aluminum tolerance in Arabidopsis thaliana. Front. Plant Sci. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.P.; de Souza, W.R.; Martins, P.K.; Vinecky, F.; Duarte, K.E.; Basso, M.F.; da Cunha, B.A.D.B.; Campanha, R.B.; de Oliveira, P.A.; Centeno, D.C.; et al. Overexpression of BdMATE gene improves aluminum tolerance in Setaria viridis. Front. Plant Sci. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Z.; Gong, L.; Chen, A.; Liu, N.; Li, S.; Sun, H.; Yang, Z.; You, J. Functional characterization of three MATE genes in relation to aluminum-induced citrate efflux from soybean root. Plant Soil 2019, 443, 121–138. [Google Scholar] [CrossRef]
- Ma, Q.; Yi, R.; Li, L.; Liang, Z.; Zeng, T.; Zhang, Y.; Huang, H.; Zhang, X.; Yin, X.; Cai, Z.; et al. GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Meng, H.; Xing, H.; Liang, L.; Zhao, X.; Luo, K. Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus. J. Exp. Bot. 2017, 68, 5669–5683. [Google Scholar] [CrossRef]
- Pooniya, V.; Palta, J.A.; Chen, Y.; Delhaize, E.; Siddique, K.H.M. Impact of the TaMATE1B gene on above and below-ground growth of durum wheat grown on an acid and Al3+-toxic soil. Plant Soil 2020, 447, 73–84. [Google Scholar] [CrossRef]
- Xu, J.M.; Lou, H.Q.; Jin, J.F.; Chen, W.W.; Wan, J.X.; Fan, W.; Yang, J.L. A half-type ABC transporter FeSTAR1 regulates Al resistance possibly via UDP-glucose-based hemicellulose metabolism and Al binding. Plant Soil 2018, 432, 303–314. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Liu, X.; Guo, C.; Yao, W.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. GmGRP-like gene confers Al tolerance in Arabidopsis. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Cai, Z.; Xian, P.; Lin, R.; Cheng, Y.; Lian, T.; Ma, Q.; Nian, H. Characterization of the Soybean GmIREG Family Genes and the Function of GmIREG3 in Conferring Tolerance to Aluminum Stress. Int. J. Mol. Sci. 2020, 21, 497. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Z.; Xu, Y.; Sun, H.; Sun, Z.; Lin, B.; Sun, W.; You, J. Soybean NADP-malic enzyme functions in malate and citrate metabolism and contributes to their efflux under Al stress. Front. Plant Sci. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Li, W.; Du, J.; Feng, H.; Wu, Q.; Xu, G.; Shabala, S.; Yu, L. Function of NHX-type transporters in improving rice tolerance to aluminum stress and soil acidity. Planta 2020, 251, 1–13. [Google Scholar] [CrossRef]
- Liu, W.; Feng, X.; Cao, F.; Wu, D.; Zhang, G.; Vincze, E.; Wang, Y.; Chen, Z.H.; Wu, F. An ATP binding cassette transporter HvABCB25 confers aluminum detoxification in wild barley. J. Hazard. Mater. 2020, 401, 123371. [Google Scholar] [CrossRef]
- Sun, G.; Zhu, H.; Wen, S.; Liu, L.; Gou, L.; Guo, Z. Citrate synthesis and exudation confer Al resistance. Plant Soil 2020, 449, 319–329. [Google Scholar] [CrossRef]
- Li, J.; Su, L.; Lv, A.; Li, Y.; Zhou, P.; An, Y. MsPG1 alleviated aluminum-induced inhibition of root growth by decreasing aluminum accumulation and increasing porosity and extensibility of cell walls in alfalfa (Medicago sativa). Environ. Exp. Bot. 2020, 175, 104045. [Google Scholar] [CrossRef]
- Kariya, K.; Sameeullah, M.; Sasaki, T.; Yamamoto, Y. Overexpression of the sucrose transporter gene NtSUT1 alleviates aluminum-induced inhibition of root elongation in tobacco (Nicotiana tabacum L.). Soil Sci. Plant Nutr. 2017, 63, 45–54. [Google Scholar] [CrossRef][Green Version]
- Wang, M.; Qiao, J.Y.; Yu, C.L.; Chen, H.; Sun, C.D.; Huang, L.Z.; Li, C.Y.; Geisler, M.; Qian, Q.; Jiang, D.A.; et al. The auxin influx carrier, OsAUX3, regulates rice root development and responses to aluminium stress. Plant Cell Environ. 2019, 42, 1125–1138. [Google Scholar] [CrossRef]
- Lou, H.Q.; Fan, W.; Xu, J.M.; Gong, Y.L.; Jin, J.F.; Chen, W.W.; Liu, L.Y.; Hai, M.R.; Yang, J.L.; Zheng, S.J. An oxalyl-CoA synthetase is involved in oxalate degradation and aluminum tolerance. Plant Physiol. 2016, 172, 1679–1690. [Google Scholar] [CrossRef]
- Lou, H.Q.; Gong, Y.L.; Fan, W.; Xu, J.M.; Liu, Y.; Cao, M.J.; Wang, M.H.; Yang, J.L.; Zheng, S.J. A formate dehydrogenase confers tolerance to aluminum and low pH. Plant Physiol. 2016, 171, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; He, C.; Han, L. Heterologous expression of ZjOMT from Zoysia japonica in Escherichia coli confers aluminum resistance through melatonin production. PLoS ONE 2018, 13, e0196952. [Google Scholar] [CrossRef]
- Du, H.; Huang, Y.; Qu, M.; Li, Y.; Hu, X.; Yang, W.; Li, H.; He, W.; Ding, J.; Liu, C.; et al. A maize ZmAT6 gene confers aluminum tolerance via reactive oxygen species scavenging. Front. Plant Sci. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lu, X.; Li, C.; Zhang, B.; Zhang, C.; Zhang, X.S.; Ding, Z. Auxin efflux carrier ZmPGP1 mediates root growth inhibition under aluminum stress. Plant Physiol. 2018, 177, 819–832. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. Molecular mechanisms for coping with al toxicity in plants. Int. J. Mol. Sci. 2019, 20, 1551. [Google Scholar] [CrossRef]
- Pin, M.A.; Magalhaes, J.V.; Alves, V.M.C.; Kochian, L.V. The physiology and biophysics of an aluminum tolerance mechanism based on root citrate exudation in maize. Plant Physiol. 2002, 129, 1194–1206. [Google Scholar]
- Ligaba, A.; Dreyer, I.; Margaryan, A.; Schneider, D.J.; Kochian, L.; Piñeros, M. Functional, structural and phylogenetic analysis of domains underlying the Al sensitivity of the aluminum-activated malate/anion transporter, TaALMT1. Plant J. 2013, 76, 766–780. [Google Scholar] [CrossRef]
- Hoekenga, O.A.; Maron, L.G.; Piñeros, M.A.; Cançado, G.M.A.; Shaff, J.; Kobayashi, Y.; Ryan, P.R.; Dong, B.; Delhaize, E.; Sasaki, T.; et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 9738–9743. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Hoekenga, O.A.; Itoh, H.; Nakashima, M.; Saito, S.; Shaff, J.E.; Maron, L.G.; Piñeros, M.A.; Kochian, L.V.; Koyama, H. Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 2007, 145, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.D.; Ryan, P.R.; Richardson, A.E.; Tyerman, S.D.; Ramesh, S.; Hebb, D.M.; Howitt, S.M.; Delhaize, E. HvALMT1 from barley is involved in the transport of organic anions. J. Exp. Bot. 2010, 61, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Ligaba, A.; Katsuhara, M.; Ryan, P.R.; Shibasaka, M.; Matsumoto, H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006, 142, 1294–1303. [Google Scholar] [CrossRef]
- Ligaba, A.; Katsuhara, M.; Sakamoto, W.; Matsumoto, H. The BnALMT1 protein that is an aluminum-activated malate transporter is localized in the plasma membrane. Plant Signal. Behav. 2007, 2, 255–257. [Google Scholar] [CrossRef]
- Ligaba, A.; Maron, L.; Shaff, J.; Kochian, L.; Piñeros, M. Maize ZmALMT2 is a root anion transporter that mediates constitutive root malate efflux. Plant Cell Environ. 2012, 35, 1185–1200. [Google Scholar] [CrossRef]
- Chen, Z.C.; Yokosho, K.; Kashino, M.; Zhao, F.J.; Yamaji, N.; Ma, J.F. Adaptation to acidic soil is achieved by increased numbers of cis-acting elements regulating ALMT1 expression in Holcus lanatus. Plant J. 2013, 76, 10–23. [Google Scholar] [PubMed]
- Park, W.; Kim, H.S.; Park, T.W.; Lee, Y.H.; Ahn, S.J. Functional characterization of plasma membrane-localized organic acid transporter (CsALMT1) involved in aluminum tolerance in Camelina sativa L. Plant Biotechnol. Rep. 2017, 11, 181–192. [Google Scholar] [CrossRef]
- Ma, X.; An, F.; Wang, L.; Guo, D.; Xie, G.; Liu, Z. Genome-wide identification of aluminum-activated malate transporter (ALMT) gene family in rubber trees (Hevea brasiliensis) highlights their involvement in aluminum detoxification. Forests 2020, 11, 142. [Google Scholar] [CrossRef]
- Upadhyay, N.; Kar, D.; Deepak Mahajan, B.; Nanda, S.; Rahiman, R.; Panchakshari, N.; Bhagavatula, L.; Datta, S. The multitasking abilities of MATE transporters in plants. J. Exp. Bot. 2019, 70, 4643–4656. [Google Scholar] [CrossRef]
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, R.; Shi, J.; Wang, J.; Sun, Q.; Zhang, H.; Xing, Y.; Qi, Y.; Zhang, N.; Guo, Y.D. Brassica oleracea MATE encodes a citrate transporter and enhances aluminum tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 1426–1436. [Google Scholar] [CrossRef] [PubMed]
- Sawaki, Y.; Kihara-Doi, T.; Kobayashi, Y.; Nishikubo, N.; Kawazu, T.; Kobayashi, Y.; Koyama, H.; Sato, S. Characterization of Al-responsive citrate excretion and citrate-transporting MATEs in Eucalyptus camaldulensis. Planta 2013, 237, 979–989. [Google Scholar] [CrossRef]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Ma, J.F. Two MATE transporters with different subcellular localization are involved in Al tolerance in buckwheat. Plant Cell Physiol. 2017, 58, 2179–2189. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Wang, W.; Gai, J.; Li, Y. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genom. 2016, 17, 1–15. [Google Scholar] [CrossRef]
- Wang, P.; Yu, W.; Zhang, J.; Rengel, Z.; Xu, J.; Han, Q.; Chen, L.; Li, K.; Yu, Y.; Chen, Q. Auxin enhances aluminium-induced citrate exudation through upregulation of GmMATE and activation of the plasma membrane H+-ATPase in soybean roots. Ann. Bot. 2016, 118, 933–940. [Google Scholar] [CrossRef]
- Iguchi, A.; Sanmiya, K.; Watanabe, K. Identification of genes encoding ALMT and MATE transporters as candidate aluminum tolerance genes from a typical acid soil plant, Psychotria rubra (Rubiaceae). PeerJ 2019, 7, e7739. [Google Scholar] [CrossRef]
- Melo, J.O.; Lana, U.G.P.; Piñeros, M.A.; Alves, V.M.C.; Guimarães, C.T.; Liu, J.; Zheng, Y.; Zhong, S.; Fei, Z.; Maron, L.G.; et al. Incomplete transfer of accessory loci influencing SbMATE expression underlies genetic background effects for aluminum tolerance in sorghum. Plant J. 2013, 73, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Chen, W.W.; Xu, J.M.; Fan, W.; Yang, J.L.; Zheng, S.J. The role of VuMATE1 expression in aluminium-inducible citrate secretion in rice bean (Vigna umbellata) roots. J. Exp. Bot. 2013, 64, 1795–1804. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Lou, H.Q.; Gong, Y.L.; Liu, M.Y.; Cao, M.J.; Liu, Y.; Yang, J.L.; Zheng, S.J. Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 2015, 208, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Yang, J.L.; Zhou, Y.; Piñeros, M.A.; Kochian, L.V.; Li, G.X.; Zheng, S.J. A de novo synthesis citrate transporter, Vigna umbellata multidrug and toxic compound extrusion, implicates in Al-activated citrate efflux in rice bean (Vigna umbellata) root apex. Plant Cell Environ. 2011, 34, 2138–2148. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Lou, H.Q.; Chen, W.W.; Piñeros, M.A.; Xu, J.M.; Fan, W.; Kochian, L.V.; Zheng, S.J.; Yang, J.L. Two citrate transporters coordinately regulate citrate secretion from rice bean root tip under aluminum stress. Plant Cell Environ. 2018, 41, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Maron, L.G.; Piñeros, M.A.; Guimarães, C.T.; Magalhaes, J.V.; Pleiman, J.K.; Mao, C.; Shaff, J.; Belicuas, S.N.J.; Kochian, L.V. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 2010, 61, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wu, J.; Jiang, Y.; Jin, J.; Zhou, W.; Wang, Y.; Han, G.; Zhao, Y.; Cheng, B. Genomewide analysis of MATE-type gene family in maize reveals microsynteny and their expression patterns under aluminum treatment. J. Genet. 2016, 95, 691–704. [Google Scholar] [CrossRef]
- Semenova, E.V.; Kosareva, I.A.; Belimov, A.A. Aluminum exclusion from root zone and maintenance of nutrient uptake are principal mechanisms of Al tolerance in Pisum sativum L. Physiol. Mol. Biol. Plants 2017, 23, 851–863. [Google Scholar]
- Klug, B.; Horst, W.J. Oxalate exudation into the root-tip water free space confers protection from aluminum toxicity and allows aluminum accumulation in the symplast in buckwheat (Fagopyrum esculentum). New Phytol. 2010, 187, 380–391. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, J.; Li, D.; Kong, X.; Rengel, Z.; Chen, L.; Yang, Y.; Cui, X.; Chen, Q. The role of the plasma membrane H+-ATPase in plant responses to aluminum toxicity. Front. Plant Sci. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Q.L.; Geng, M.J.; Guo, Z.H.; Zhao, Z. Rhizosphere ph difference regulated by plasma membrane H+-ATPase is related to differential Al-tolerance of two wheat cultivars. Plant Soil Environ. 2011, 57, 201–206. [Google Scholar] [CrossRef]
- Zhang, F.; Yan, X.; Han, X.; Tang, R.; Chu, M.; Yang, Y.; Yang, Y.H.; Zhao, F.; Fu, A.; Luan, S.; et al. A defective vacuolar proton pump enhances aluminum tolerance by reducing vacuole sequestration of organic acids. Plant Physiol. 2019, 181, 743–761. [Google Scholar] [CrossRef]
- Kidd, P.S.; Llugany, M.; Poschenrieder, C.; Gunsé, B.; Barceló, J. The role of root exudates in aluminium resistance and silicon-induced amelioration of aluminium toxicity in three varieties of maize (Zea mays L.). J. Exp. Bot. 2001, 52, 1339–1352. [Google Scholar] [PubMed]
- Ma, Z.; Lin, S. Transcriptomic revelation of phenolic compounds involved in aluminum toxicity responses in roots of cunninghamia lanceolata (lamb.) hook. Genes 2019, 10, 835. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, L.; Liang, X.; Dai, P.; Zhang, Y.; Li, B.; Lin, X.; Sun, C. Enhancement of polyphenolic metabolism as an adaptive response of lettuce (Lactuca sativa) roots to aluminum stress. Environ. Pollut. 2020, 261, 114230. [Google Scholar] [CrossRef]
- Fu, Z.; Jiang, X.; Li, W.W.; Shi, Y.; Lai, S.; Zhuang, J.; Yao, S.; Liu, Y.; Hu, J.; Gao, L.; et al. Proanthocyanidin-aluminum complexes improve aluminum resistance and detoxification of Camellia sinensis. J. Agric. Food Chem. 2020, 68, 7861–7869. [Google Scholar] [CrossRef]
- Yang, J.L.; Li, Y.Y.; Zhang, Y.J.; Zhang, S.S.; Wu, Y.R.; Wu, P.; Zheng, S.J. Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol. 2008, 146, 602–611. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef]
- Che, J.; Yamaji, N.; Yokosho, K.; Shen, R.F.; Ma, J.F. Two genes encoding a bacterial-type ATP-binding cassette transporter are implicated in aluminum tolerance in buckwheat. Plant Cell Physiol. 2018, 59, 2502–2511. [Google Scholar] [CrossRef]
- Fan, W.; Xu, J.M.; Wu, P.; Yang, Z.X.; Lou, H.Q.; Chen, W.W.; Jin, J.F.; Zheng, S.J.; Yang, J.L. Alleviation by abscisic acid of Al toxicity in rice bean is not associated with citrate efflux but depends on ABI5-mediated signal transduction pathways. J. Integr. Plant Biol. 2019, 61, 140–154. [Google Scholar] [CrossRef]
- Yang, T.-Y.; Huang, W.-T.; Zhang, J.; Yang, L.-T.; Huang, Z.-R.; Wu, B.-S.; Lai, N.-W.; Chen, L.-S. Raised pH conferred the ability to maintain a balance between production and detoxification of reactive oxygen species and methylglyoxal in aluminum-toxic Citrus sinensis leaves and roots. Environ. Pollut. 2020, 268, 115676. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Zeng, Z.H.; Yan, J.Y.; Fan, W.; Bian, H.W.; Zhu, M.Y.; Yang, J.L.; Zheng, S.J. Association of specific pectin methylesterases with Al-induced root elongation inhibition in rice. Physiol. Plant. 2013, 148, 502–511. [Google Scholar] [CrossRef]
- Baylis, A.D.; Gragopoulou, C.; Davidson, K.J.; Birchall, J.D. Effects of silicon on the toxicity of aluminium to soybean. Commun. Soil Sci. Plant Anal. 1994, 25, 537–546. [Google Scholar] [CrossRef]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Yang, J.L.; Zhu, X.F.; Peng, Y.X.; Zheng, C.; Li, G.X.; Liu, Y.; Shi, Y.Z.; Zheng, S.J. Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 2011, 155, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.Y.; Cai, L.Y.; Qi, Y.P.; Yang, L.T.; Lai, N.W.; Chen, L.S. Increasing nutrient solution ph alleviated aluminum-induced inhibition of growth and impairment of photosynthetic electron transport chain in Citrus sinensis seedlings. BioMed Res. Int. 2019, 2019, 9058715. [Google Scholar] [CrossRef]
- Yang, T.Y.; Qi, Y.P.; Huang, H.Y.; Wu, F.L.; Huang, W.T.; Deng, C.L.; Yang, L.T.; Chen, L.S. Interactive effects of pH and aluminum on the secretion of organic acid anions by roots and related metabolic factors in Citrus sinensis roots and leaves. Environ. Pollut. 2020, 262, 114303. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Peng, Z.; Li, D.; Ma, W.; An, R.; Khan, D.; Wang, X.; Liu, Y.; Yang, E.; He, Y.; et al. Magnesium decreases aluminum accumulation and plays a role in protecting maize from aluminum-induced oxidative stress. Plant Soil 2020, 457, 71–81. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, D.; Meng, H.; Li, S.; Wang, S.; Xiao, Z.; Sun, J.; Chang, L.; Luo, K.; Li, N. Magnesium alleviates aluminum toxicity by promoting polar auxin transport and distribution and root alkalization in the root apex in populus. Plant Soil 2020, 448, 565–585. [Google Scholar] [CrossRef]
- Li, J.Y.; Liu, J.; Dong, D.; Jia, X.; McCouch, S.R.; Kochian, L.V. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA 2014, 111, 6503–6508. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Yamaji, N.; Kasai, T.; Ma, J.F. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 18381–18385. [Google Scholar] [CrossRef] [PubMed]
- Nhan, P. Amelioration of aluminum toxicity on OM4900 rice seedlings by sodium silicate. Afr. J. Plant Sci. 2013, 7, 208–212. [Google Scholar] [CrossRef]
- Carr, H.P.; Lombi, E.; Küpper, H.; McGrath, S.P.; Wong, M.H. Accumulation and distribution of aluminium and other elements in tea (Camellia sinensis) leaves. Agronomie 2003, 23, 705–710. [Google Scholar] [CrossRef]
- Negishi, T.; Oshima, K.; Hattori, M.; Kanai, M.; Mano, S.; Nishimura, M.; Yoshida, K. Tonoplast-and plasma membrane-localized aquaporin-family transporters in Blue Hydrangea Sepals of Aluminum Hyperaccumulating Plant. PLoS ONE 2012, 7, e43189. [Google Scholar] [CrossRef]
- Lu, M.; Yang, G.; Li, P.; Wang, Z.; Fu, S.; Zhang, X.; Chen, X.; Shi, M.; Ming, Z.; Xia, J. Bioinformatic and functional analysis of a key determinant underlying the substrate selectivity of the al transporter, Nrat1. Front. Plant Sci. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.B.; Geisler, M.J.B.; Jones, C.A.; Williams, K.M.; Cancel, J.D. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005, 41, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Fujii-Kashino, M.; Ma, J.F. Functional characterization of two half-size ABC transporter genes in aluminium-accumulating buckwheat. New Phytol. 2017, 215, 1080–1089. [Google Scholar] [CrossRef]
- Ezaki, B.; Takahashi, K.; Utsumi, K.; Higashi, A. A half-type AvABCG1 transporter derived from Andropogon virginicus L. confers aluminum tolerance. Environ. Exp. Bot. 2015, 118, 21–31. [Google Scholar] [CrossRef]
- Wang, Y.; Li, R.; Li, D.; Jia, X.; Zhou, D.; Li, J.; Lyi, S.M.; Hou, S.; Huang, Y.; Kochian, L.V.; et al. NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 5047–5052. [Google Scholar] [CrossRef]
- Hazak, O.; Mamon, E.; Lavy, M.; Sternberg, H.; Behera, S.; Schmitz-Thom, I.; Bloch, D.; Dementiev, O.; Gutman, I.; Danziger, T.; et al. A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth. PLoS Boil. 2019, 17, e3000085. [Google Scholar] [CrossRef] [PubMed]
- Kar, D.; Pradhan, A.A.; Datta, S. The role of solute transporters in aluminium toxicity and tolerance. Physiol. Plant. 2020. [Google Scholar] [CrossRef]
- Lan, T.; You, J.; Kong, L.; Yu, M.; Liu, M.; Yang, Z. The interaction of salicylic acid and Ca2+ alleviates aluminum toxicity in soybean (Glycine max L.). Plant Physiol. Biochem. 2016, 98, 146–154. [Google Scholar] [CrossRef]
- Su, L.; Lv, A.; Wen, W.; Zhou, P.; An, Y. Auxin is involved in magnesium-mediated photoprotection in photosystems of alfalfa seedlings under aluminum stress. Front. Plant Sci. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Chen, Z.C.; Yamaji, N.; Motoyama, R.; Nagamura, Y.; Ma, J.F. Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol. 2012, 159, 1624–1633. [Google Scholar] [CrossRef]
- Li, D.; Ma, W.; Wei, J.; Mao, Y.; Peng, Z.; Zhang, J.; Kong, X.; Han, Q.; Fan, W.; Yang, Y.; et al. Magnesium promotes root growth and increases aluminum tolerance via modulation of nitric oxide production in Arabidopsis. Plant Soil 2019, 1–13. [Google Scholar] [CrossRef]
- Vera-Villalobos, H.; Lunario-Delgado, L.; Pérez-Retamal, D.; Román, D.; Leiva, J.C.; Zamorano, P.; Mercado-Seguel, A.; Gálvez, A.S.; Benito, C.; Wulff-Zottele, C. Sulfate nutrition improves short-term Al3+-stress tolerance in roots of Lolium perenne L. Plant Physiol. Biochem. 2020, 148, 103–113. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kobayashi, Y.; Devi, S.R.; Rikiishi, S.; Matsumoto, H. Oxidative stress triggered by aluminum in plant roots. Plant Soil 2003, 255, 239–243. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Zhou, W.; Lu, L.; Jin, C.; Lin, X. Aluminum induces distinct changes in the metabolism of reactive oxygen and nitrogen species in the roots of two wheat genotypes with different aluminum resistance. J. Agric. Food Chem. 2017, 65, 9419–9427. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Good, A.G.; Taylor, G.J. Transgenic Brassica napus plants overexpressing aluminium-induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminium. Plant Cell Environ. 2001, 24, 1269–1278. [Google Scholar] [CrossRef]
- Ezaki, B.; Gardner, R.C.; Ezaki, Y.; Matsumoto, H. Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol. 2000, 122, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiao, D.; Yang, C.; Chen, J.; Li, Y.; Liang, S.; Lin, K.; Chen, Z. Genome-wide identification and expression analysis of SABATH methyltransferases in tea plant (Camellia sinensis): Insights into their roles in plant defense responses. Plant Signal. Behav. 2020, 15, 1804684. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, J.; Guo, S.; Yuan, X.; Zhao, S.; Tian, H.; Dai, S.; Kong, X.; Ding, Z. AtHB7/12 regulate root growth in response to aluminum stress. Int. J. Mol. Sci. 2020, 21, 4080. [Google Scholar] [CrossRef]
- Li, C.X.; Yan, J.Y.; Ren, J.Y.; Sun, L.; Xu, C.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 2019, 62, 1176–1192. [Google Scholar] [CrossRef]
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F.; et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Arun, A.; Yuriko, D.; Sanjib, K.; Panda, K. Characterization of CcSTOP1; a C2H2-type transcription factor regulates Al tolerance gene in pigeonpea. Planta 2017, 247, 201–214. [Google Scholar]
- Kundu, A.; Das, S.; Basu, S.; Kobayashi, Y.; Kobayashi, Y.; Koyama, H.; Ganesan, M. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 2019, 21, 35–44. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Z.M.; Gong, L.; Liu, R.K.; Sun, H.R.; You, J.F. Molecular characterization of GmSTOP1 homologs in soybean under Al and proton stress. Plant Soil 2018, 427, 213–230. [Google Scholar] [CrossRef]
- Liu, Y.T.; Shi, Q.H.; Cao, H.J.; Ma, Q.B.; Nian, H.; Zhang, X.X. Heterologous expression of a Glycine soja C2H2 zinc finger gene improves aluminum tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 2754. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Guoa, Y.; Caia, S.; Kuanga, L.; Shena, Q.; Wu, D.; Zhanga, G. A zinc finger transcription factor HvATF1 regulates aluminum tolerance in barley. J. Exp. Bot. 2020, 71, 6512–6523. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Kobayashi, Y.; Yamamoto, Y.Y.; Koyama, H. Characterization of NtSTOP1-regulating genes in tobacco under aluminum stress. Soil Sci. Plant Nutr. 2019, 65, 251–258. [Google Scholar] [CrossRef]
- Che, J.; Tsutsui, T.; Yokosho, K.; Yamaji, N.; Ma, J.F. Functional characterization of an aluminum (Al)-inducible transcription factor, ART2, revealed a different pathway for Al tolerance in rice. New Phytol. 2018, 220, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gao, J.; You, J.; Liang, Y.; Guan, K.; Yan, S.; Zhan, M.; Yang, Z. Identification of STOP1-like proteins associated with aluminum tolerance in sweet sorghum (Sorghum bicolor L.). Front. Plant Sci. 2018, 9, 258. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Yang, C.; Cheng, Y.; Han, Z.; Cai, Z.; Nian, H.; Ma, Q. GsMAS1 encoding a MADS-box transcription factor enhances the tolerance to aluminum stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 2004. [Google Scholar] [CrossRef]
- Feng, X.; Liu, W.; Dai, H.; Qiu, Y.; Zhang, G.; Chen, Z.-H.; Wu, F. HvHOX9, a novel homeobox leucine zipper transcription factor revealed by root miRNA and RNA sequencing in tibetan wild barley, positively regulates Al tolerance. J. Exp. Bot. 2020, 71, 6057–6073. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.N.; Yao, J.F.; Ren, Y.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple MdMYC2 reduces aluminum stress tolerance by directly regulating MdERF3 gene. Plant Soil 2017, 418, 255–266. [Google Scholar] [CrossRef]
- Lou, H.Q.; Fan, W.; Jin, J.F.; Xu, J.M.; Chen, W.W.; Yang, J.L.; Zheng, S.J. A NAC-type transcription factor confers aluminium resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant. Cell Environ. 2020, 43, 463–478. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Guo, J.; Zhou, F.; Singh, S.; Xu, X.; Xie, Q.; Yang, Z.; Huang, C.F. F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 319–327. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, Y.; Gao, H.; Li, S.; Wang, Z.-Y.; Huang, C.-F. Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminium resistance in Arabidopsis thaliana. New Phytol. 2020, 228, 179–193. [Google Scholar] [CrossRef]
- Fang, Q.; Zhang, J.; Zhang, Y.; Fan, N.; van den Burg, H.A.; Huang, C.F. Regulation of aluminum-resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell 2020, 32, 3921–3938. [Google Scholar] [CrossRef]
- Tsutsui, T.; Yamaji, N.; Feng Ma, J. Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 2011, 156, 925–931. [Google Scholar] [CrossRef]
- Qin, H.; He, L.; Huang, R. The coordination of ethylene and other hormones in primary root development. Front. Plant Sci. 2019, 10, 874. [Google Scholar] [CrossRef]
- Yang, Z.B.; Geng, X.; He, C.; Zhang, F.; Wang, R.; Horst, W.J.; Ding, Z. TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 2014, 26, 2889–2904. [Google Scholar] [CrossRef]
- Lv, B.; Yan, Z.; Tian, H.; Zhang, X.; Ding, Z. Local auxin biosynthesis mediates plant growth and development. Trends Plant Sci. 2019, 24, 6–9. [Google Scholar] [CrossRef]
- Liu, G.; Gao, S.; Tian, H.; Wu, W.; Robert, H.S.; Ding, Z. Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PLoS Genet. 2016, 12, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, G.; Liu, J.; Zhang, B.; Meng, W.; Müller, B.; Hayashi, K.; Zhang, X.; Zhao, Z.; De Smet, I.; et al. Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep. 2017, 18, 1213–1230. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Bian, H.; Zeng, Z.; Hou, N.; Shi, B.; Wang, J.; Zhu, M.; Han, N. MiR393-mediated auxin signaling regulation is involved in root elongation inhibition in response to toxic aluminum stress in barley. Plant Cell Physiol. 2017, 58, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Nian, F.; Zhao, L.; Li, F.; Yang, H.; Yang, Y. Exogenous indole-3-acetic acid could reduce the accumulation of aluminum in root apex of wheat (Triticum aestivum L.) under Al stress. J. Soil Sci. Plant Nutr. 2013, 13, 534–543. [Google Scholar] [CrossRef]
- Wang, S.; Yuan, S.; Su, L.; Lv, A.; Zhou, P.; An, Y. Aluminum toxicity in alfalfa (Medicago sativa) is alleviated by exogenous foliar IAA inducing reduction of Al accumulation in cell wall. Environ. Exp. Bot. 2017, 139, 1–13. [Google Scholar] [CrossRef]
- Tan, D.-X.; Hardeland, R.; Manchester, L.C.; Korkmaz, A.; Ma, S.; Rosales-Corral, S.; Reiter, R.J. Functional roles of melatonin in plants, and perspectives in nutritional and agricultural science. J. Exp. Bot. 2012, 63, 577–597. [Google Scholar] [CrossRef]
- Zhang, J.; Zeng, B.; Mao, Y.; Kong, X.; Wang, X.; Yang, Y.; Zhang, J.; Xu, J.; Rengel, Z.; Chen, Q. Melatonin alleviates aluminium toxicity through modulating antioxidative enzymes and enhancing organic acid anion exudation in soybean. Funct. Plant Biol. 2017, 44, 961–968. [Google Scholar] [CrossRef]
- Zhang, J.; Li, D.; Wei, J.; Ma, W.; Kong, X.; Rengel, Z.; Chen, Q. Melatonin alleviates aluminum-induced root growth inhibition by interfering with nitric oxide production in Arabidopsis. Environ. Exp. Bot. 2019, 161, 157–165. [Google Scholar] [CrossRef]
- Sun, C.; Lv, T.; Huang, L.; Liu, X.; Jin, C.; Lin, X. Melatonin ameliorates aluminum toxicity through enhancing aluminum exclusion and reestablishing redox homeostasis in roots of wheat. J. Pineal Res. 2020, 68, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Tian, Q.Y.; Chen, J.; Zhang, W.H. Aluminium-induced inhibition of root elongation in Arabidopsis is mediated by ethylene and auxin. J. Exp. Bot. 2010, 61, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, Q.; Nian, H.; Yang, C.; Li, X.; Cheng, Y.; Cai, Z. GsERF enhances aluminum tolerance through an ethylene-mediated pathway in Arabidopsis thaliana. BioRxiv 2020, 1–21. [Google Scholar] [CrossRef]
- Reyna-Llorens, I.; Corrales, I.; Poschenrieder, C.; Barcelo, J.; Cruz-Ortega, R. Both aluminum and ABA induce the expression of an ABC-like transporter gene (FeALS3) in the Al-tolerant species Fagopyrum esculentum. Environ. Exp. Bot. 2015, 111, 74–82. [Google Scholar] [CrossRef]
- Salazar-Chavarría, V.; Sánchez-Nieto, S.; Cruz-Ortega, R. Fagopyrum esculentum at early stages copes with aluminum toxicity by increasing ABA levels and antioxidant system. Plant Physiol. Biochem. 2020, 152, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Emamverdian, A.; Ding, Y.; Xie, Y.; Sangari, S. Silicon mechanisms to ameliorate heavy metal stress in plants. BioMed Res. Int. 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.; Stass, A.; Horst, W.J. Apoplastic binding of aluminum is involved in silicon-induced amelioration of aluminum toxicity in maize. Plant Physiol. 2004, 136, 3762–3770. [Google Scholar] [CrossRef]
- Prabagar, S.; Hodson, M.J.; Evans, D.E. Silicon amelioration of aluminium toxicity and cell death in suspension cultures of Norway spruce (Picea abies L. Karst.). Environ. Exp. Bot. 2011, 70, 266–276. [Google Scholar] [CrossRef]
- Qian, L.; Chen, B.; Chen, M. Novel alleviation mechanisms of aluminum phytotoxicity via released biosilicon from rice straw-derived biochars. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- De Freitas, L.B.; Fernandes, D.M.; Maia, S.C.M.; Fernandes, A.M. Effects of silicon on aluminum toxicity in upland rice plants. Plant Soil 2017, 420, 263–275. [Google Scholar] [CrossRef]
- Pontigo, S.; Godoy, K.; Jiménez, H.; Gutiérrez-Moraga, A.; Mora, M.D.L.L.; Cartes, P. Silicon-mediated alleviation of aluminum toxicity by modulation of Al/Si uptake and antioxidant performance in ryegrass plants. Front. Plant Sci. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Kopittke, P.M.; Gianoncelli, A.; Kourousias, G.; Green, K.; McKenna, B.A. Alleviation of Al toxicity by Si is associated with the formation of Al–Si complexes in root tissues of sorghum. Front. Plant Sci. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Cocker, K.M.; Evans, D.E.; Hodson, M.J. The amelioration of aluminium toxicity by silicon in wheat (Triticum aestivum L.): Malate exudation as evidence for an in planta mechanism. Planta 1998, 204, 318–323. [Google Scholar] [CrossRef]
- Yan, L.; Riaz, M.; Wu, X.; Du, C.; Liu, Y.; Lv, B.; Jiang, C. Boron inhibits aluminum-induced toxicity to citrus by stimulating antioxidant enzyme activity. J. Environ. Sci. Health Part C 2018, 36, 145–163. [Google Scholar] [CrossRef]
- Riaz, M.; Yan, L.; Wu, X.; Hussain, S.; Aziz, O.; Wang, Y.; Imran, M.; Jiang, C. Boron alleviates the aluminum toxicity in trifoliate orange by regulating antioxidant defense system and reducing root cell injury. J. Environ. Manag. 2018, 208, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.; Yan, L.; Wu, X.; Hussain, S.; Aziz, O.; Imran, M.; Rana, M.S.; Jiang, C. Boron reduces aluminum-induced growth inhibition, oxidative damage and alterations in the cell wall components in the roots of trifoliate orange. Ecotoxicol. Environ. Saf. 2018, 153, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Stass, A.; Kotur, Z.; Horst, W.J. Effect of boron on the expression of aluminium toxicity in Phaseolus vulgaris. Physiol. Plant. 2007, 131, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Shen, R.; Xiao, H.; Xu, M.; Wang, H.; Wang, H.; Zeng, Q.; Bian, J. Boron alleviates aluminum toxicity in pea (Pisum sativum). Plant Soil 2009, 314, 87–98. [Google Scholar] [CrossRef]
- Li, X.W.; Liu, J.Y.; Fang, J.; Tao, L.; Shen, R.F.; Li, Y.L.; Xiao, H.D.; Feng, Y.M.; Wen, H.X.; Guan, J.H.; et al. Boron supply enhances aluminum tolerance in root border cells of pea (Pisum sativum) by interacting with cell wall pectins. Front. Plant Sci. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Mai, J.; Tao, L.; Qu, M.; Liu, J.; Shen, R.; Xu, G.; Feng, Y.; Xiao, H.; et al. Boron alleviates aluminum toxicity by promoting root alkalization in transition zone via polar auxin transport. Plant Physiol. 2018, 177, 1254–1266. [Google Scholar] [CrossRef]
- Riaz, M.; Wu, X.; Yan, L.; Hussain, S.; Aziz, O.; Shah, A.; Jiang, C. Boron supply alleviates al-induced inhibition of root elongation and physiological characteristics in rapeseed (Brassica napus L.). J. Plant Interact. 2018, 13, 270–276. [Google Scholar] [CrossRef]
- Ruiz, J.M.; Rivero, R.M.; Romero, L. Boron increases synthesis of glutathione in sunflower plants subjected to aluminum stress. Plant Soil 2006, 279, 25–30. [Google Scholar] [CrossRef]
- Hossain, A.K.M.Z.; Hossain, M.A.; Koyama, H.; Hara, T. Effects of aluminum and boron supply on growth of seedlings among 15 cultivars of wheat (Triticum aestivum L.) grown in Bangladesh. Soil Sci. Plant Nutr. 2004, 50, 189–195. [Google Scholar] [CrossRef]
- Zhu, C.Q.; Cao, X.C.; Zhu, L.F.; Hu, W.J.; Hu, A.Y.; Abliz, B.; Bai, Z.G.; Huang, J.; Liang, Q.D.; Sajid, H.; et al. Boron reduces cell wall aluminum content in rice (Oryza sativa) roots by decreasing H2O2 accumulation. Plant Physiol. Biochem. 2019, 138, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Riaz, M.; Liu, J.; Liu, Y.; Zeng, Y.; Jiang, C. Boron reduces aluminum deposition in alkali-soluble pectin and cytoplasm to release aluminum toxicity. J. Hazard. Mater. 2020, 401, 123388. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhou, W.; Liang, X.; Zhou, K.; Lin, X. Increased bound putrescine accumulation contributes to the maintenance of antioxidant enzymes and higher aluminum tolerance in wheat. Environ. Pollut. 2019, 252, 941–949. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, W.; Zhou, K.; Liu, W.; Liang, X.; Chen, Y.; Sun, D.; Lin, X. Polyamines modulate aluminum-induced oxidative stress differently by inducing or reducing H2O2 production in wheat. Chemosphere 2018, 212, 645–653. [Google Scholar] [CrossRef]
- Yu, Y.; Jin, C.; Sun, C.; Wang, J.; Ye, Y.; Zhou, W.; Lu, L.; Lin, X. Inhibition of ethylene production by putrescine alleviates aluminium-induced root inhibition in wheat plants. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.Q.; Cao, X.C.; Bai, Z.G.; Zhu, L.F.; Hu, W.J.; Hu, A.Y.; Abliz, B.; Zhong, C.; Liang, Q.D.; Huang, J.; et al. Putrescine alleviates aluminum toxicity in rice (Oryza sativa) by reducing cell wall Al contents in an ethylene-dependent manner. Physiol. Plant. 2019, 167, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Hou, J.; Gao, W.; Tong, X.; Li, M.; Chu, X.; Chen, G. Exogenous spermidine alleviates the adverse effects of aluminum toxicity on photosystem II through improved antioxidant system and endogenous polyamine contents. Ecotoxicol. Environ. Saf. 2020, 207, 111265. [Google Scholar] [CrossRef] [PubMed]
- Nahar, K.; Hasanuzzaman, M.; Suzuki, T.; Fujita, M. Polyamines-induced aluminum tolerance in mung bean: A study on antioxidant defense and methylglyoxal detoxification systems. Ecotoxicology 2017, 26, 58–73. [Google Scholar] [CrossRef]
- Qu, X.; Zhou, J.; Masabni, J.; Yuan, J. Phosphorus relieves aluminum toxicity in oil tea seedlings by regulating the metabolic profiling in the roots. Plant Physiol. Biochem. 2020, 152, 12–22. [Google Scholar] [CrossRef]
- Yao, H.; Zhang, S.; Zhou, W.; Liu, Y.; Liu, Y.; Wu, Y. The effects of exogenous malic acid in relieving aluminum toxicity in Pinus massoniana. Int. J. Phytoremediation 2020, 22, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Shetty, R.; Vidya, C.S.N.; Prakash, N.B.; Lux, A.; Vaculík, M. Aluminum toxicity in plants and its possible mitigation in acid soils by biochar: A review. Sci. Total Environ. 2021, 765, 142744. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zhang, L.; Riaz, M.; Zhang, M.; Xia, H.; Lv, B.; Jiang, C. Assessing the potential of biochar and aged biochar to alleviate aluminum toxicity in an acid soil for achieving cabbage productivity. Ecotoxicol. Environ. Saf. 2018, 161, 290–295. [Google Scholar] [CrossRef]
- Xia, H.; Riaz, M.; Zhang, M.; Liu, B.; El-Desouki, Z.; Jiang, C. Biochar increases nitrogen use efficiency of maize by relieving aluminum toxicity and improving soil quality in acidic soil. Ecotoxicol. Environ. Saf. 2020, 196, 110531. [Google Scholar] [CrossRef]
- Meena, V.S.; Meena, S.K.; Verma, J.P.; Kumar, A.; Aeron, A.; Mishra, P.K.; Bisht, J.K.; Pattanayak, A.; Naveed, M.; Dotaniya, M.L. Plant beneficial rhizospheric microorganism (PBRM) strategies to improve nutrients use efficiency: A review. Ecol. Eng. 2017, 107, 8–32. [Google Scholar] [CrossRef]
- Shailendra Singh, G.G. Plant growth promoting rhizobacteria (PGPR): Current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 2015, 07, 96–102. [Google Scholar] [CrossRef]
- Zerrouk, I.Z.; Benchabane, M.; Khelifi, L.; Yokawa, K.; Ludwig-Müller, J.; Baluska, F. A Pseudomonas strain isolated from date-palm rhizospheres improves root growth and promotes root formation in maize exposed to salt and aluminum stress. J. Plant Physiol. 2016, 191, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Zerrouk, I.Z.; Rahmoune, B.; Khelifi, L.; Mounir, K.; Baluska, F.; Ludwig-Müller, J. Algerian Sahara PGPR confers maize root tolerance to salt and aluminum toxicity via ACC deaminase and IAA. Acta Physiol. Plant. 2019, 41, 1–10. [Google Scholar] [CrossRef]
- Zerrouk, I.Z.; Rahmoune, B.; Auer, S.; Rößler, S.; Lin, T.; Baluska, F.; Dobrev, P.I.; Motyka, V.; Ludwig-Müller, J. Growth and aluminum tolerance of maize roots mediated by auxin- and cytokinin-producing Bacillus toyonensis requires polar auxin transport. Environ. Exp. Bot. 2020, 176, 104064. [Google Scholar] [CrossRef]
- Silambarasan, S.; Logeswari, P.; Cornejo, P.; Kannan, V.R. Role of plant growth–promoting rhizobacterial consortium in improving the Vigna radiata growth and alleviation of aluminum and drought stresses. Environ. Sci. Pollut. Res. 2019, 26, 27647–27659. [Google Scholar] [CrossRef]
- Silambarasan, S.; Logeswari, P.; Cornejo, P.; Abraham, J.; Valentine, A. Simultaneous mitigation of aluminum, salinity and drought stress in Lactuca sativa growth via formulated plant growth promoting Rhodotorula mucilaginosa CAM4. Ecotoxicol. Environ. Saf. 2019, 180, 63–72. [Google Scholar] [CrossRef]
- Panhwar, Q.A.; Naher, U.A.; Radziah, O.; Shamshuddin, J.; Razi, I.M. Eliminating aluminum toxicity in an acid sulfate soil for rice cultivation using plant growth promoting bacteria. Molecules 2015, 20, 3628–3646. [Google Scholar] [CrossRef]
- Farh, M.E.A.; Kim, Y.J.; Sukweenadhi, J.; Singh, P.; Yang, D.C. Aluminium resistant, plant growth promoting bacteria induce overexpression of aluminium stress related genes in Arabidopsis thaliana and increase the ginseng tolerance against aluminium stress. Microbiol. Res. 2017, 200, 45–52. [Google Scholar] [CrossRef]
- Mora, M.d.l.L.; Demanet, R.; Acuña, J.J.; Viscardi, S.; Jorquera, M.; Rengel, Z.; Durán, P. Aluminum-tolerant bacteria improve the plant growth and phosphorus content in ryegrass grown in a volcanic soil amended with cattle dung manure. Appl. Soil Ecol. 2017, 115, 19–26. [Google Scholar] [CrossRef]
- Kang, J.P.; Huo, Y.; Yang, D.U.; Yang, D.C. Influence of the plant growth promoting rhizobium panacihumi on aluminum resistance in Panax ginseng. J. Ginseng Res. 2020, 1–8. [Google Scholar] [CrossRef]
- Anwar, A.; Kim, J.K. Transgenic breeding approaches for improving abiotic stress tolerance: Recent progress and future perspectives. Int. J. Mol. Sci. 2020, 21, 2695. [Google Scholar] [CrossRef]
- Wang, H.; Ji, F.; Zhang, Y.; Hou, J.; Liu, W.; Huang, J.; Liang, W. Interactions between hydrogen sulphide and nitric oxide regulate two soybean citrate transporters during the alleviation of aluminium toxicity. Plant Cell Environ. 2019, 42, 2340–2356. [Google Scholar] [CrossRef]
- Yokosho, K.; Yamaji, N.; Mitani-Ueno, N.; Shen, R.F.; Ma, J.F. An aluminum-inducible IREG gene is required for internal detoxification of aluminum in buckwheat. Plant Cell Physiol. 2016, 57, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Chen, Z.; Ma, J.F. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012, 69, 857–867. [Google Scholar] [CrossRef]
- Xia, J.; Yamaji, N.; Che, J.; Shen, R.F.; Ma, J.F. Differential expression of Nrat1 is responsible for Al-tolerance QTL on chromosome 2 in rice. J. Exp. Bot. 2014, 65, 4297–4304. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wang, Z.; Fu, S.; Yang, G.; Shi, M.; Lu, Y.; Wang, X.; Xia, J. Functional characterization of the SbNrat1 gene in sorghum. Plant Sci. 2017, 262, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Chi, Y.; Jiang, Z.; Xu, Y.; Xie, L.; Huang, F.; Wan, D.; Ni, J.; Yuan, F.; Wu, X.; et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 2020, 578, 577–581. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef]
| Genes | Description | Plant Species | Functions | References |
|---|---|---|---|---|
| AhFRDL1 | Ferric reductase defective-like 1 protein | Arachis hypogaea | Transport citrate | [28] |
| AtPrx64 | Class III peroxidase | Arabidopsis thaliana | Peroxidase | [29] |
| AvSAMS1 | S-adenosyl methionine synthetase | Andropogon virginicus | Alterations of methylation status | [30] |
| BoALMT1 | Aluminum induced malate transporter | Brassica oleracea | Transport malate | [31] |
| BdMATE | Multidrug and toxic compound extrusion | Brachypodium distachyon | Transport citrate | [32] |
| GmMATE75 | Glycine max | [33] | ||
| GmMATE79 | Glycine max | [33] | ||
| GmMATE87 | Glycine max | [33] | ||
| GsMATE | Glycine soja | [34] | ||
| PtrMATE1 | Populus tomentosa | [35] | ||
| PtrMATE2 | Populus tomentosa | [35] | ||
| TaMATE1B | Triticum aestivum | [36] | ||
| FeSTAR1 | Half-type ABC transporter | Fagopyrum esculentum | Affect cell wall hemicellulose metabolism | [37] |
| GmGRPL | Glycine-rich protein-like protein | Glycine max | Regulating the level of indole-3-acetic acid (IAA) and ethylene | [38] |
| GmIREG3 | Iron regulated/ferroportin | Glycine max | Sequestrating Al into the vacuoles | [39] |
| GmME1 | NADP-malic enzyme | Glycine max | NADP-malic enzyme activity | [40] |
| HtNHX1 | Sodium (potassium)/proton antiporters | Helianthus tuberosus | Na+/H+ antiporter | [41] |
| HtNHX2 | Helianthus tuberosus | [41] | ||
| HvABCB25 | ATP binding cassette transporters | Hordeum vulgare | Vacuolar Al sequestration | [42] |
| MsCS | Citrate synthase | Medicago sativa | Citrate synthesis | [43] |
| MsPG | Polygalacturonase | Medicago sativa | Decreasing Al accumulation and increasing porosity and extensibility of cell walls | [44] |
| NtSUT1 | Sucrose transporter | Nicotiana tabacum | Sucrose uptake | [45] |
| OsAUX3 | Auxin carrier | Oryza sativa | Auxin influx carrier | [46] |
| VuAAE3 | Acyl activating enzyme | Vigna umbellata | Oxalyl-CoA synthetase | [47] |
| VuFDH | Formate dehydrogenase | Vigna umbellata | Catalyze the oxidation of formate | [48] |
| ZjOMT | Methyltransferase | Zoysia japonica | Melatonin synthesis | [49] |
| ZmAT6 | Aluminum tolerance protein | Zea mays | Scavenging reactive oxygen species | [50] |
| ZmPGP1 | P-glycoprotein | Zea mays | Auxin efflux carrier | [51] |
| TFs | Categories | Plant Species | Functions | References |
|---|---|---|---|---|
| AtHB7 | HD-Zip I transcription factor | Arabidopsis thaliana | Regulate the capacity of the cell wall to bind Al | [124] |
| AtHB12 | Arabidopsis thaliana | [124] | ||
| AtWRKY47 | WRKY transcription factor | Arabidopsis thaliana | Regulating genes responsible for cell wall modification | [125] |
| OsWRKY22 | Oryza sativa | Activation of OsFRDL4 expression and enhancement of citrate secretion | [126] | |
| CcSTOP1 | C2H2-type zinc finger transcription factor | Cajanus cajan | Regulate genes for OA transporters Regulate GhMATE and GhALMT1 expression Regulate the downstream Al or low pH resistance genes | [127] |
| GhSTOP1 | Gossypium hirsutum L. | [128] | ||
| GmSTOP1a | Glycine max | [129] | ||
| GsGIS3 | Glycine soja | Regulating Al-tolerance-related genes | [130] | |
| HvATF1 | Hordeum vulgare L. | Regulating multiple downstream genes involved in Al resistance | [131] | |
| NtSTOP1 | Nicotiana tabacum | Activation of NtMATE expression | [132] | |
| OsART2 | Oryza sativa | Regulate at least four genes implicated in Al tolerance | [133] | |
| SbSTOP1 | Sorghum bicolor L. | Regulate SbMATE and SbSTAR2 expression | [134] | |
| GsMAS1 | MADS-box transcription factor | Glycine Soja | Accumulation of Al-activated citrate and malate | [135] |
| HvHOX9 | Homeobox-leucine zipper transcription factor | Hordeum vulgare L. | Regulate the capacity of the cell wall to bind Al | [136] |
| MdMYC2 | bHLH transcription factor | Malus domestica | Activation of ethylene biosynthesis | [137] |
| VuABI5 | Basic-leucine zipper transcription factor | Vigna umbellata | Regulate genes involved cell wall modification and osmoregulation | [91] |
| VuNAR1 | NAC-type transcription factor | Vigna umbellata | Regulate cell wall pectin metabolism | [138] |
| Strains | Function | Target Plant | Effect of Aluminum Imposed on Plant | References |
|---|---|---|---|---|
| Bacillus megaterium CAM12 | Produce siderophore | Vigna radiata | Reduced Al uptake | [195] |
| Bacillus sp. PSB16 | Produce OAs and polysaccharides | Oryza sativa | Chelated the Al, increased solution pH, and enhanced rice growth | [197] |
| Bacillus toyonensis Bt04 | Produce auxin and cytokinin | Zea mays | Reduced Al accumulation in the young maize roots, promotes maize growth, and enhances root development | [194] |
| Burkholderia ginsengiterrae N11–2 | Produce auxins and siderophores and phosphate solubilization | Arabidopsis thaliana and Panax ginseng | Showed the higher expression level of Al-stress related genes and higher biomass and higher chlorophyll content | [198] |
| Burkholderia seminalis ASB21 | Produce OAs and polysaccharides | Oryza sativa | Chelated the Al, increased solution pH, and enhanced rice growth | [197] |
| Burkholderia thailandensis ASB7 | Produce OAs and polysaccharides | Oryza sativa | Chelated the Al, increased solution pH, and enhanced rice growth | [197] |
| Chryseobacterium polytrichastri N10 | Produce auxins and siderophores and phosphate solubilization | Arabidopsis thaliana and Panax ginseng | Showed the higher expression level of Al-stress related genes and higher biomass and higher chlorophyll content | [198] |
| Enterobacter sp. RJAL6 | Secrete oxalate, citrate, succinate and siderophores | Lolium perenne | Promote ryegrass growth by forming Al-siderophore complexes | [199] |
| Klebsiella sp. RC3 | Secrete oxalate, malate, citrate, succinate and siderophores | Lolium perenne | Promote ryegrass growth by forming Al-siderophore complexes | [199] |
| Klebsiella sp. RCJ4 | Secrete malate, citrate, succinate and siderophores | Lolium perenne | Promote ryegrass growth by forming Al-siderophore complexes | [199] |
| Pantoea agglomerans CAH6 | Produce siderophore | Vigna radiata | Reduced Al uptake | [195] |
| Pseudomonas fragi N8 | Produce auxins and siderophores and phosphate solubilization | Arabidopsis thaliana and Panax ginseng | Showed the higher expression level of Al-stress related genes and higher biomass and higher chlorophyll content | [198] |
| Pseudomonas plecoglossicida Pp20 | Produce ACC deaminase and IAA | Zea mays | Increased in lengths of seminal roots and root dry mass | [193] |
| Pseudomonas simiae N3 | Produce auxins and siderophores and phosphate solubilization | Arabidopsis thaliana and Panax ginseng | Showed the higher expression level of Al-stress related genes and higher biomass and higher chlorophyll content | [198] |
| Stenotrophomonas maltophilla Sb16 | Produce OAs and polysaccharides | Oryza sativa | Chelated the Al, increased solution pH, and enhanced rice growth | [197] |
| Pseudomonas fluorescens 002 | Releases IAA | Zea mays | increased primary, lateral, and seminal root lengths and numbers, as well as root dry mass | [182] |
| Rhodotorula mucilaginosa CAM4 | Bioaccumulation of Al | Lactuca sativa | Reduce proline and MDA contents, and enhance accumulation of antioxidant enzymes | [196] |
| Rhizobium panacihumi DCY116T | Produce IAA | Panax ginseng | Produced higher proline, phenolic, sugar contents and related gene expressions to induce ROS scavenging activity | [200] |
| Stenotrophomonas sp. RC5, | Secrete malate, citrate, succinate and siderophores | Lolium perenne | Promote ryegrass growth by forming Al-siderophore complexes | [199] |
| Serratia sp. RCJ6 | Secrete malate, citrate and succinate | Lolium perenne | Promote ryegrass growth by forming Al-siderophore complexes | [199] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Han, R.; Xie, Y.; Jiang, C.; Yu, Y. Recent Advances in Understanding Mechanisms of Plant Tolerance and Response to Aluminum Toxicity. Sustainability 2021, 13, 1782. https://doi.org/10.3390/su13041782
Wei Y, Han R, Xie Y, Jiang C, Yu Y. Recent Advances in Understanding Mechanisms of Plant Tolerance and Response to Aluminum Toxicity. Sustainability. 2021; 13(4):1782. https://doi.org/10.3390/su13041782
Chicago/Turabian StyleWei, Yunmin, Rongrong Han, Yonghong Xie, Caode Jiang, and Yongxiong Yu. 2021. "Recent Advances in Understanding Mechanisms of Plant Tolerance and Response to Aluminum Toxicity" Sustainability 13, no. 4: 1782. https://doi.org/10.3390/su13041782
APA StyleWei, Y., Han, R., Xie, Y., Jiang, C., & Yu, Y. (2021). Recent Advances in Understanding Mechanisms of Plant Tolerance and Response to Aluminum Toxicity. Sustainability, 13(4), 1782. https://doi.org/10.3390/su13041782






