Responses of Bacterial Community, Root-Soil Interaction and Tomato Yield to Different Practices in Subsurface Drip Irrigation
Abstract
1. Introduction
2. Materials and Methods
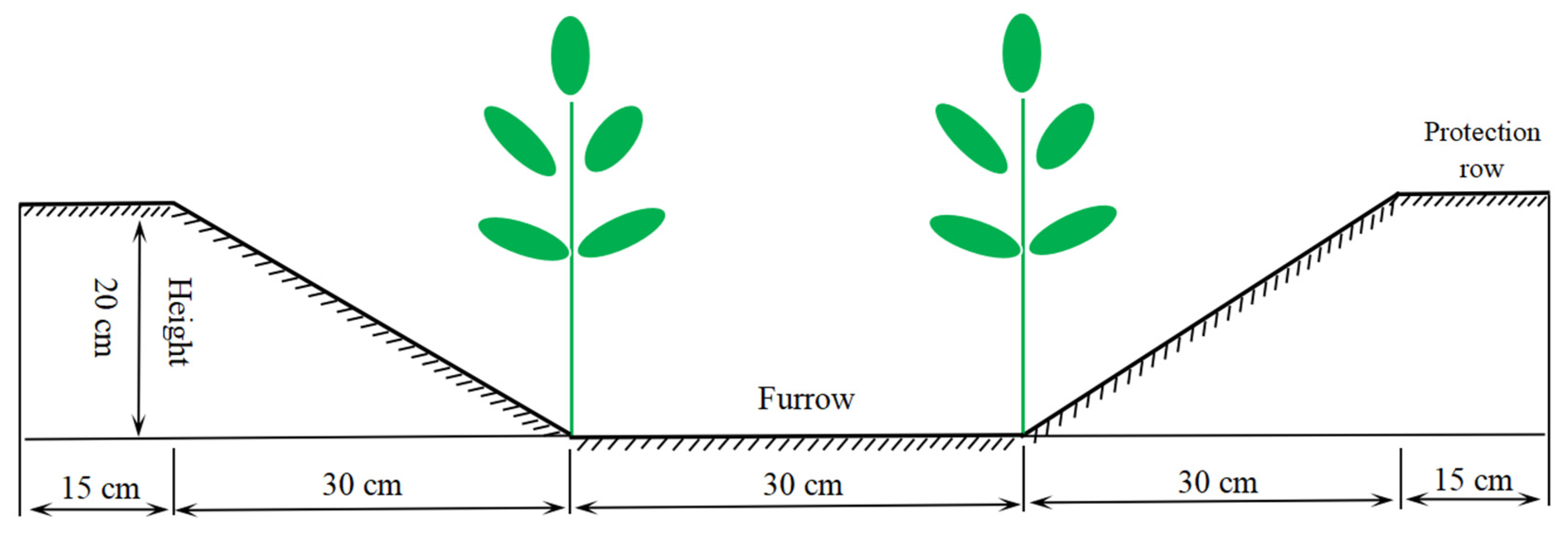
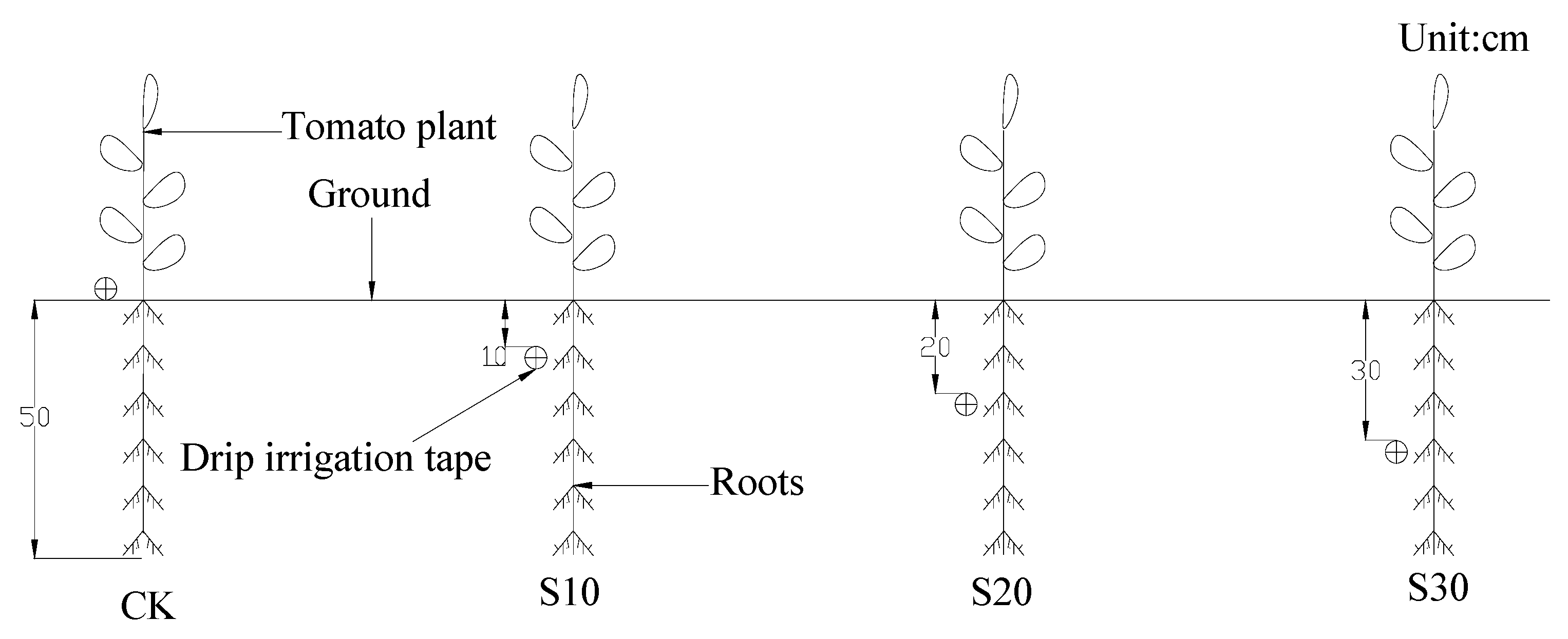
2.1. Experimental Design
2.2. Measurement of Response Variables
2.2.1. Sample Collection
2.2.2. Soil Bacterial Community Analysis
2.2.3. Tomato Roots
2.2.4. Soil Temperature, Porosity, Enzymes, and CO2 Flux
2.3. Data Processing
3. Results
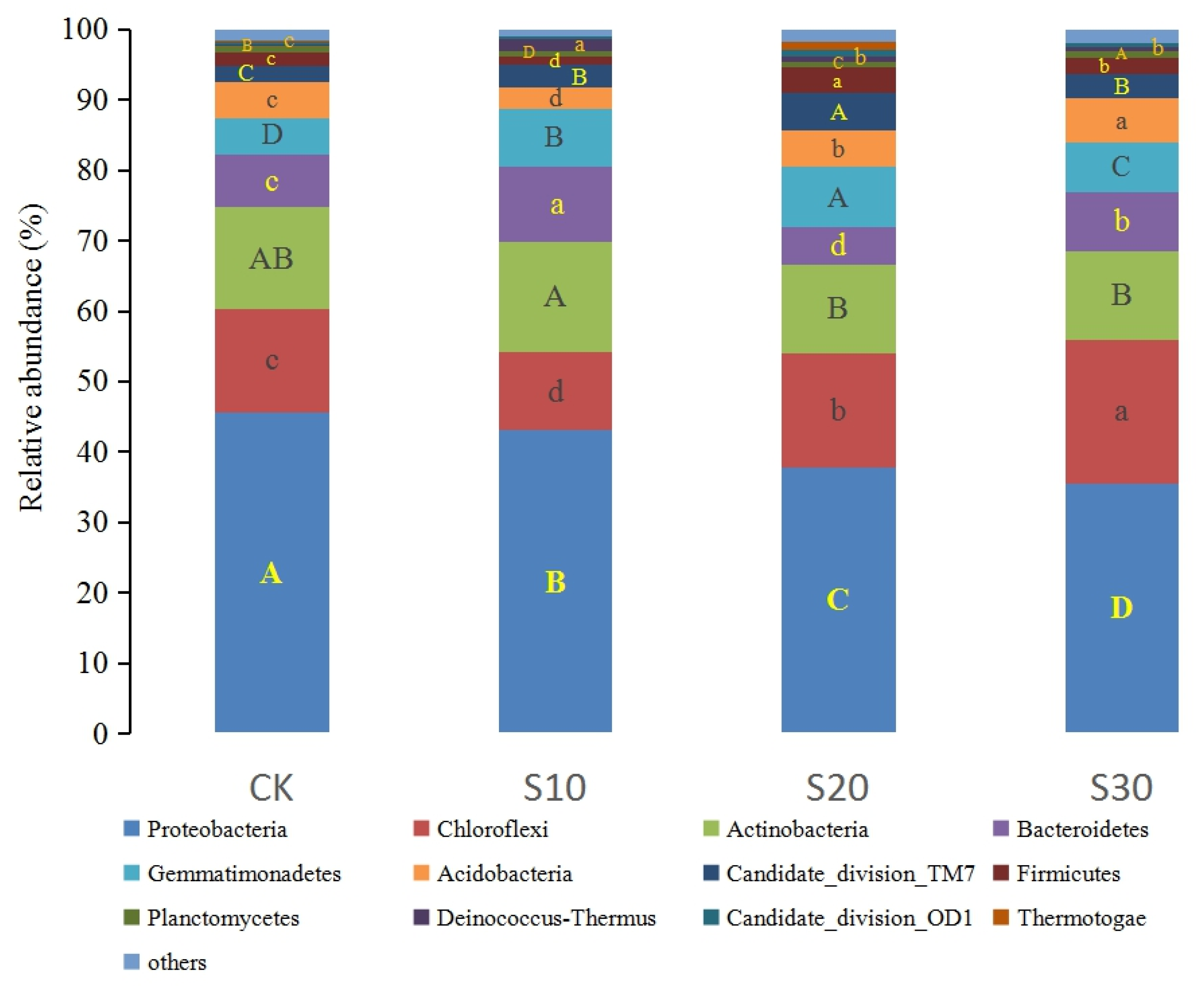
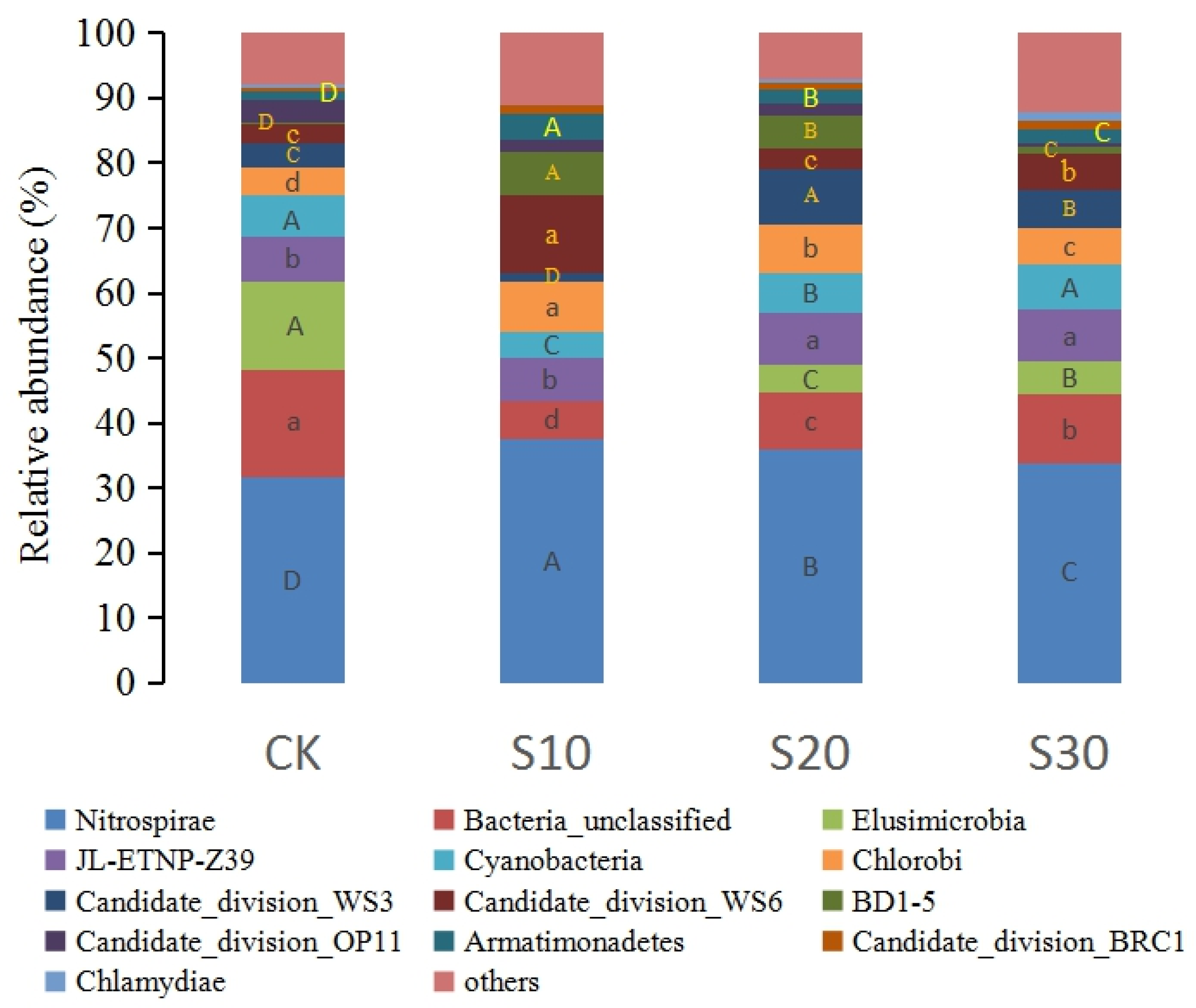
3.1. Soil Bacterial Sequence, Abundance, and Diversity of SDI
3.2. Soil Bacterial Community Composition in SDI
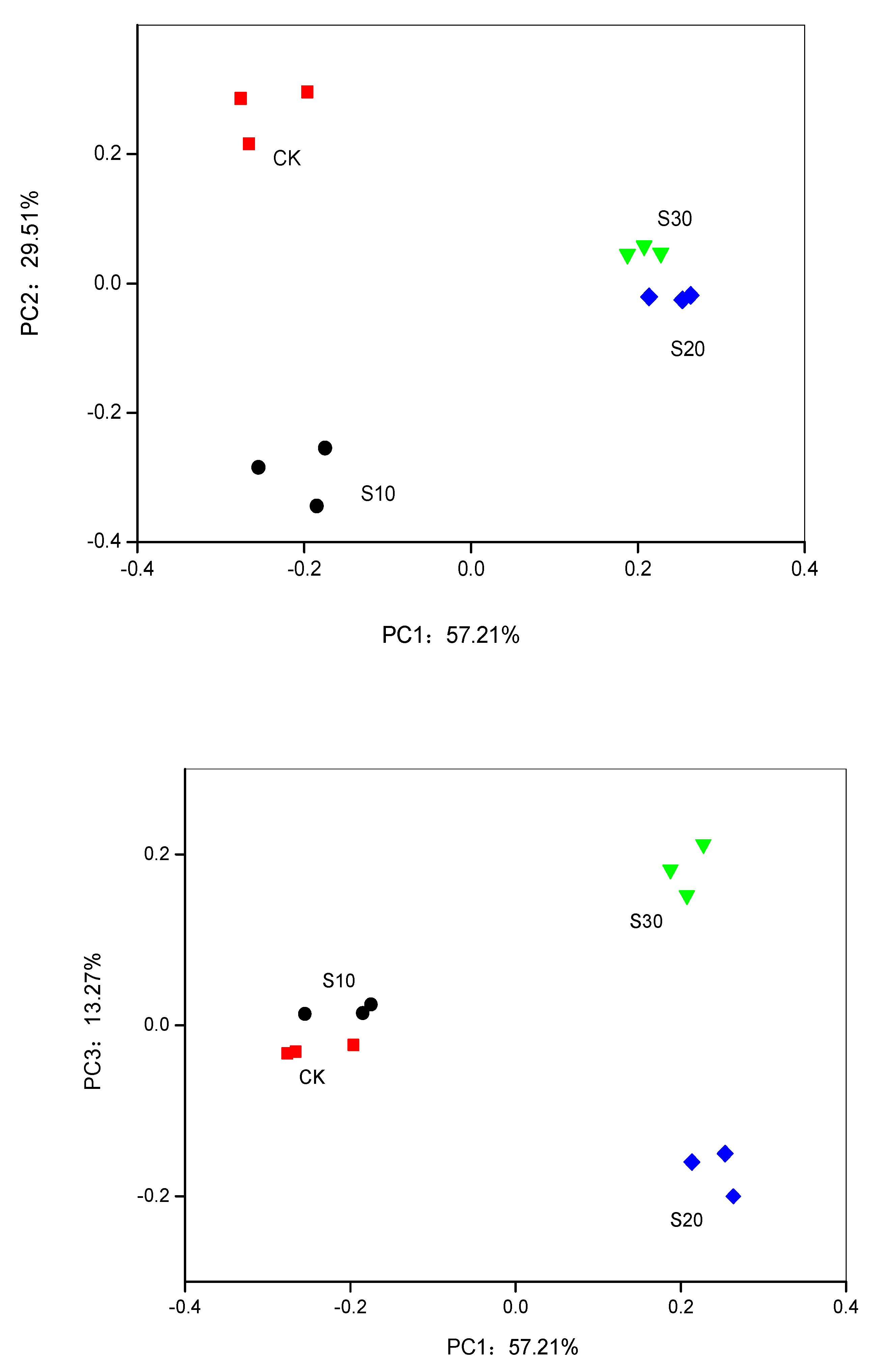
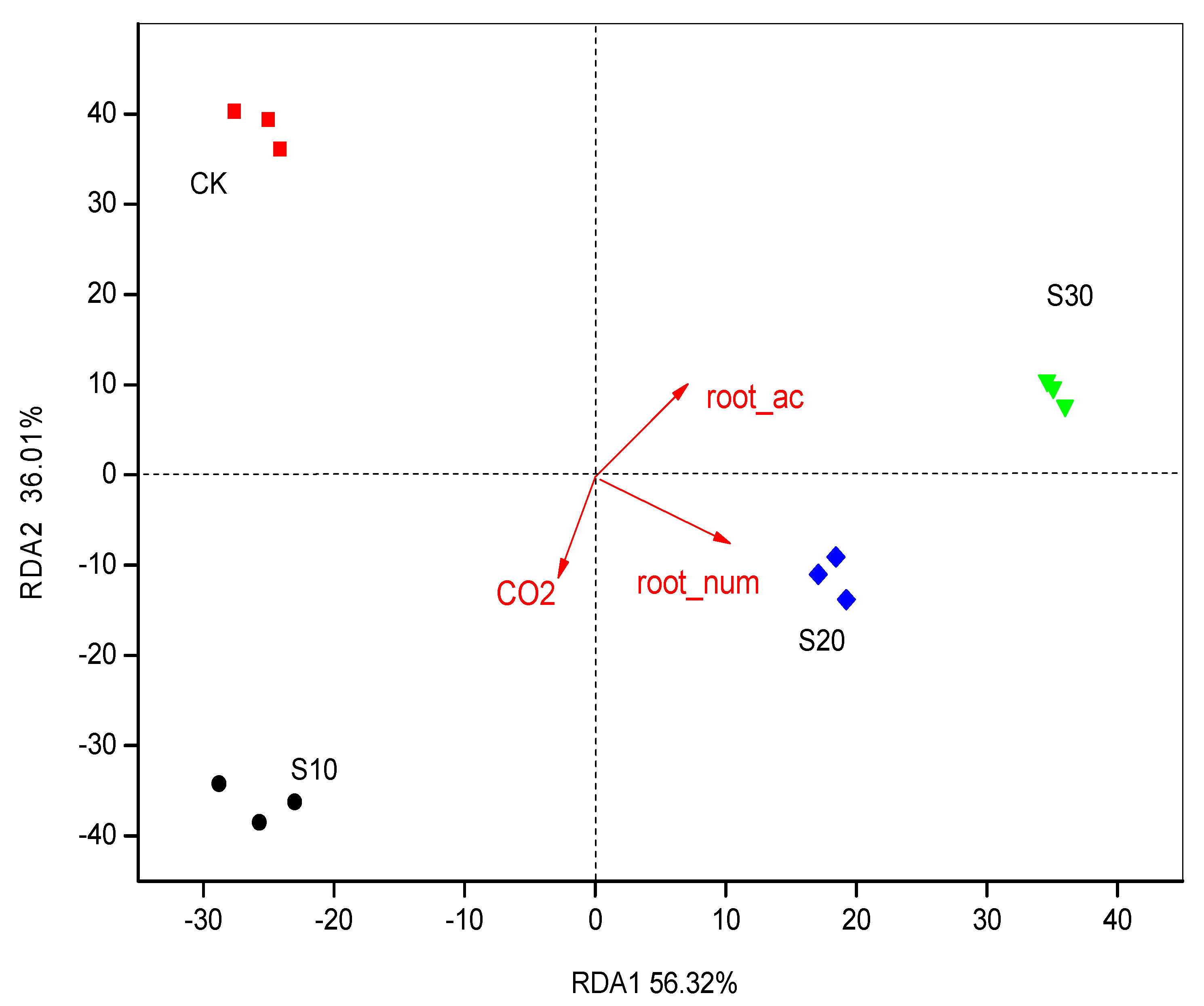
3.3. PCA and RDA of Soil Bacterial Community
3.4. Tomato Yield and Root Growth under SDI
3.5. The Abundance of Metabolism Based on PICRUSt and its Effect on Tomato Growh
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhong, W.H.; Gu, T.; Wei, W.; Zhang, B.; Lin, X.G.; Huang, Q.R.; Shen, W.H. The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant Soil 2010, 326, 511–522. [Google Scholar] [CrossRef]
- Zhalnina, K.; Dias, R.; de Quadros, P.D.; Davisrichardson, A.; Camargo, F.A.; Clark, I.M.; McGrath, S.P.; Hirsch, P.R.; Triplett, E.W. Soil pH determines microbial diversity and composition in the park grass experiment. Microb. Ecol. 2015, 69, 395–406. [Google Scholar] [CrossRef]
- Heijden, M.G.A.V.D.; Bardgett, R.D.; Straalen, N.M.V. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Torsvik, V.; Ovreas, L. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Lu, R. Microzone soil science-a possible new branch of soil science. Acta Pedol. Sin. 1999, 36, 287–288. [Google Scholar]
- Camp, C.R. Subsurface drip irrigation: A review. Trans. Chin. Soc. Agric. Eng. 1998, 41, 1353–1367. [Google Scholar] [CrossRef]
- He, H.; Kang, S.; Cao, H. Effect of Latera depth on root and seedling growth and water use efficiency of winter wheat. Trans. Chin. Soc. Agric. Eng. 2001, 17, 31–33. [Google Scholar]
- Songmei, Z.; Feng, W.; Ji, W.; Wang, Z.H. Evaluation method of soil water distribution uniformity under conditions of field subsurface drip irrigation. Trans. Chin. Soc. Agric. Eng. 2009, 25, 51–57. [Google Scholar]
- Attia, A.; Rajan, N.; Ritchie, G.; Cui, S.; Ibrahim, A.; Hays, D.; Xue, Q.W.; Wilborn, J. Yield, quality, and spectral reflectance responses of cotton under subsurface drip irrigation. Agron. J. 2015, 107, 1355–1364. [Google Scholar] [CrossRef]
- Sharma, S.P.; Leskovar, D.I.; Crosby, K.M.; Volder, A.; Ibrahim, A.M.H. Root growth, yield, and fruit quality responses of reticulatus, and inodorus, melons (cucumis melo, L.) to deficit subsurface drip irrigation. Agric. Water Manag. 2014, 136, 75–85. [Google Scholar] [CrossRef]
- Egamberdiyeva, D. The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Appl. Soil Ecol. 2007, 36, 184–189. [Google Scholar] [CrossRef]
- Wang, J.W.; Niu, W.Q.; Zhang, M.Z.; Li, Y. Effect of alternate partial root-zone drip irrigation on soil bacterial communities and tomato yield. Appl. Soil Ecol. 2017, 119, 250–259. [Google Scholar] [CrossRef]
- Chen, S.N.; Shang, P.L.; Kang, P.L.; Du, M.M. Metabolic Functional Community Diversity of Associated Bacteria during the Degradation of Phytoplankton from a Drinking Water Reservoir. Int. J. Environ. Res. Public Health 2020, 17, 1687. [Google Scholar] [CrossRef]
- Tang, B.; Yin, C.; Wang, Y.; Sun, Y.; Liu, Q. Positive effects of night warming on physiology of coniferous trees in late growing season: Leaf and root. Acta Oecologica 2016, 73, 21–30. [Google Scholar] [CrossRef]
- Franzluebbers, A.J.; Hons, F.M.; Zuberer, D.A. Tillage and crop effects on seasonal dynamics of soil CO2 evolution, water content, temperature, and bulk density. Appl. Soil Ecol. 1995, 2, 95–109. [Google Scholar] [CrossRef]
- Yan, M.M.; Chen, S.N.; Huang, T.L.; Li, B.Q.; Li, N.; Liu, K.W.; Zong, R.R.; Miao, Y.T.; Huang, X. Community Compositions of Phytoplankton and Eukaryotes during the Mixing Periods of a Drinking Water Reservoir: Dynamics and Interactions. Int. J. Environ. Res. Public Health 2020, 17, 1128. [Google Scholar] [CrossRef]
- Jiang, X.T.; Peng, X.; Deng, G.H.; Sheng, H.F.; Wang, Y.; Zhou, H.W.; Tam, F.Y. Illumina sequencing of 16s rrna tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb. Ecol. 2013, 66, 96–104. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.F.; He, Y.; Wu, J.Y.; Jiang, Y.X.; Tam, N.F.Y.; Zhou, H.W. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 2012, 78, 8264–8827. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ. Microb. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; Mcdonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Gao, J.; Teng, Y.; Feng, C.; Tian, M. Temporal variations in soil moisture for three typical vegetation types in inner mongolia, northern china. PLoS ONE 2015, 10, e0118964. [Google Scholar] [CrossRef] [PubMed]
- Haling, R.E.; Brown, L.K.; Bengough, A.G.; Young, I.M.; Hallett, P.D.; White, P.J.; George, T.S. Root hairs improve root penetration, root–soil contact, and phosphorus acquisition in soils of different strength. J. Exp. Bot. 2013, 64, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Vivanco, J.M.; Shen, Q. The unseen rhizosphere root-soil-microbe interactions for crop production. Curr. Opin. Microbiol. 2017, 37, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.D.; Risgaard-Petersen, N.; Sørensen, J.; Brandt, K.K. Rapid and sensitive nitrosomonas europaea biosensor assay for quantification of bioavailable ammonium sensu strictu in soil. Environ. Sci. Technol. 2011, 45, 1048–1054. [Google Scholar]
- Ningthoujam, D.S.; Sanasam, S.; Tamreihao, K.; Nimaichand, S. Antagonistic activities of local actinomycete isolates against rice fungal pathogens. Afr. J. Microbiol. Res. 2009, 3, 737–742. [Google Scholar]
- Buckley, D.H.; Huangyutitham, V.; Nelson, T.A.; Rumberger, A.; Thies, J.E. Diversity of Planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl. Environ. Microbiol. 2006, 72, 4522–4531. [Google Scholar] [CrossRef]
- Ishii, S.; Ikeda, S.; Minamisawa, K.; Senoo, K. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ. 2011, 26, 282–292. [Google Scholar] [CrossRef]
- Bryant, D.A.; Liu, Z.; Li, T.; Zhao, F.; Costas, A.M.G.; Klatt, C.G.; Ward, N.U.F.; Overmann, J. Comparative and functional genomics of anoxygenic green bacteria from the taxa chlorobi, chloroflexi, and acidobacteria. Adv. Photosynth. Respir. 2012, 33, 47–102. [Google Scholar]
- Hanada, S.; Sekiguchi, Y. The Phylum Gemmatimonadetes. Prokaryotes 2014, 11, 677–681. [Google Scholar]
- Debruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Radosevich, M. Global biogeography and quantitative seasonal dynamics of gemmatimonadetes in soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar] [CrossRef] [PubMed]
- Eichorst, S.A.; Kuske, C.R.; Schmidt, T.M. Influence of plant polymers on the distribution and cultivation of Vbacteria in the phylum acidobacteria. Appl. Environ. Microbiol. 2011, 77, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Rappé, M.S.; Giovannoni, S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [PubMed]
- Mowlick, S.; Inoue, T.; Takehara, T.; Tonouchi, A.; Kaku, N.; Ueki, K.; Ueki, A. Usefulness of japanese-radish residue in biological soil disinfestation to suppress spinach wilt disease accompanying with proliferation of soil bacteria in the firmicutes. Crop Prot. 2014, 61, 64–73. [Google Scholar] [CrossRef]
- Lin, W.; Deng, A.; Wang, Z.; Li, Y.; Wen, T.; Wu, L.F.; Wu, M.; Pan, Y.G. Genomic insights into the uncultured genus ‘Candidatus Magnetobacterium’ in the phylum Nitrospirae. Isme J. 2015, 8, 2463–2477. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Klatt, C.G.; Ludwig, M.; Rusch, D.B.; Jensen, S.I.; Kühl, M.; Ward, D.M.; Bryant, D.A. ‘Candidatus Thermochlorobacter aerophilum’: An aerobic chlorophotoheterotrophic member of the phylum chlorobi defined by metagenomics and metatranscriptomics. Isme J. 2012, 6, 1869–1882. [Google Scholar] [CrossRef]
- Valverde, A.; Makhalanyane, T.P.; Seely, M.; Cowan, D.A. Cyanobacteria drive community composition and functionality in rock-soil interface communities. Mol. Ecol. 2015, 24, 812–821. [Google Scholar] [CrossRef]
- Lee, K.C.; Dunfield, P.F.; Morgan, X.C.; Crowe, M.A.; Houghton, K.M.; Vyssotski, M.; Ryan, J.L.J.; Lagutin, K.; McDonald, I.R.; Stott, M.B. Chthonomonas calidirosea gen. nov. sp. nov. an aerobic, pigmented, thermophilic microorganism of a novel bacterial class, chthonomonadetes classis nov. of the newly described phylum armatimonadetes originally designated candidate division OP10. Int. J. Syst. Evol. Microbiol. 2011, 61, 2482–2490. [Google Scholar] [CrossRef]
| Treatments | DNA Sequence | OTU | Ace | Shannon Diversity | Coverage |
|---|---|---|---|---|---|
| CK | 19428 ± 301b | 1181 ± 17b | 1378 ± 21c | 5.96 ± 0.015a | 0.989 ± a |
| S10 | 14889 ± 284c | 1201 ± 23ab | 1387 ± 26bc | 6.03 ± 0.015a | 0.983 ± a |
| S20 | 26424 ± 304a | 1256 ± 14a | 1478 ± 17a | 6.13 ± 0.012a | 0.993 ± a |
| S30 | 27345 ± 279a | 1255 ± 12a | 1450 ± 14ab | 6.13 ± 0.0088a | 0.994 ± a |
| Treatments | Fruit Numbers per Strains | Single Fruit Weight (g) | Yield per Plant (kg) | Root Length (cm) | Root Forks |
|---|---|---|---|---|---|
| CK | 13.83 ± 0.35b | 164.24 ± 4.81a | 2.27 ± 0.05b | 1473.69 ± 10.17b | 3969 ± 91c |
| S10 | 13.57 ± 0.07b | 178.80 ± 11.28a | 2.43 ± 0.16b | 1365.85 ± 116.51b | 7348 ± 80b |
| S20 | 14.91 ± 0.42ab | 186.24 ± 1.83a | 2.78 ± 0.06a | 2110.57 ± 123.67a | 10978 ± 615a |
| S30 | 15.47 ± 0.74a | 176.11 ± 9.02a | 2.71 ± 0.03a | 2153.12 ± 24.39a | 8825 ± 49ab |
| Treatments | Energy Metabolism | Nitrogen Metabolism | Phosphonate and Phosphinate Metabolism |
|---|---|---|---|
| CK | 148,068 ± 326c | 128,564 ± 221c | 8646 ± 163c |
| S10 | 117,363 ± 640d | 100,414 ± 119d | 6826 ± 49d |
| S20 | 193,694 ± 121b | 167,050 ± 182b | 11,014 ± 129b |
| S30 | 210,976 ± 263 a | 180,157 ± 86a | 12,225 ± 189a |
| Energy Metabolism | Nitrogen Metabolism | Phosphonate and Phosphinate Metabolism | Root Length | Root Forks | |
|---|---|---|---|---|---|
| Yield perplant | 0.799 | 0.791 | 0.778 | 0.914 | 0.957 * |
| Single fruit weight | 0.309 | 0.3 | 0.274 | 0.518 | 0.947 |
| Fruit numbers | 0.980 * | 0.974 * | 0.980 * | 0.977 * | 0.667 |
| Root length | 0.974 * | 0.971 * | 0.964 * | 1 | 0.764 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Li, Y.; Niu, W. Responses of Bacterial Community, Root-Soil Interaction and Tomato Yield to Different Practices in Subsurface Drip Irrigation. Sustainability 2020, 12, 2338. https://doi.org/10.3390/su12062338
Wang J, Li Y, Niu W. Responses of Bacterial Community, Root-Soil Interaction and Tomato Yield to Different Practices in Subsurface Drip Irrigation. Sustainability. 2020; 12(6):2338. https://doi.org/10.3390/su12062338
Chicago/Turabian StyleWang, Jingwei, Yuan Li, and Wenquan Niu. 2020. "Responses of Bacterial Community, Root-Soil Interaction and Tomato Yield to Different Practices in Subsurface Drip Irrigation" Sustainability 12, no. 6: 2338. https://doi.org/10.3390/su12062338
APA StyleWang, J., Li, Y., & Niu, W. (2020). Responses of Bacterial Community, Root-Soil Interaction and Tomato Yield to Different Practices in Subsurface Drip Irrigation. Sustainability, 12(6), 2338. https://doi.org/10.3390/su12062338






