Exploring Rice Root Microbiome; The Variation, Specialization and Interaction of Bacteria and Fungi In Six Tropic Savanna Regions in Ghana
Abstract
:1. Introduction
2. Materials and Methods
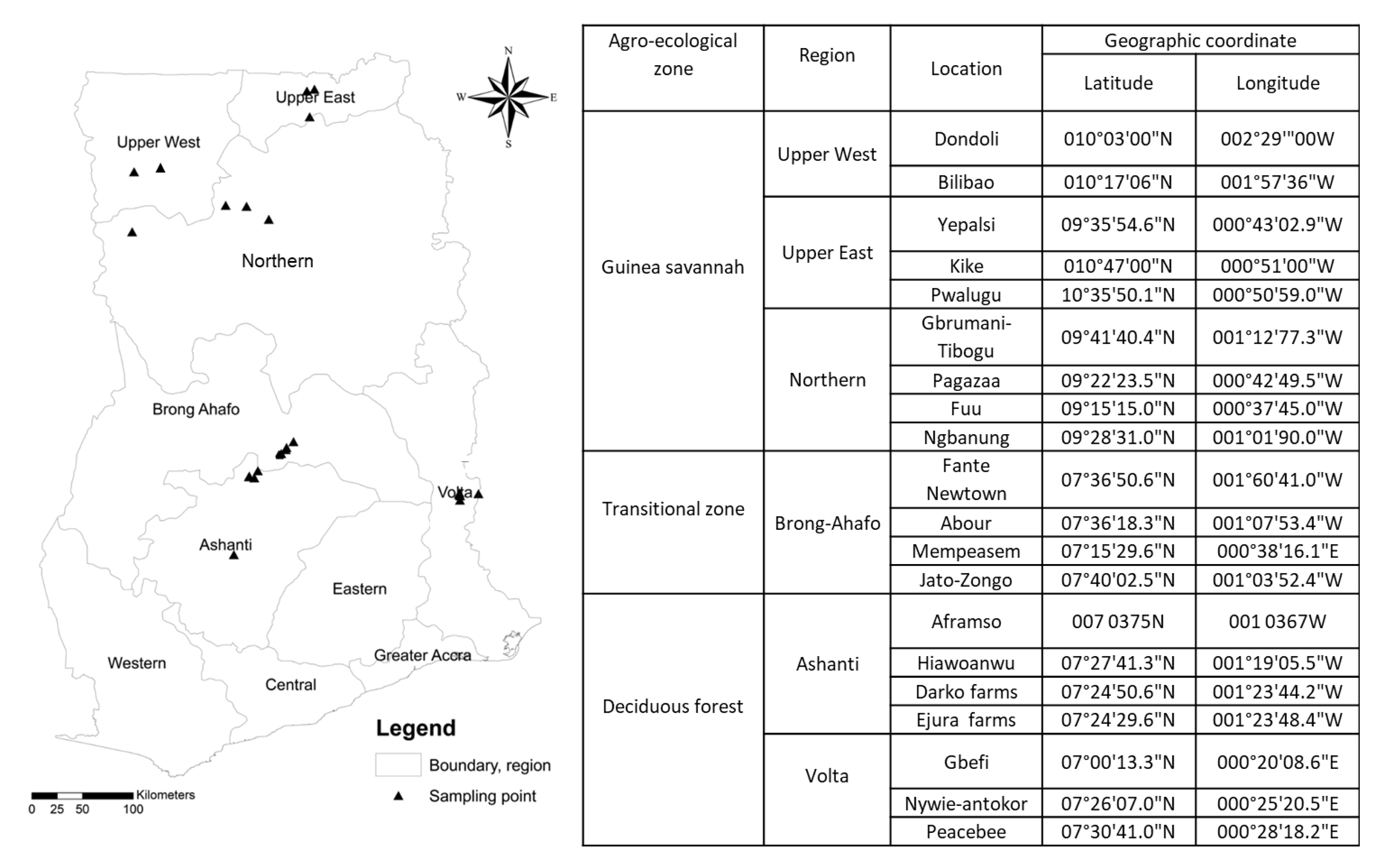
2.1. Study Site
2.2. Root and Soil Sampling
2.3. Soil Property Analysis
2.4. Root DNA Isolation and PCR
2.5. Statistical Analysis
3. Results
4. Discussion
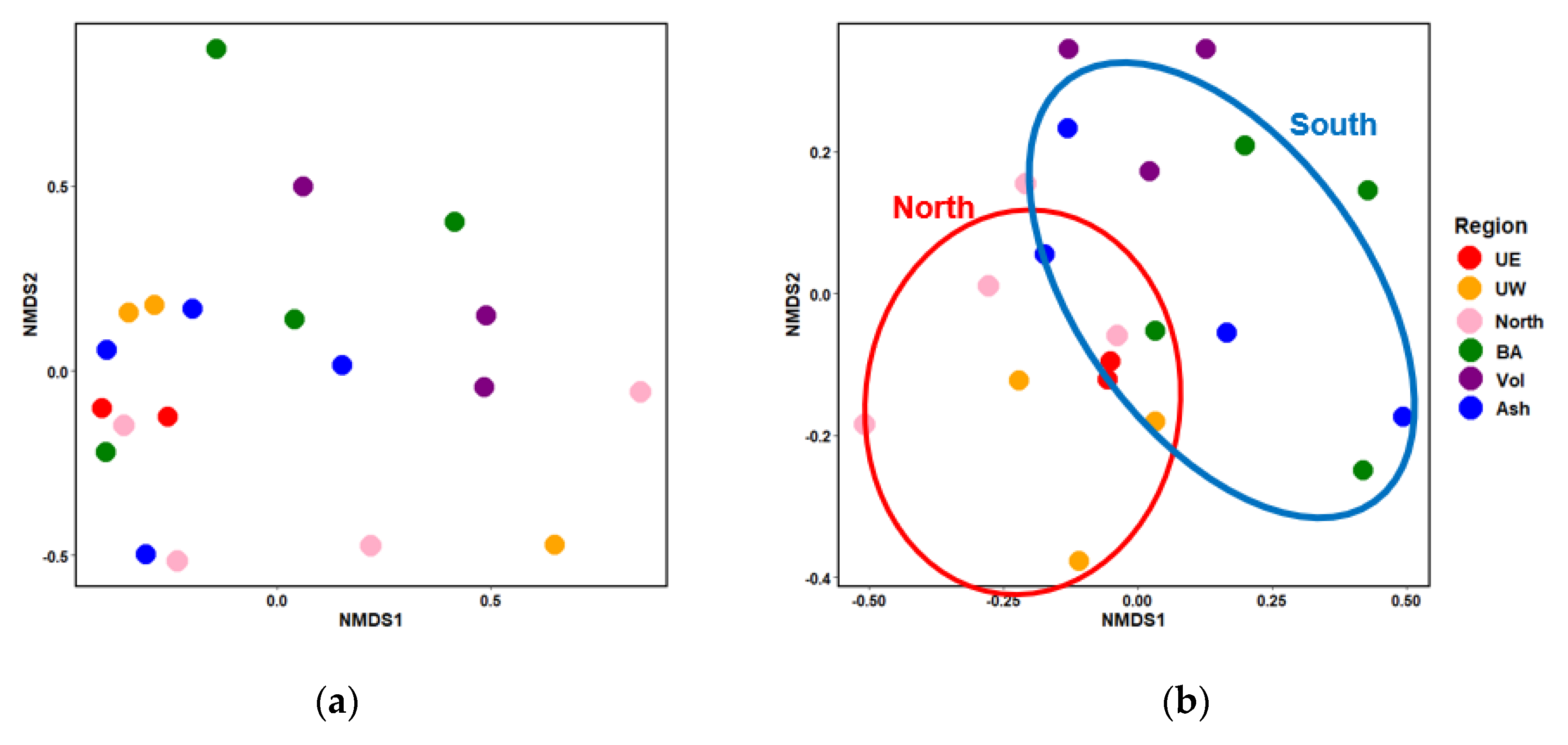
4.1. Variations in Rice Root Microbiome Composition and the Influence of Environmental Factors.
4.2. Abundantly Presenting Bacterial and Fungal Taxa and Their Potential Functions in the Root-Associated Microbiome in the Six Regions
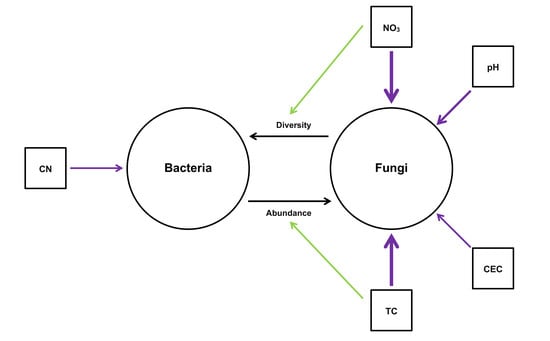
4.3. Potential Interactions Between Bacteria and Fungi in the Root Microbiome in Six Regions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Samir, K.; Luts, W. The human core of the shared socioeconomic pathways: Population scenarios by age, sex and level of education for all countries to 2100. Glob. Environ. Chang. 2014, 42, 181–192. [Google Scholar]
- Tietenberg, T.; Lewis, L. Environmental and Natural Resource Economics, 10th ed.; Routledge: New York, NY, USA, 2016. [Google Scholar]
- Crutzen, P.J.; Mosier, A.R.; Smith, K.A.; Winiwarter, W. N2O release from agro-biofuel production negates global warming reduction by replacing fossil fuels. Atmos Chem. Phys. 2008, 8, 389–395. [Google Scholar] [CrossRef] [Green Version]
- Cordell, D.; Drangert, J.O.; White, S. The story of phosphorus: Global food security and food for thought. Glob. Environ. Chang. 2009, 19, 292–305. [Google Scholar] [CrossRef]
- Raja, N. Biopesticides and Biofertilizers: Ecofriendly Sources for Sustainable Agriculture. J. Biofertil. Biopestic. 2013, 4, e112. [Google Scholar] [CrossRef] [Green Version]
- Archibald, D.; Ashitey, E. Gain Report: Ghana. U.S.D.A. Foreign Agricultural Service. 2018. Available online: https://www.fas.usda.gov/data/ghana-grain-and-feed-annual-0 (accessed on 21 May 2019).
- Buri, M.M.; Issaka, R.N.; Wakatsuki, T.; Kawano, N. Improving the productivity of lowland soils for rice cultivation in Ghana: The role of the ‘Sawah’ system. J. Soil Sci. 2012, 3, 56–62. [Google Scholar]
- Tkacz, A.; Poole, P. Role of root microbiota in plant productivity. J. Exp. Bot. 2015, 66, 2167–2175. [Google Scholar] [CrossRef] [Green Version]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, L. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- Barrow, J.R.; Lucero, M.E.; Reyes-Vera, I.; Havstad, K.M. Do symbiotic microbes have a role in regulating plant performance and response to stress? Commun. Integr. Biol. 2008, 1, 69–73. [Google Scholar] [CrossRef]
- Van Rhijn, R.; Vanderleyden, J. The Rhizobium-Plant Symbiosis. Microbiol. Rev. 1995, 59, 124–142. [Google Scholar] [CrossRef] [Green Version]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA 2013, 110, 17131–17136. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: London, UK, 2008. [Google Scholar]
- Powell, J.R.; Rillig, M.C. Biodiversity of arbuscular mycorrhizal fungi and ecosystem Function. New Phytol. 2018, 220, 1059–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiruma, K.; Gerlach, N.; Sacristán, S.; Nakano, R.T.; Hacquard, S.; Kracher, B.; Neumann, U.; Ramírez, D.; Bucher, M.; O’Connell, R.J.; et al. Root Endophyte Colletotrichum tofieldiae Confers Plant Fitness Benefits that Are Phosphate Status Dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saunders, M.; Glenn, A.E.; Kohn, L.M. Exploring the evolutionary ecology of fungal endophytes in agricultural systems: Using functional traits to reveal mechanisms in community processes. Evol. Appl. 2010, 3, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Dastogeer, K.M.G.; Wylie, S.J. Plant–Fungi Association: Role of fungal endophytes in improving plant tolerance to water stress. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Singapore, 2017; pp. 143–159. [Google Scholar]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adair, L.K.; Douglas, A.E. Making a microbiome: The many determinants of host-associated microbial community composition. Curr. Opin. Microbiol. 2017, 35, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Joseph, E.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; et al. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.; Jonson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Jonathan, A.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, 911–920. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, S.; Sasaki, K.; Okubo, T.; Yamashita, A.; Terasawa, K.; Bao, Z.; Liu, D.; Watanabe, T.; Murase, J.; Asakawa, S.; et al. Low Nitrogen Fertilization Adapts Rice Root Microbiome to Low Nutrient Environment by Changing Biogeochemical Functions. Microbes Environ. 2011, 29, 50–59. [Google Scholar] [CrossRef] [Green Version]
- Yaish, M.W.; Al-Lawati, A.; Jana, G.A.; Patankar, H.V.; Glick, B.R. Impact of Soil Salinity on the Structure of the Bacterial Endophytic Community Identified from the Roots of Caliph Medic (Medicago truncatula). PLoS ONE 2016, 11, e0159007. [Google Scholar] [CrossRef]
- Beck, S.; Powell, J.R.; Drigo, B.; Cairney, J.W.G.; Anderson, I.C. The role of stochasticity differs in the assembly of soil- and root-associated fungal communities. Soil Biol. Biochem. 2015, 80, 18–25. [Google Scholar] [CrossRef]
- Scherlach, K.; Graupner, K.; Hertweck, C. Molecular Bacteria-Fungi Interactions: Effects on Environment, Food, and Medicine. Annl. Rev. Microbial. 2013, 67, 375–397. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C.; Banba, M.; Croset, V.; An, K.; Miyao, A.; An, G.; Hirochika, H.; Imaizumi-Anraku, H.; Paszkowskia, U. Arbuscular Mycorrhiza–Specific Signaling in Rice Transcends the Common Symbiosis Signaling Pathway. Plant Cell 2009, 20, 2989–3005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Pan, Q.; Chen, F.; Yan, X.; Liao, H. Effects of co-inoculation with arbuscular mycorrhizal fungi and rhizobia on soybean growth as related to root architecture and availability of N and P. Mycorrhiza 2011, 21, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Bandara, W.M.M.S.; Seneviratne, G.; Kulasooriya, S.A. Interactions among endophytic bacteria and fungi: Effects and potentials. J. Biosci. 2006, 3, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Purahong, W.; Wubet, T.; Lentendu, G.; Schloter, M.; Pecyna, M.J.; Kapturska, D.; Hofrichter, M.; Kruger, D.; Buscot, F. Life in leaf litter: Novel insights into community dynamics of bacteria and fungi during litter decomposition. Moleculer Ecol. 2016, 25, 4059–4074. [Google Scholar] [CrossRef]
- De Menezes, A.B.; Prendergast-Miller, M.T.; Richardson, A.E.; Toscas, P.; Farrell, M.; Macdonald, L.M.; Baker, G.; Wark, T.; Thrall, P.H. Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Enviol Microbiol. 2015, 17, 2677–2689. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [Green Version]
- Pessoa-Filho, M.; Barreto, C.; dos Reis, C.C., Jr.; Fragoso, R.R.; Costa, F.S.; Mendes, L.D.C.; de Andrade, L.R.M. Microbiological functioning, diversity, and structure of bacterial communities in ultramafic soils from a tropical savanna. Antonie Van Leeuwenhoek 2015, 107, 935–949. [Google Scholar] [CrossRef]
- Lau, J.A.; Lennon, J.T. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl. Acad. Sci. USA 2012, 109, 14058–14062. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Cai, Y.; Li, X.; Christie, P.; Zhang, J.; Gai, J. Temperature-mediated phylogenetic assemblage of fungal communities and local adaptation in mycorrhizal symbioses. Enviol. Micol. Rep. 2019, 11, 215–226. [Google Scholar] [CrossRef]
- Sarkodee-Addo, E.; Yasuda, M.; Lee, C.G.; Kanasugi, M.; Fujii, Y.; Omari, R.A.; Abebrese, S.O.; Bam, R.; Asuming-Brempong, S.; Dastogeer, K.M.G.; et al. Arbuscular Mycorrhizal Fungi Associated with Rice (Oryza sativa L.) in Ghana: Effect of Regional Locations and Soil Factors on Diversity and Community Assembly. Agronomy 2020, 10, 559. [Google Scholar] [CrossRef]
- Aryee, J.N.A.; Amekudzi, L.K.; Quansah, E.; Klutse, N.A.B.; Atiah, W.A.; Yorke, C. Development of high spatial resolution rainfall data for Ghana. Int. J. Climatol. 2018, 38, 1201–1215. [Google Scholar] [CrossRef]
- Asare-Nuamah, P.; Botchway, E. Understanding climate variability and change: Analysis of temperature and rainfall across agroecological zones in Ghana. Heliyon 2019, 5, e02654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.M. Tropical Soil Biology and Fertility: A Handbook of Methods, 2nd ed.; Anderson, J.M., Ingram, J.S.I., Eds.; International Union of Biological Sciences: Paris, France; International Society of Soil Science: Vienna, Austria; CAB International: Wallingford, UK, 1993; ISBN 978-0-85198-821-4. [Google Scholar]
- Truog, E. The Determination of the Readily Available Phosphorus of Soils 1. Agron. J. 1930, 22, 874–882. [Google Scholar] [CrossRef] [Green Version]
- Parsons, T.R.; Maita, Y.; Lalli, C.M. Manual of Chemical and Biological Methods for Seawater Analysis; Pergamon Press: Oxford, UK, 1984; ISBN 978-0-08-030287-4. [Google Scholar]
- Schollenberger, C.J.; Simon, R.H. Determination of Exchange Capacity and Exchangeable bases in soil-Ammonium acetate method. Soil Sci. 1945, 59, 13–24. [Google Scholar] [CrossRef]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 2015, 1, e00009–e00015. [Google Scholar] [CrossRef] [Green Version]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Karina, E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal R.N.A. Genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. Available online: https://www.r-project.org/ (accessed on 1 October 2019).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Package ‘Vegan’: Community Ecology Package. R Package Version 2.5-6. Available online: https://cran.r-project.org/package=vegan (accessed on 1 October 2019).
- Hsieh, T.C.; Ma, K.H.; Chao, A. Package ‘iNEXT’ Interpolation and Extrapolation for Species Diversity. Version 2.0.19. 2019. Available online: https://cran.r-project.org/web/packages/iNEXT/index.html (accessed on 10 December 2019).
- Kassambara, A. Package ‘ggpubr’. ‘ggplot2’ Based Publication Ready Plots R package. R Package Version 0. 2. 4. Available online: https://cran.r-project.org/web/packages/ggpubr/ (accessed on 21 May 2019).
- De Caceres, M.; Jansen, F. Package ‘indicspecies’: Relationship Between Species and Groups of Sites. Version 1.7. 6. 2016. Available online: https://cran.r-project.org/web/packages/indicspecies/index.html (accessed on 29 October 2019).
- Hubbard, C.J.; Brock, M.T.; van Diepen, L.T.; Maignien, L.; Ewers, B.E.; Weinig, C. The plant circadian clock influences rhizosphere community structure and function. ISME J. 2018, 12, 400–410. [Google Scholar] [CrossRef] [Green Version]
- Faust, K.; Raes, J. CoNet app: Inference of biological association networks using Cytoscape. F1000Research 2016, 5, 1519. [Google Scholar] [CrossRef]
- Faust, K.; Sathirapongsasuti, S.; Izard, J.; Segata, N.; Gevers, D.; Raes, J.; Huttenhower, C. Microbial Co-occurrence Relationships in the Human Microbiome. PLoS Comput. Biol. 2012, 8, e1002606. [Google Scholar] [CrossRef]
- Mandakovic, D.; Rojas, C.; Maldonado, J. Structure and co-occurrence patterns in microbial communities under acute environmental stress reveal ecological factors fostering resilience. Sci Rep. 2018, 8, 5875. [Google Scholar] [CrossRef]
- Hodge, A.; Robinson, D.; Fitter, A. Are microorganisms more effective than plants at competing for nitrogen? Trends Plant Sci. 2000, 5, 304–308. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Mark, A.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Lan, Z.; Hu, S.; Bai, Y. Effects of nitrogen enrichment on belowground communities in grassland: Relative role of soil nitrogen availability vs. soil acidification. Soil Biol. Biochem. 2015, 89, 99–108. [Google Scholar] [CrossRef]
- Williams, A.; Manoharan, L.; Rosenstock, N.P.; Olsson, P.A.; Hedlund, K. Long-term agricultural fertilization alters arbuscular mycorrhizal fungal community composition and barley (Hordeum vulgare) mycorrhizal carbon and phosphorus exchange. New Phytol. 2017, 213, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; Hanson, C.A.; Treseder, K.K. Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol. Biochem. 2007, 39, 1878–1887. [Google Scholar] [CrossRef] [Green Version]
- Al-Hassan, R.; Poulton, C. Agriculture and Social Protection in Ghana. F.A.C. Working Paper 09; Future Agricultures Consortium: Brighton, UK, 2009. [Google Scholar]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.T.; Vitousek, P.M.; Zhang, F.S. Significant Acidification in Major Chinese Croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, R.; Redman, R. More than 400 million years of evolution and some plants still can’t make it on their own: Plant stress tolerance via fungal symbiosis. J. Exp. Bot. 2008, 59, 1109–1114. [Google Scholar] [CrossRef]
- Hartman, K.; Tringe, S.G. Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 2019, 476, 2705–2724. [Google Scholar] [CrossRef] [Green Version]
- Devictor, V.; Clavel, J.; Julliard, R. Defining and measuring ecological specialization. J. Appl. Ecol. 2010, 47, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Ayayee, P.A.; Barrantes, O.J.V.; Blackwood, C.B.; Royer, T.V.; Leff, L.G. Initial nitrogen enrichment conditions determines variations in nitrogen substrate utilization by heterotrophic bacterial isolates. BMC Microbiol. 2017, 17, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janda, J.M.; Abbott, S.L. The Genus Aeromonas: Taxonomy, Pathogenicity, and Infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunda, P.; Dhal, P.K.; Mukherjee, A. Endophytic bacterial community of rice (Oryza sativa L.) from coastal saline zone of West Bengal: 16S rRNA gene-based metagenomics approach. Meta Gene 2018, 18, 79–86. [Google Scholar] [CrossRef]
- Miyamoto, T.; Kawahara, M.; Minamisawa, K. Novel Endophytic Nitrogen-Fixing Clostridia from the Grass Miscanthus sinensis as Revealed by Terminal Restriction Fragment Length Polymorphism Analysis. Appl. Envirol. Microbiol. 2004, 70, 6580–6586. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.H.; Yokota, A. Pleomorphomonas oryzae gen. nov., sp. nov., a nitrogen-fixing bacterium isolated from paddy soil of Oryza sativa. Int. J. Syst. Evol. Microbiol. 2005, 55, 1233–1237. [Google Scholar] [CrossRef] [PubMed]
- Mhedbi-Hajri, N.; Jacques, M.A.; Koebnik, R. Adhesion Mechanisms of Plant-Pathogenic Xanthomonadaceae. Adv. Exp. Med. Biol. 2011, 715, 71–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Rangaraj, N.; Sonti, R.V. Multiple Adhesin-Like Functions of Xanthomonas oryzae pv. oryzae Are Involved in Promoting Leaf Attachment, Entry, and Virulence on Rice. Mol. Plant Microbe Interact. 2011, 22, 73–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naik, B.S.; Shashikala, J.; Krishnamurthy, Y.L. Study on the diversity of endophytic communities from rice (Oryza sativa L.) and their antagonistic activities in vitro. Mycol. Res. 2009, 164, 290–296. [Google Scholar] [CrossRef]
- Fatima, M.; Khan, I.; Qazi, M.A.; Shahzadi, I.; Mumtaz, A.; Hashmi, M.A.; Khan, A.K.; Ismail, T. Chaetomium endophytes: A repository of pharmacologically active metabolites. Acta Physiol. Plant. 2016, 38, 136. [Google Scholar] [CrossRef]
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Hyde, K.D. Pleosporales. Fungal Divers. 2011, 53, 1–221. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Castlebury, L.A.; Miller, A.N.; Huhndorf, S.M.; Schoch, C.L.; Seifert, K.A.; Rossman, A.Y.; Rogers, J.D.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; et al. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 2006, 98, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.G.; dos Reis, S.; Costa, M.A.F.; de Souza, C.G.M.; Peralta, R.M. Production of hydrolytic enzymes by the plant pathogenic fungus myrothecium verrucaria in submerged cultures. Braz. J. Microbiol. 2005, 36, 7–11. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.S.; Haque, M.S.; Islam, M.M.; Emdad, E.M.; Halim, A.; Hossen, Q.M.M.; Hossain, M.Z.; Ahmed, B.; Rahim, S.; Rahman, M.S.; et al. Tools to kill: Genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genom. 2012, 13, 493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Herv´e, V.; Labb´e, J.; Lastovetsky, O.A.; et al. Bacterial–fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [Green Version]
- Stopnisek, N.; Zühlke, D.; Carlier, A.; Barberán, A.; Fierer, N.; Becher, D.; Riedel, K.; Eberl, L.; Weisskopf, L. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 2016, 10, 253–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cugini, C.; Calfee, M.W.; Farrow, J.M.; Morales, D.K.; Pesci, E.C.; Hogan, D.A. Farnesol, a common sesquiterpene, inhibits P.Q.S. production in Pseudomonas aeruginosa. Mol. Microbiol. 2007, 65, 896–906. [Google Scholar] [CrossRef] [PubMed]
- De Boer, W. Upscaling of fungal-bacterial interactions: From the lab to the field. Curr. Opin. Microbiol. 2017, 37, 35–41. [Google Scholar] [CrossRef]
- Van Overbeek, L.S.; Saikkonen, K. Impact of Bacterial–Fungal Interactions on the Colonization of the Endosphere. Trends Plant Sci. 2016, 21, 230–242. [Google Scholar] [CrossRef]
- Singh, B.K.; Nunan, N.; Ridgway, K.P.; McNicol, J.; Young, J.P.W.; Daniell, T.J.; Prosser, J.I.; Millard, P. Relationship between assemblages of mycorrhizal fungi and bacteria on grass roots. Enviol. Microbiol. 2008, 10, 534–541. [Google Scholar] [CrossRef]
- Cheung, M.K.; Wong, C.K.; Chu, K.H.; Kwan, H.S. Community Structure, Dynamics and Interactions of Bacteria, Archaea and Fungi in Subtropical Coastal Wetland Sediments. Sci. Rep. 2018, 8, 14397. [Google Scholar] [CrossRef] [Green Version]




| Site | Region | pH | Moisture | NO3- | P | TC | TN | CN | C.E.C. |
|---|---|---|---|---|---|---|---|---|---|
| Don | UW | 4.70 ± 0.20b | 7.000 ± 0.655k | 25.77 ± 1.28i | 0.3603 ± 0.1107gh | 5.216 ± 0.058k | 0.2633 ± 0.0042k | 10.39 ± 0.01l | 5.466 ± 0.042n |
| Bib | UW | 5.10 ± 0.24ab | 13.14 ± 1.74i | 44.20 ± 1.14h | 0.2560 ± 0.0307gh | 6.084 ± 0.039k | 0.6703 ± 0.8110i | 11.69 ± 0.01h | 2.577 0.291m |
| Yep | UE | 5.00 ± 0.16ab | 13.22 ± 1.15e | 58.25 ± 0.35g | 0.1520 ± 0.0272h | 12.90 ± 0.01h | 2.097 ± 0.011e | 15.08 ± 0.01a | 14.76 ± 0.05h |
| Kike | UE | 5.10 ± 0.10ab | 11.66 ± 0.65h | 67.78 ± 1.07f | 0.5370 ± 0.0450g | 13.09 ± 0.03h | 1.424 ± 0.014h | 13.02 ± 0.01c | 14.56 ± 0.11h |
| Pw | UE | 4.80 ± 0.12b | 14.60 ± 1.21d | 13.27 ± 0.34j | 1.221 ± 0.121d | 12.08 ± 0.03h | 2.500 ± 0.101d | 12.37 ± 0.02f | 10.87 ± 0.06k |
| GT | NO | 4.70 ± 0.20b | 18.62 ± 0.69jk | 24.89 ± 0.57i | 2.146 ± 0.054b | 5.837 ± 0.029k | 0.3593 ±0.0149jk | 9.840 ± 0.021n | 9.917 0.121l |
| PAGA | NO | 5.10 ± 0.10ab | 8.440 ± 0.287jk | 24.22 ± 0.98i | 4.602 ± 0.090a | 8.833 ± 0.028j | 0.3510 ± 0.0045jk | 9.337 ± 0.026p | 12.45 ±0.09i |
| FUU | NO | 4.70 ± 0.20b | 15.40 ± 0.49gh | 26.52 ± 1.29i | 1.183 ± 0.033d | 10.84 ± 0.059i | 1.446 ± 0.003gh | 10.61 ± 0.02k | 11.73 ± 0.07j |
| Ngb2 | NO | 5.00 ± 0.10ab | 9.280 ± 0.591j | 44.79 ± 0.63h | 1.633 ± 0.036c | 7.789 ± 0.055j | 0.4733 ± 0.0244j | 9.517 ± 0.017° | 10.79 ± 0.08k |
| Aframso | Ash | 5.80 ± 0.16a | 15.58 ± 0.99h | 106.6 ± 2.0d | 1.017 ± 0.027de | 28.36 ± 0.04d | 1.374 ± 0.014h | 12.89 ± 0.02d | 22.43 ± 0.08d |
| Hia | Ash | 5.00 ± 0.16ab | 23.24 ± 1.89cd | 126.6 ± 0.7b | 0.2540 ± 0.146gh | 29.64 ± 0.03c | 2.638 ± 0.025cd | 10.85 ± 0.03j | 20.84 ± 0.09e |
| Dar | Ash | 5.00 ± 0.16ab | 10.82 ± 1.21f | 96.50 ± 2.32e | 0.7487 ± 0.0373ef | 24.84 ± 0.04e | 1.839 ± 0.025f | 10.17 ±0.03 m | 22.94 ± 0.03d |
| Ejura | Ash | 5.30 ± 0.3ab | 9.840 ± 0.52g | 148.8 ± 1.04a | 0.8963 ±0.0103de | 25.87 ±0.06e | 1.608 ± 0.037g | 11.81 ± 0.01h | 22.96 ± 0.04d |
| FanteN | BA | 5.10 ± 0.10ab | 17.22 ±1.02e | 129.5 ± 0.27b | 0.5313 ± 0.0218fg | 20.23 ± 0.04f | 2.191 ± 0.020e | 12.19 ± 0.02g | 15.54 ± 0.17g |
| Abour | BA | 5.50 ± 0.16ab | 6.784 ± 0.54c | 126.7 ± 1.87b | 0.5093 ± 0.0124fg | 17.69 ± 1.01g | 2.715 ± 0.043c | 12.88 ± 0.004d | 14.45 ± 0.03h |
| Mem | BA | 5.60 ± 0.10ab | 11.88 ± 0.91a | 146.9 ± 0.7a | 0.4500 ± 0.0271fgh | 25.93 ± 0.03e | 3.917 ± 0.010a | 12.66 ± 0.004e | 16.85 ± 0.03f |
| Jato | BA | 5.00 ± 0.16ab | 16.80 ± 2.71f | 149.0 ± 0.4a | 1.072 ± 0.042de | 24.86 ± 0.07e | 1.845 ± 0.029f | 11.63 ± 0.02i | 15.95 ± 0.03g |
| Gbe | Vol | 5.60 ± 0.24ab | 13.40 ± 1.13a | 126.9 ± 2.0b | 0.5037 ± 0.0088fg | 39.92 ± 0.02a | 3.918 ± 0.010a | 12.88 0.01d | 24.94 ± 0.01c |
| Ny1 | Vol | 5.10 ± 0.10ab | 18.76 ± 0.88b | 108.3 ± 0.8d | 0.4027 ± 0.0096gh | 38.44 ± 0.02b | 3.486 ± 0.028b | 13.97 ± 0.01b | 29.97 ± 0.09a |
| Pea2 | Vol | 5.60 ± 0.29ab | 19.60 ± 0.32e | 118.9 ± 0.8c | 0.5177 ± 0.0110fg | 38.31 ± 0.02b | 2.229 ± 0.034e | 12.47 ± 0.03f | 27.55 ± 0.05b |
| Kingdom | PermANOVA | ANOSIM | ||
|---|---|---|---|---|
| Pseudo-F | P | R | P | |
| Bacteria | 2.436 | <0.001 | 0.2114 | 0.0338 |
| Fungi | 1.589 | 0.0092 | −0.0228 | 0.5786 |
| Kingdom | Factor | Variable | NMDS1 | NMDS2 | r2 | P |
|---|---|---|---|---|---|---|
| Bacteria | abiotic | Moisture | 0.9773 | 0.2118 | 0.3112 | 0.0405 |
| TN | 0.2311 | 0.9729 | 0.2428 | 0.0915 | ||
| biotic | ReadFun | 0.7245 | −0.6893 | 0.4304 | 0.0078 | |
| ObsFun | 0.15572 | −0.9878 | 0.2625 | 0.0740 | ||
| ShnFun | −0.8061 | −0.5918 | 0.3374 | 0.0292 | ||
| SimFun | −0.7407 | −0.6718 | 0.3150 | 0.0404 | ||
| Fungi | abiotic | pH | 0.45549 | 0.8903 | 0.4114 | 0.0104 |
| NO3- | 0.7563 | 0.6543 | 0.5797 | <0.001 | ||
| TC | 0.3870 | 0.9221 | 0.5601 | <0.001 | ||
| TN | 0.6247 | 0.7809 | 0.2796 | 0.0629 | ||
| CEC | 0.38942 | 0.92106 | 0.3546 | 0.0226 | ||
| biotic | ReadBac | −0.08821 | −0.9961 | 0.3546 | 0.0251 |
| OTU.ID. | Probability | Indicator Value Index | P | Characteristic | Reference | Host | |
|---|---|---|---|---|---|---|---|
| A | B | ||||||
| Sporomusa_sp. OTU_B4896 | 0.9971 | 1.0000 | 0.999 | 0.001 | Free living or unknown | [54] | |
| Aeromonadaceae OTU_B18812 | 0.9760 | 1.0000 | 0.988 | 0.020 | Endophyte | [55] | rice |
| Clostridium sp. OTU_B44691 | 0.9752 | 1.0000 | 0.988 | 0.011 | Endophyte | [56] | maiden silver grass |
| Micromonosporaceae OTU_B6546 | 0.9312 | 1.0000 | 0.965 | 0.015 | Endophyte | [57] | Leguminous and actinorhizal plants |
| Pleomorphomonas sp. OTU_B24241 | 0.9698 | 0.8889 | 0.928 | 0.012 | Endophyte | [58] | rice |
| Achromobacter sp. OTU_B36942 | 0.9506 | 0.8889 | 0.919 | 0.046 | Endophyte | [59] | wheat |
| Xanthomonadaceae OTU_B14074 | 0.7998 | 1.0000 | 0.894 | 0.032 | Endophyte and Pathogen | [60] | rice and other plants |
| OTU.ID. | Probability | Indicator Value Index | P | Characteristic | Reference | Host | |
|---|---|---|---|---|---|---|---|
| A | B | ||||||
| Pleosporales OTU_4099 | 0.9971 | 1.0000 | 0.999 | 0.001 | Endophyte and Pathogen | [61] | rice and other plants |
| Curvularia_prasadii OTU_6231 | 0.9963 | 1.0000 | 0.998 | 0.001 | Pathogen | [62] | Achyranthes aspera |
| Pleosporales OTU_3272 | 0.9651 | 1.0000 | 0.982 | 0.003 | Both endophyte and Pathogen | [61] | rice and other plants |
| Sebacinales OTU_1840 | 0.9631 | 1.0000 | 0.981 | 0.002 | Endophyte | [63] | barley |
| Stachybotryaceae OTU_3669 | 0.9448 | 1.0000 | 0.972 | 0.031 | Endophyte | [64,65] | black cottonwood, wild rice |
| Myrothecium_verrucaria OTU_7577 | 0.9996 | 0.8889 | 0.943 | 0.002 | Pathogen | [66] | rice and other plants |
| Bambusicolaceae OTU_3283 | 0.9925 | 0.8889 | 0.939 | 0.012 | Endophyte and Pathogen | [67] | bamboo |
| Macrophomina_phaseolina OTU_4358 | 0.8767 | 1.0000 | 0.936 | 0.003 | Pathogen | [68] | rice and other plants |
| Xylariales OTU_8174 | 0.9755 | 0.8889 | 0.931 | 0.032 | Endophyte | [65,69] | wild rice and other plants |
| Thielavia_terrestris OTU_8307 | 1.0000 | 0.7778 | 0.882 | 0.002 | Free living or unknown | [70] | |
| Sordariomycetes OTU_2298 | 1.0000 | 0.7778 | 0.882 | 0.002 | Endophyte and Pathogen | [71] | rice and other plants |
| Chaetomium OTU_10228 | 0.8635 | 0.7778 | 0.819 | 0.014 | Endophyte | [72] | rice |
| Dothideomycetes OTU_7194 | 1.0000 | 0.6667 | 0.816 | 0.006 | Endophyte and Pathogen | [73,74] | Lycopodium annotinum, wheat |
| Pleosporales OTU_11327 | 1.0000 | 0.5556 | 0.745 | 0.004 | Endophyte and Pathogen | [61] | rice and other plants |
| Chaetomium OTU_4724 | 0.9999 | 0.5556 | 0.745 | 0.030 | Endophyte | [72] | rice |
| Pezizacea OTU_11633 | 1.0000 | 0.4444 | 0.667 | 0.032 | Endophyte | [75] | Quercus, Pinus, Populus |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanasugi, M.; Sarkodee-Addo, E.; Ansong Omari, R.; Mohammad Golam Dastogeer, K.; Fujii, Y.; Oppong Abebrese, S.; Bam, R.; Asuming-Brempong, S.; Okazaki, S. Exploring Rice Root Microbiome; The Variation, Specialization and Interaction of Bacteria and Fungi In Six Tropic Savanna Regions in Ghana. Sustainability 2020, 12, 5835. https://doi.org/10.3390/su12145835
Kanasugi M, Sarkodee-Addo E, Ansong Omari R, Mohammad Golam Dastogeer K, Fujii Y, Oppong Abebrese S, Bam R, Asuming-Brempong S, Okazaki S. Exploring Rice Root Microbiome; The Variation, Specialization and Interaction of Bacteria and Fungi In Six Tropic Savanna Regions in Ghana. Sustainability. 2020; 12(14):5835. https://doi.org/10.3390/su12145835
Chicago/Turabian StyleKanasugi, Makoto, Elsie Sarkodee-Addo, Richard Ansong Omari, Khondoker Mohammad Golam Dastogeer, Yoshiharu Fujii, Samuel Oppong Abebrese, Ralph Bam, Stella Asuming-Brempong, and Shin Okazaki. 2020. "Exploring Rice Root Microbiome; The Variation, Specialization and Interaction of Bacteria and Fungi In Six Tropic Savanna Regions in Ghana" Sustainability 12, no. 14: 5835. https://doi.org/10.3390/su12145835
APA StyleKanasugi, M., Sarkodee-Addo, E., Ansong Omari, R., Mohammad Golam Dastogeer, K., Fujii, Y., Oppong Abebrese, S., Bam, R., Asuming-Brempong, S., & Okazaki, S. (2020). Exploring Rice Root Microbiome; The Variation, Specialization and Interaction of Bacteria and Fungi In Six Tropic Savanna Regions in Ghana. Sustainability, 12(14), 5835. https://doi.org/10.3390/su12145835