Pakchoi Antioxidant Improvement and Differential Rhizobacterial Community Composition under Organic Fertilization
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil and Material Preparation
2.2. Greenhouse Pot Trial
2.3. Plant and Soil Sampling
2.4. Analysis of Plant and Soil Properties
2.5. Rhizobacterial Analysis
2.6. Statistical Analysis
3. Results
3.1. The Effects of Fertilizer on Plant and Soil Properties
3.2. Correlations between Plant and Soil Properties
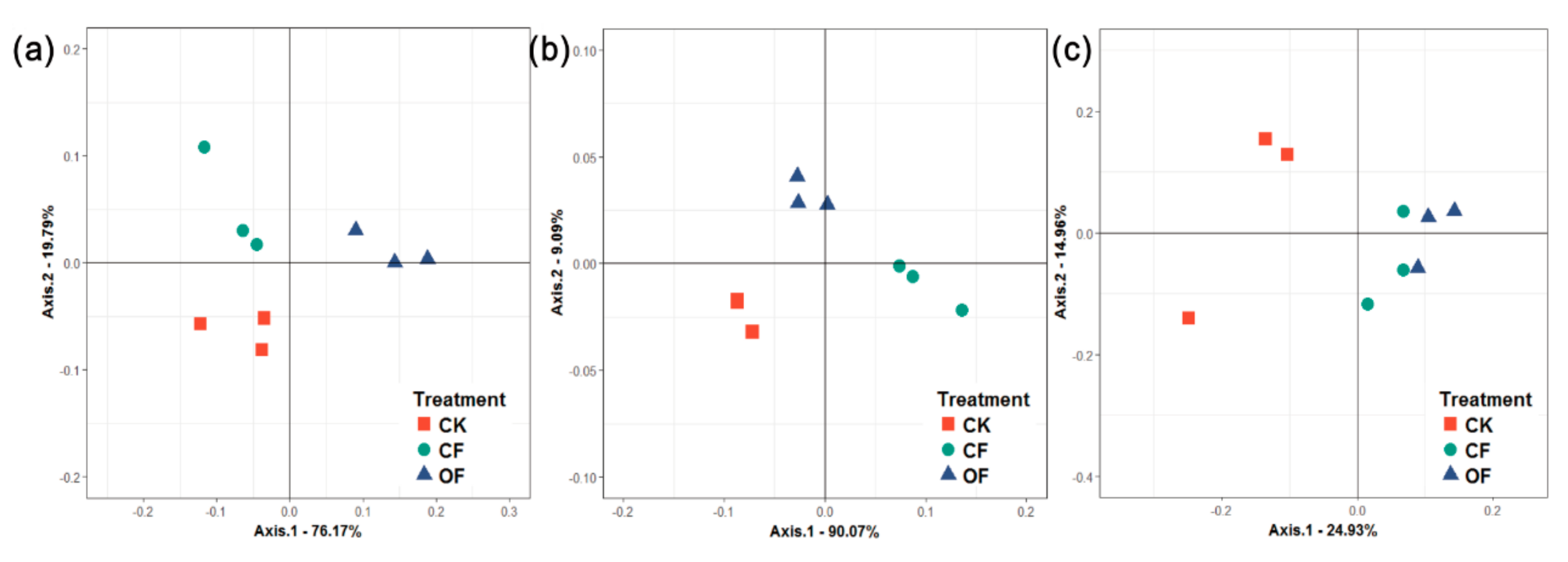
3.3. Rhizobacterial Community Composition
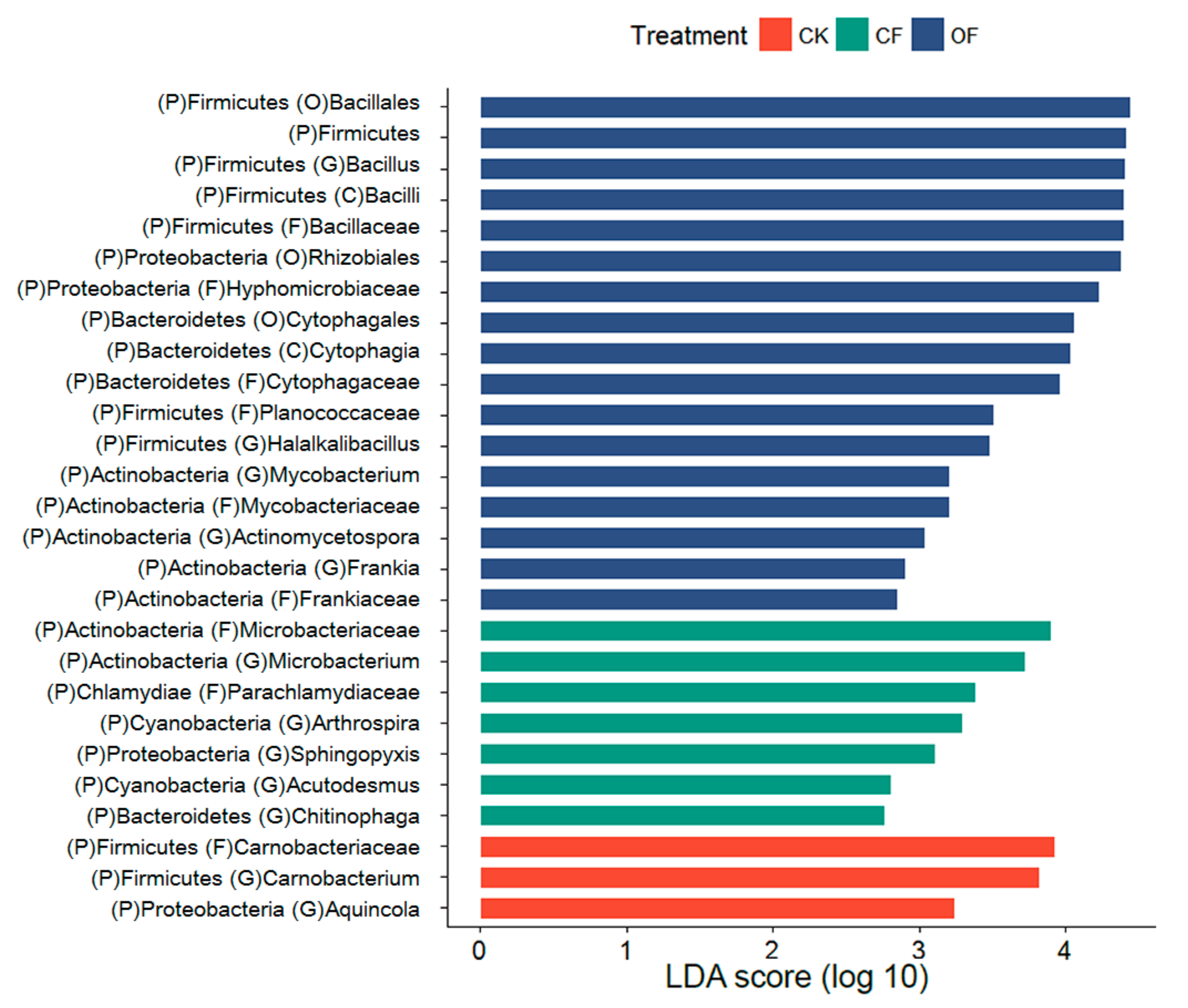
3.4. Fertilizer-Associated Rhizobacteria
3.5. Correlations between Plant Parameters and Rhizobacteria
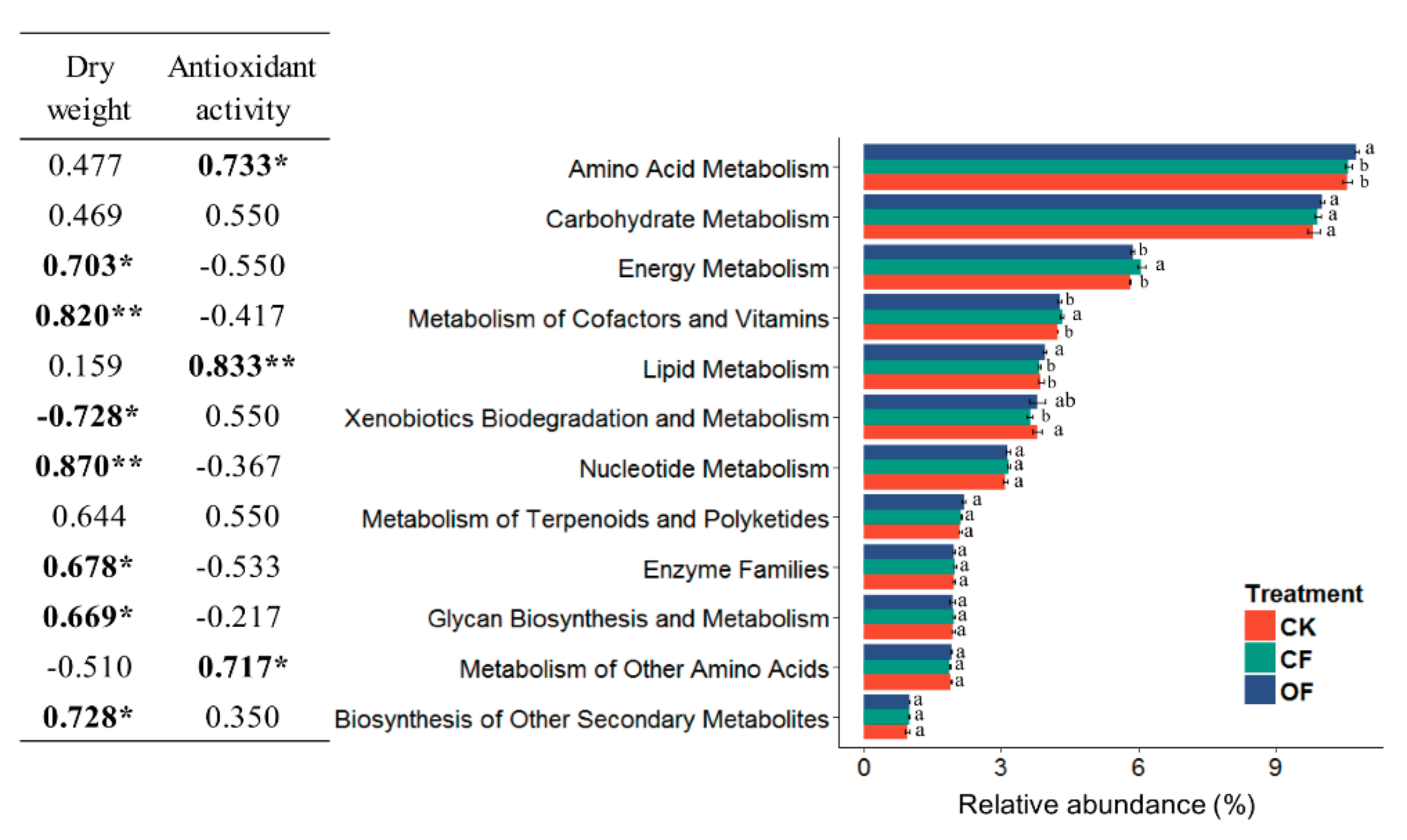
3.6. Correlation between the Predicted Functional Metabolism Profiles and Plant Traits
4. Discussion
4.1. The Impact of Fertilizers on Soil and Plant Properties and Rhizobacterial Community
4.2. Links between Rhizobacteria and Plant Performance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reganold, J.P.; Wachter, J.M. Organic agriculture in the twenty–first century. Nat. Plants 2016, 2, 15221. [Google Scholar] [CrossRef] [PubMed]
- Lehesranta, S.J.; Koistinen, K.M.; Massat, N.; Davies, H.V.; Shepherd, L.V.T.; McNicol, J.W.; Cakmak, I.; Cooper, J.; Lück, L.; Kärenlampi, S.O.; et al. Effects of agricultural production systems and their components on protein profiles of potato tubers. Proteomics 2007, 7, 597–604. [Google Scholar] [CrossRef]
- Wojtaszek, P.; Stobiecki, M.; Gulewicz, K. Role of nitrogen and plant growth regulators in the exudation and accumulation of lsoflavonoids by roots of intact white lupin (Lupinus albus L.) plants. J. Plant Physiol. 1993, 142, 689–694. [Google Scholar] [CrossRef]
- Kong, L.; Xie, Y.; Hu, L.; Si, J.; Wang, Z. Excessive nitrogen application dampens antioxidant capacity and grain filling in wheat as revealed by metabolic and physiological analyses. Sci. Rep. 2017, 7, 43363. [Google Scholar] [CrossRef] [PubMed]
- Coronado, C.; Esnault, R.; Kondorosi, A.; Ratet, P. Alfalfa root flavonoid production is nitrogen regulated. Plant Physiol. 1995, 108, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Tartoura, K.A.H.; Youssef, S.A.G. Effect of compost on the antioxidant defense systems of cucumber (Cucumis sativus L.) against cadmium toxicity. Ann. Agric. Sci. 2010, 191–203. [Google Scholar]
- Yang, X.; Feng, L.; Zhao, L.; Liu, X.; Hassani, D.; Huang, D. Effect of glycine nitrogen on lettuce growth under soilless culture: A metabolomics approach to identify the main changes occurred in plant primary and secondary metabolism. J. Sci. Food Agric. 2018, 98, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Gans, J.; Wolinsky, M.; Dunbar, J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 2005, 309, 1387–1390. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Hinsinger, P. Structure and function of the rhizosphere: Mechanisms at the soil-root interface. Ocl-Ol. Corps Gras Lipides 1998, 5, 340–341. [Google Scholar]
- Wang, Q.; Jiang, X.; Guan, D.; Wei, D.; Zhao, B.; Ma, M.; Chen, S.; Li, L.; Cao, F.; Li, J. Long-term fertilization changes bacterial diversity and bacterial communities in the maize rhizosphere of Chinese Mollisols. Appl. Soil Ecol. 2018, 125, 88–96. [Google Scholar] [CrossRef]
- Pineda, A.; Kaplan, I.; Bezemer, T.M. Steering soil microbiomes to suppress aboveground insect pests. Trends Plant Sci. 2017, 22, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Saharan, B.S. Plant growth promoting rhizobacteria: A critical review. Life Sci. Med. Res. 2011, 21, 30. [Google Scholar]
- Bacon, C.W.; Palencia, E.R.; Hinton, D.M. Abiotic and Biotic Plant Stress—Tolerant and Beneficial Secondary Metabolites Produced by Endophytic Bacillus Species. In Plant Microbes Symbiosis: Applied Facets; Arora, N.K., Ed.; Springer India: Uttar Pradesh, India, 2015; pp. 163–177. ISBN 978-81-322-2068-8. [Google Scholar]
- Singhai, P.K.; Sarma, B.K.; Srivastava, J.S. Biological management of common scab of potato through Pseudomonas species and vermicompost. Biol. Control 2011, 57, 150–157. [Google Scholar] [CrossRef]
- Kavamura, V.N.; Hayat, R.; Clark, I.M.; Rossmann, M.; Mendes, R.; Hirsch, P.R.; Mauchline, T.H. Inorganic nitrogen application affects both taxonomical and predicted functional structure of wheat rhizosphere bacterial communities. Front Microbiol. 2018, 9, 1074. [Google Scholar] [CrossRef]
- Wolters, B.; Jacquiod, S.; Sørensen, S.J.; Widyasari–Mehta, A.; Bech, T.B.; Kreuzig, R.; Smalla, K. Bulk soil and maize rhizosphere resistance genes, mobile genetic elements and microbial communities are differently impacted by organic and inorganic fertilization. FEMS Microbiol. Ecol. 2018, 94, fiy027. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Kumari, H.; Kumar, M.; Verma, M.; Kumari, K.; Malhotra, S.; Khurana, J.; Lal, R. From bacterial genomics to metagenomics: Concept, tools and recent advances. Indian J. Microbiol. 2008, 48, 173–194. [Google Scholar] [CrossRef]
- Acosta-Martinez, V.; Dowd, S.; Sun, Y.; Allen, V. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 2008, 40, 2762–2770. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Ding, J.; Zhao, L.; Wang, L.; Zhao, W.; Zhai, Z.; Leng, L.; Wang, Y.; He, C.; Zhang, Y.; Zhang, H.; et al. Divergent selection-induced obesity alters the composition and functional pathways of chicken gut microbiota. Genet. Sel. Evol. 2016, 48, 93. [Google Scholar] [CrossRef]
- Abia, A.L.K.; Alisoltani, A.; Keshri, J.; Ubomba-Jaswa, E. Metagenomic analysis of the bacterial communities and their functional profiles in water and sediments of the Apies river, South Africa, as a function of land use. Sci. Total Environ. 2018, 616, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Bonanomi, G.; De Filippis, F.; Cesarano, G.; La Storia, A.; Ercolini, D.; Scala, F. Organic farming induces changes in soil microbiota that affect agro-ecosystem functions. Soil Biol. Biochem. 2016, 103, 327–336. [Google Scholar] [CrossRef]
- Xiao, X.; Fan, M.; Wang, E.; Chen, W.; Wei, G. Interactions of plant growth-promoting rhizobacteria and soil factors in two leguminous plants. Appl. Microbiol. Biotechnol. 2017, 101, 8485–8497. [Google Scholar] [CrossRef] [PubMed]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2006; World Soil Resources Report; FAO: Rome, Italy, 2010. [Google Scholar]
- Tariq Rafiq, M.; Aziz, R.; Yang, X.; Xiao, W.; Stoffella, P.J.; Saghir, A.; Azam, M.; Li, T. Phytoavailability of cadmium (Cd) to pak choi (Brassica chinensis L.) grown in chinese soils: A model to evaluate the impact of soil Cd pollution on potential dietary toxicity. PLoS ONE 2014, 9, e111461. [Google Scholar]
- Eghball, B.; Wienhold, B.J.; Gilley, J.E.; Eigenberg, R.A. Mineralization of manure nutrients. J. Soil Water Conserv. 2002, 57, 470–473. [Google Scholar]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Orthofer, R.; Lamuela–Raventós, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzym. 1999, 299, 152–178. [Google Scholar]
- Szaufer-Hajdrych, M. Phenolic acids in leaves of species of the Aquilegia L. genus. Herba Pol. 2004, 50, 50–54. [Google Scholar]
- Lamaison, J.L.; Carnat, A. The amount of main flavonoids in flowers and leaves of Crataegus monogyna Jacq. and Crataegus laevigata (Poiret) DC. (Rosaceae). Pharm. Acta Helv. 1990, 65, 315–320. [Google Scholar]
- Złotek, U.; Świeca, M.; Jakubczyk, A. Effect of abiotic elicitation on main health–promoting compounds, antioxidant activity and commercial quality of butter lettuce (Lactuca sativa L.). Food Chem. 2014, 148, 253–260. [Google Scholar]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Khalid, M.; Hassani, D.; Bilal, M.; Huang, D. Elevation of secondary metabolites synthesis in Brassica campestris ssp. chinensis L. via exogenous inoculation of Piriformospora indica with appropriate fertilizer. PLoS ONE 2017, 12, e0177185. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- R–Core–Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Ghani, A.; Dexter, M.; Carran, R.A.; Theobald, P.W. Dissolved organic nitrogen and carbon in pastoral soils: The New Zealand experience. Eur. J. Soil Sci. 2007, 58, 832–843. [Google Scholar] [CrossRef]
- Näsholm, T.; Kielland, K.; Ganeteg, U. Uptake of organic nitrogen by plants. New Phytol. 2009, 182, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. organic amendments: Microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long–term fertilization strategies. Front Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef]
- Ren, G.; Ma, Y.; Guo, D.; Gentry, T.J.; Hu, P.; Pierson, E.A.; Gu, M. Soil bacterial community was changed after Brassicaceous seed meal application for suppression of Fusarium wilt on pepper. Front Microbiol. 2018, 9, 185. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zheng, A.P.; Bromfield, E.S.P.; Zhu, J.; Li, S.C.; Wang, S.Q.; Deng, Q.M.; Li, P. 16S rRNA gene sequence analysis of halophilic and halotolerant bacteria isolated from a hypersaline pond in Sichuan, China. Ann. Microbiol. 2011, 61, 375–381. [Google Scholar] [CrossRef]
- Pershina, E.V.; Ivanova, E.A.; Nagieva, A.G.; Zhiengaliev, A.T.; Chirak, E.L.; Andronov, E.E.; Sergaliev, N.K. A comparative analysis of microbiomes in natural and anthropogenically disturbed soils of northwestern Kazakhstan. Eurasian Soil Sci. 2016, 49, 673–684. [Google Scholar] [CrossRef]
- Andersson, A.; Höglander, H.; Karlsson, C.; Huseby, S. Key role of phosphorus and nitrogen in regulating cyanobacterial community composition in the northern Baltic Sea. Estuar. Coast. Shelf Sci. 2015, 164, 161–171. [Google Scholar] [CrossRef]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Nitrogen starvation-induced cellular crosstalk of ROS–scavenging antioxidants and phytohormone enhanced the biofuel potential of green microalga. Biotechnol. Biofuels 2017, 10, 60. [Google Scholar] [CrossRef]
- Kaewkla, O.; Franco, C.M.M. Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb. Ecol. 2013, 65, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, R.; Nielsen, S.; Joseph, S.D.; Huang, D.; Thomas, T. A combination of biochar-mineral complexes and compost improves soil bacterial processes, soil quality, and plant properties. Front Microbiol. 2016, 7, 372. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Perez, P.G.; Zhang, R.; Nielsen, S.; Huang, D.; Thomas, T. Effects of different C/N ratios on bacterial compositions and processes in an organically managed soil. Biol. Fert. Soils 2017, 54, 137–147. [Google Scholar] [CrossRef]
- Gong, W.; Yan, X.; Wang, J. The effect of chemical fertilizer on soil organic carbon renewal and CO2 emission—A pot experiment with maize. Plant Soil 2011, 353, 85–94. [Google Scholar] [CrossRef]
- Bacon, C.W.; White, J.F., Jr. Functions, mechanisms and regulation of endophytic and epiphytic microbial communities of plants. Symbiosis 2016, 68, 87–98. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, Y.; Lu, S.; Xiang, Q.; Yu, X.; Zhao, K.; Zou, L.; Chen, Q.; Tu, S.; Zhang, X. Long-term fertilization structures bacterial and archaeal communities along soil depth gradient in a paddy soil. Front Microbiol. 2017, 8, 1516. [Google Scholar] [CrossRef] [PubMed]
- Gluszek, S.; Sas–Paszt, L.; Sumorok, B.; Kozera, R. Biochar–Rhizosphere interactions—A review. Pol. J. Microbiol. 2017, 66, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Ikoyi, I.; Fowler, A.; Schmalenberger, A. One-time phosphate fertilizer application to grassland columns modifies the soil microbiota and limits its role in ecosystem services. Sci. Total Environ. 2018, 630, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yi, M.; Yi, H.; Guo, E.; Zhang, A. Manure and mineral fertilization change enzyme activity and bacterial community in millet rhizosphere soils. World J. Microbiol. Biotechnol. 2018, 34, 8. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Su, J.; Sun, G.; Wu, J.; Wei, W. Increased microbial functional diversity under long-term organic and integrated fertilization in a paddy soil. Appl. Microbiol. Biotechnol. 2018, 102, 1969–1982. [Google Scholar] [CrossRef] [PubMed]

| Parameter | CK | CF | OF |
|---|---|---|---|
| Plant | |||
| Fresh weight (g) | 2.10 ± 0.20 b | 4.11 ± 1.66 a | 3.84 ± 1.05 a |
| Dried weight (g) | 0.15 ± 0.02 b | 0.23 ± 0.02 a | 0.25 ± 0.05 a |
| Leaf number | 7.67 ± 0.58 a | 9.00 ± 1.00 a | 9.00 ± 1.00 a |
| Leaf area (cm2) | 13.78 ± 2.05 a | 16.75 ± 1.25 a | 15.63 ± 4.93 a |
| Phenolic content (mg GAE/g FW) | 0.88 ± 0.03 b | 0.68 ± 0.08 c | 1.22 ± 0.06 a |
| Phenolic acid (mg CAE/g FW) | 52.72 ± 8.28 b | 53.08 ± 7.92 b | 145.71 ± 12.80 a |
| Flavonoids (mg QE/g FW) | 18.40 ± 2.53 a | 11.78 ± 2.45 b | 18.17 ± 1.12 a |
| Antioxidant activity (mM Trolox/g FW) | 1.13 ± 0.06 b | 0.86 ± 0.12 c | 1.38 ± 0.05 a |
| Soil | |||
| pH | 8.02 ± 0.03 b | 8.01 ± 0.01 b | 8.07 ± 0.01 a |
| EC (μS/cm) | 223.00 ± 6.24 b | 465.33 ± 31.82 a | 432.00 ± 8.19 a |
| Available P (mg/kg) | 107.18 ± 10.77 b | 164.18 ± 5.84 a | 180.57 ± 10.51 a |
| Total C (%) | 2.05 ± 0.07 b | 2.19 ± 0.08 b | 2.44 ± 0.13 a |
| Total N (%) | 0.22 ± 0.00 c | 0.24 ± 0.01 b | 0.27 ± 0.01 a |
| NH4+–N (mg/kg) | 12.61 ± 1.10 a | 10.47 ± 0.26 b | 4.81 ± 0.60 c |
| NO3−–N (mg/kg) | 14.37 ± 4.29 b | 111.09 ± 32.89 a | 39.11 ± 8.46 b |
| TIN (mg/kg) | 26.98 ± 3.77 b | 121.56 ± 33.00 a | 43.92 ± 8.23 b |
| TON (%) | 0.21 ± 0.00 b | 0.23 ± 0.00 b | 0.27 ± 0.01 a |
| C/N ratio | 9.55 ± 0.15 a | 9.03 ± 0.13 b | 8.92 ± 0.35 b |
| Parameter | Leaf Number | Leaf Area | Fresh Weight | Dry Weight | Phenolic Content | Phenolic Acid | Flavonoids | Antioxidant Activity |
|---|---|---|---|---|---|---|---|---|
| pH | 0.154 | −0.105 | 0.250 | 0.533 | 0.774 * | 0.853 ** | 0.359 | 0.618 |
| EC | 0.651 | 0.349 | 0.654 | 0.786 * | 0.010 | 0.393 | −0.512 | −0.161 |
| Available P | 0.617 | 0.257 | 0.582 | 0.779 * | 0.343 | 0.645 | −0.300 | 0.190 |
| Total C | 0.235 | 0.190 | 0.399 | 0.482 | 0.615 | 0.825 ** | 0.281 | 0.527 |
| Total N | 0.486 | 0.072 | 0.431 | 0.717 * | 0.600 | 0.840 ** | 0.024 | 0.472 |
| NH4+-N | −0.457 | −0.260 | −0.458 | −0.722 * | −0.765 * | −0.928 ** | −0.213 | −0.630 |
| NO3−-N | 0.391 | 0.386 | 0.517 | 0.359 | −0.601 | −0.243 | −0.698 * | −0.665 |
| TIN | 0.355 | 0.365 | 0.481 | 0.303 | −0.657 | −0.312 | −0.712 * | −0.710 * |
| TON | 0.426 | 0.008 | 0.349 | 0.669 * | 0.722 * | 0.903 ** | 0.151 | 0.603 |
| C/N ratio | −0.746 * | 0.029 | −0.374 | −0.878 ** | −0.300 | −0.549 | 0.426 | −0.090 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, J.; Ye, J.; Liang, Y.; Khalid, M.; Huang, D. Pakchoi Antioxidant Improvement and Differential Rhizobacterial Community Composition under Organic Fertilization. Sustainability 2019, 11, 2424. https://doi.org/10.3390/su11082424
Liao J, Ye J, Liang Y, Khalid M, Huang D. Pakchoi Antioxidant Improvement and Differential Rhizobacterial Community Composition under Organic Fertilization. Sustainability. 2019; 11(8):2424. https://doi.org/10.3390/su11082424
Chicago/Turabian StyleLiao, Jianli, Jun Ye, Yun Liang, Muhammad Khalid, and Danfeng Huang. 2019. "Pakchoi Antioxidant Improvement and Differential Rhizobacterial Community Composition under Organic Fertilization" Sustainability 11, no. 8: 2424. https://doi.org/10.3390/su11082424
APA StyleLiao, J., Ye, J., Liang, Y., Khalid, M., & Huang, D. (2019). Pakchoi Antioxidant Improvement and Differential Rhizobacterial Community Composition under Organic Fertilization. Sustainability, 11(8), 2424. https://doi.org/10.3390/su11082424





