Natural Farming Improves Soil Quality and Alters Microbial Diversity in a Cabbage Field in Japan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site
2.2. Soil Sampling and Analysis
2.3. Soil DNA Extraction, Sequencing, and Processing
2.4. Statistical Analysis
3. Results
3.1. Edaphic Properties
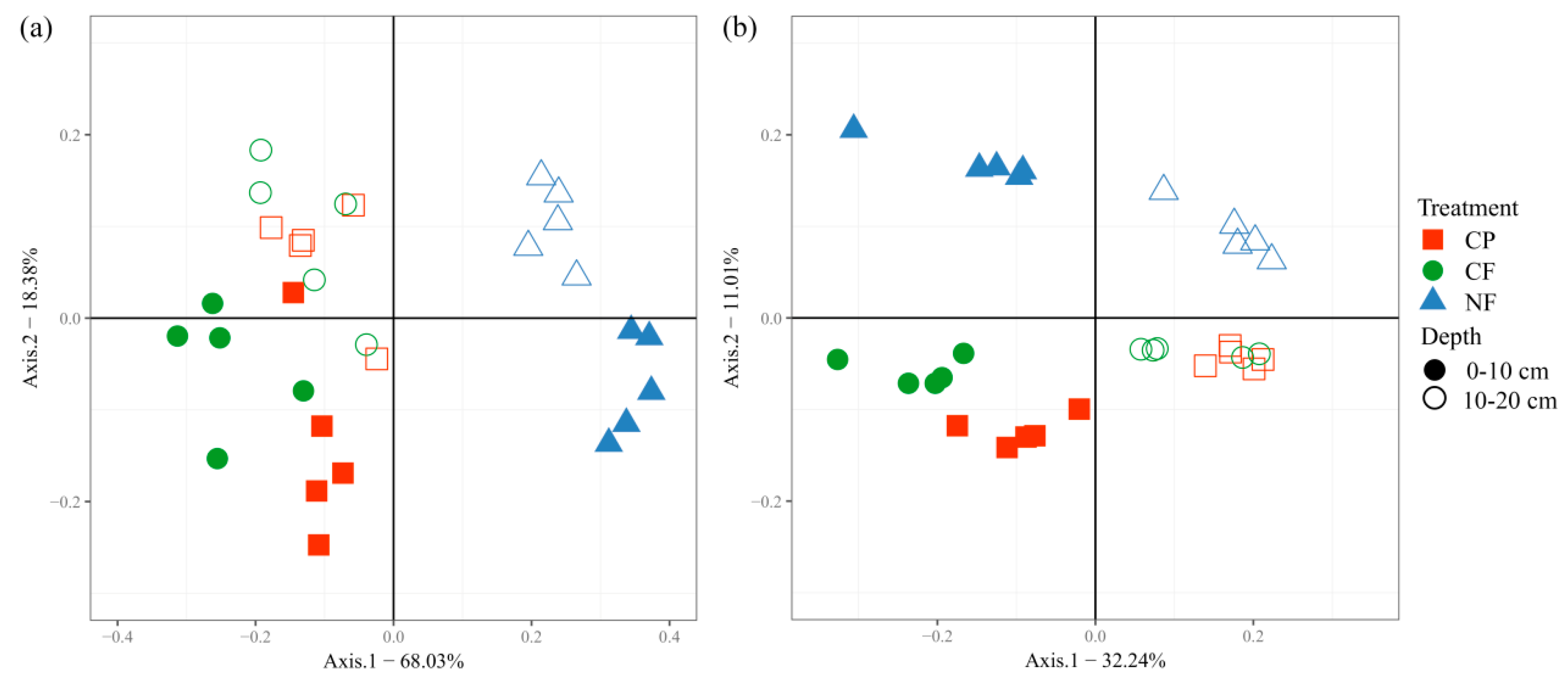
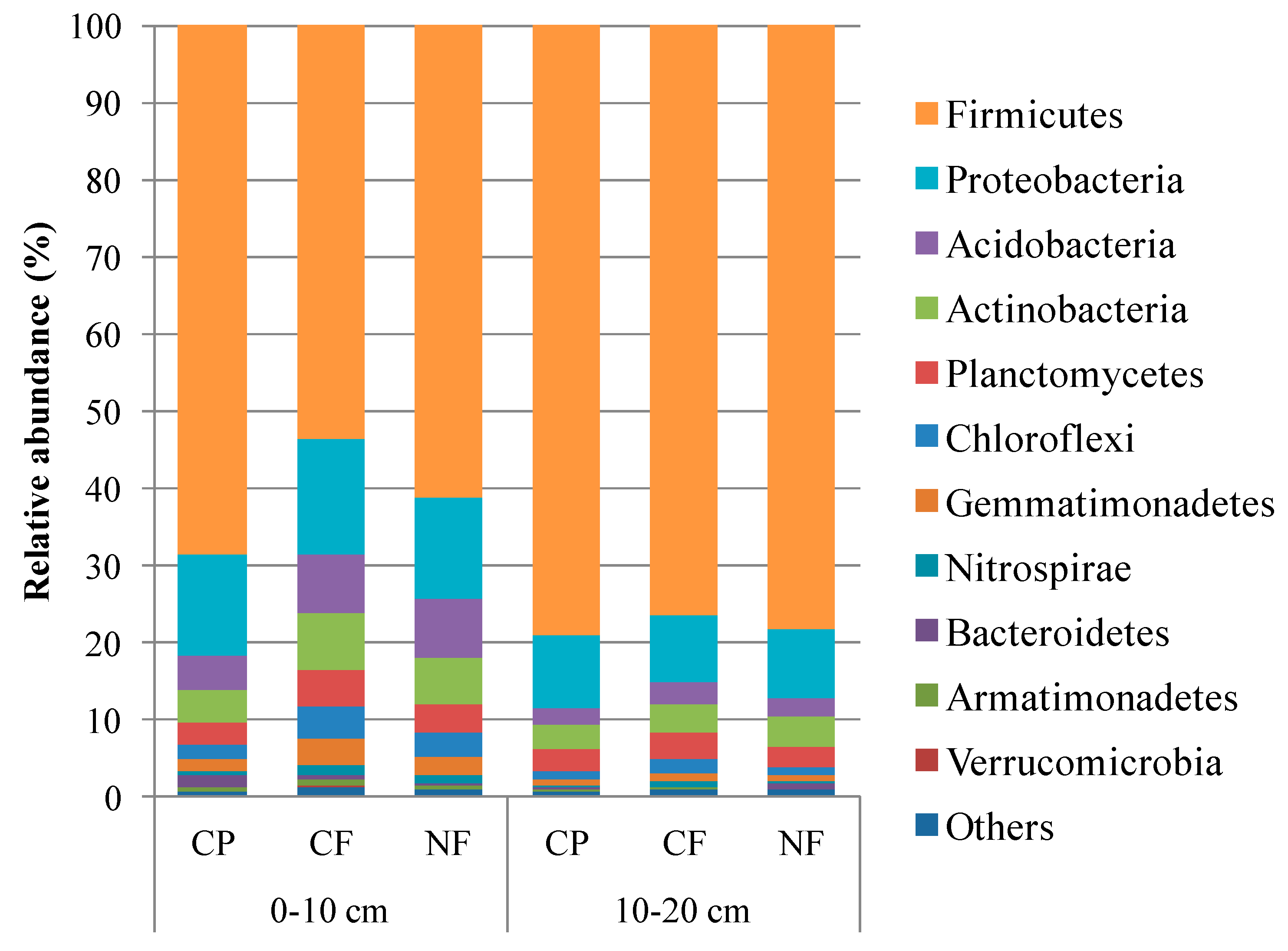
3.2. Microbial Community Structure
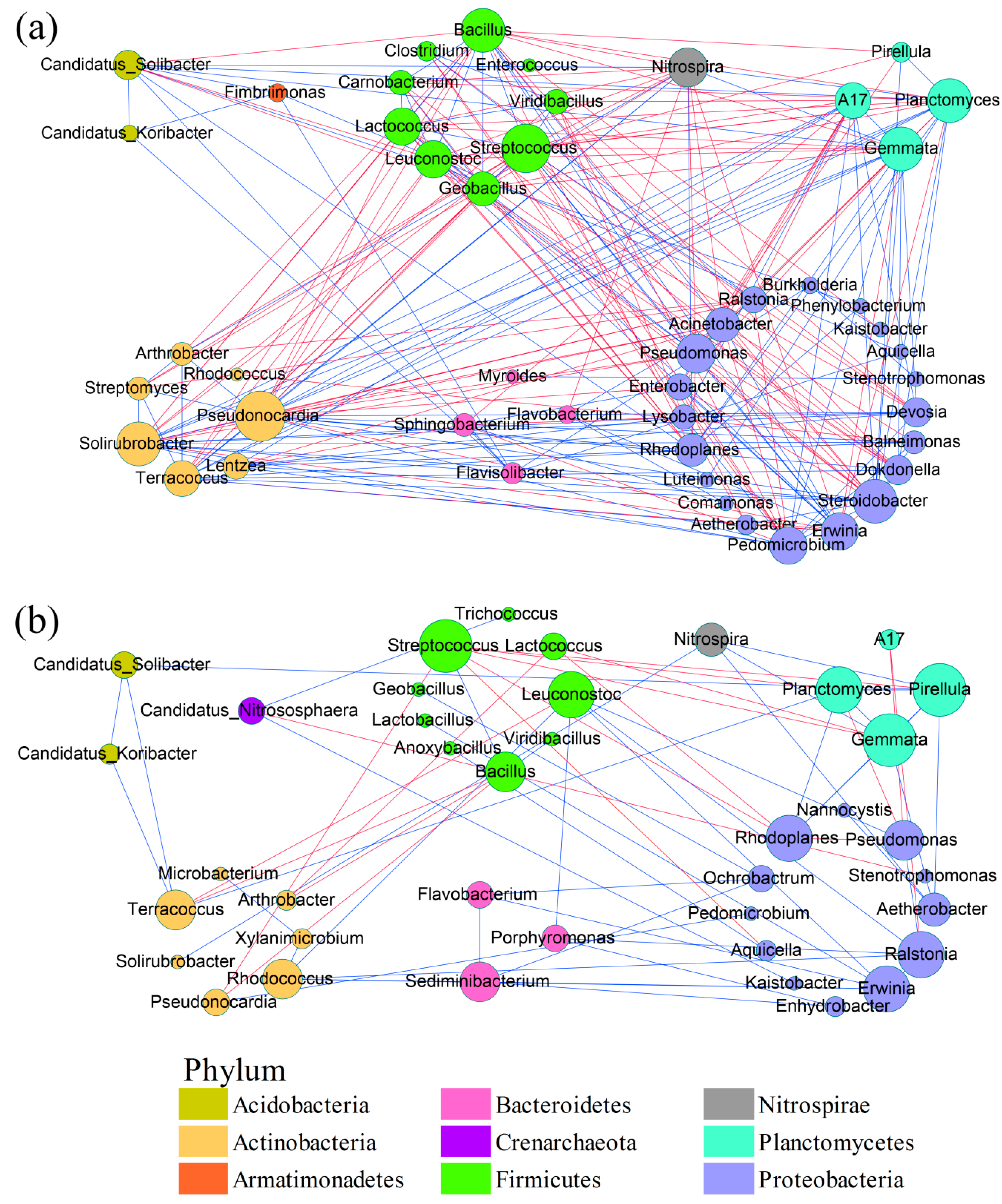
3.3. Microbial Network Analysis
4. Discussion
4.1. Edaphic Properties Vary in Response to Agricultural Management and at Different Soil Depths
4.2. Characteristics of the Soil Microbial Community under Different Agricultural Managements
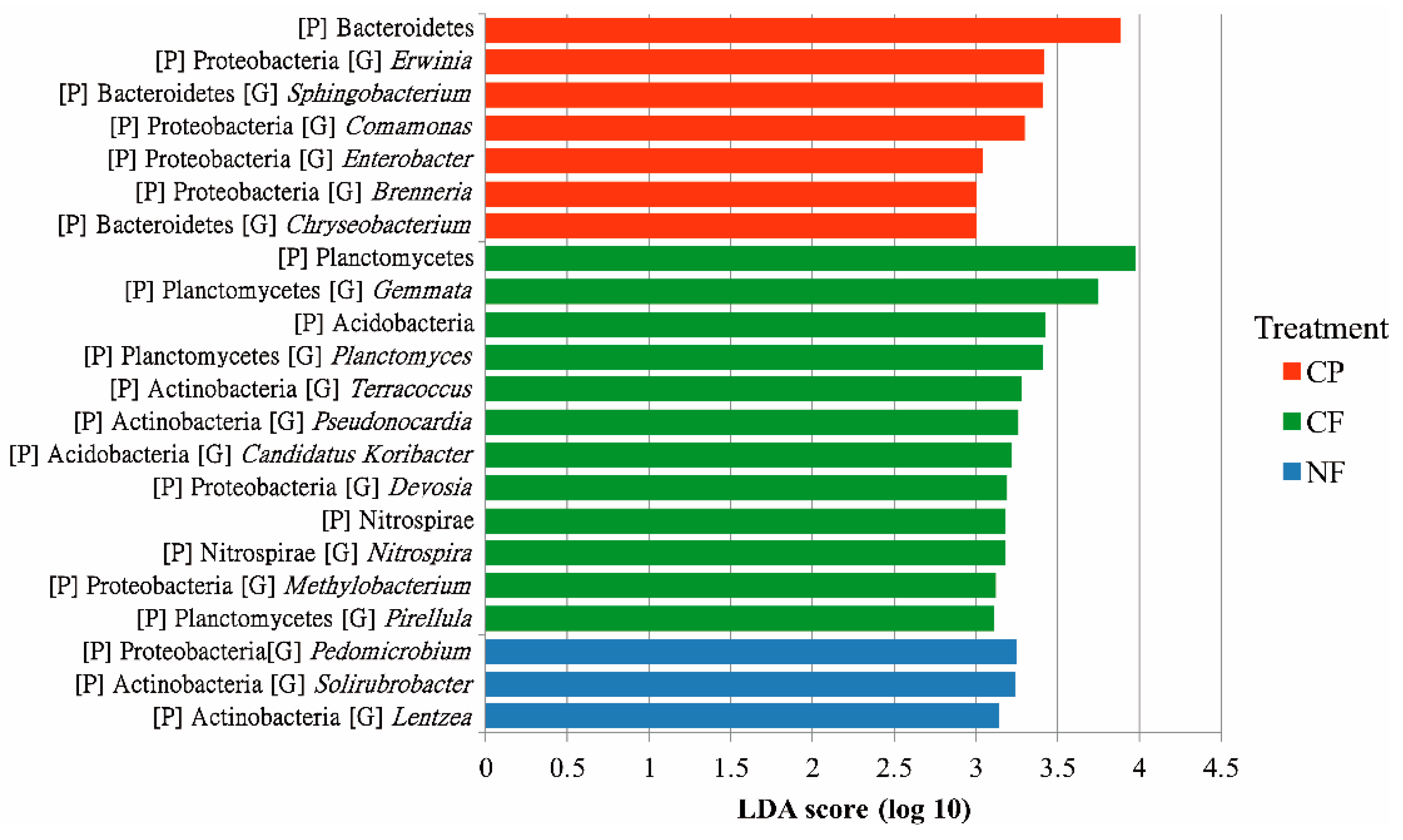
4.3. Agricultural Management-Associated Microbial Taxa
4.4. Robustness of and Key Genera in Microbial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stokstad, E. Organic farms reap many benefits. Science 2002, 296, 1589. [Google Scholar] [CrossRef] [PubMed]
- Doran, J.W.; Zeiss, M.R.; Zeiss, M.R. Soil health and sustainability: Managing the biotic component of soil quality. Appl. Soil. Ecol. 2000, 15, 3–11. [Google Scholar] [CrossRef]
- Mouazen, A.M.; Palmqvist, M. Development of a framework for the evaluation of the environmental benefits of controlled traffic farming. Sustainability 2015, 7, 8684–8708. [Google Scholar] [CrossRef]
- Pezzuolo, A.; Dumont, B.; Sartori, L.; Marinello, F.; Migliorati, M.; Basso, B. Evaluating the impact of soil conservation measures on soil organic carbon at the farm scale. Comput. Electron. Agri. 2017, 175–182. [Google Scholar] [CrossRef]
- Liao, J.; Liang, Y.; Huang, D. Organic Farming Improves Soil Microbial Abundance and Diversity under Greenhouse Condition: A Case Study in Shanghai (Eastern China). Sustainability 2018, 10, 3825. [Google Scholar] [CrossRef]
- Organic Agriculture Worldwide 2016: Current Statistics. Available online: http://orgprints.org/32677/19/Willer-2018-global-data-biofach.pdf (accessed on 14 February 2018).
- Xu, H.L. Nature Farming in Japan; Research Signpost Press: Kerala, India, 2006. [Google Scholar]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct soil microbial diversity under long–term organic and conventional farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef] [PubMed]
- Bonanomi, G.; De Filippis, F.; Cesarano, G.; La Storia, A.; Ercolini, D.; Scala, F. Organic farming induces changes in soil microbiota that affect agro–ecosystem functions. Soil Biol. Biochem. 2016, 103, 327–336. [Google Scholar] [CrossRef]
- Li, R.; Khafipour, E.; Krause, D.O.; Entz, M.H.; de Kievit, T.R.; Fernando, W.G.D. Pyrosequencing Reveals the Influence of Organic and Conventional Farming Systems on Bacterial Communities. PLoS ONE 2012, 7, e51897. [Google Scholar] [CrossRef]
- Wang, W.; Wang, H.; Feng, Y.; Wang, L.; Xiao, X.; Xi, Y.; Luo, X.; Sun, R.; Ye, X.; Huang, Y.; et al. Consistent responses of the microbial community structure to organic farming along the middle and lower reaches of the Yangtze River. Sci. Rep. 2016, 6, 35046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Zhang, R.; Nielsen, S.; Joseph, S.D.; Huang, D.; Thomas, T. A combination of biochar–mineral complexes and compost improves soil bacterial processes, soil quality, and plant properties. Front. Microbiol. 2016, 7, 372. [Google Scholar] [CrossRef] [PubMed]
- Tejeda–Agredano, M.C.; Gallego, S.; Vila, J.; Grifoll, M.; Ortega-Calvo, J.J.; Cantos, M. Influence of the sunflower rhizosphere on the biodegradation of PAHs in soil. Soil Biol. Biochem. 2013, 57, 830–840. [Google Scholar] [CrossRef] [Green Version]
- Lian, T.X.; Wang, G.H.; Yu, Z.H.; Li, Y.S.; Liu, X.B.; Zhang, S.Q.; Herbert, S.J.; Jin, J. Bacterial communities incorporating plant–derived carbon in the soybean rhizosphere in Mollisols that differ in soil organic carbon content. Appl. Soil Ecol. 2017, 119, 375–383. [Google Scholar] [CrossRef]
- Tao, R.; Liang, Y.C.; Wakelin, S.A.; Chu, G.X. Supplementing chemical fertilizer with an organic component increases soil biological function and quality. Appl. Soil Ecol. 2015, 96, 42–51. [Google Scholar] [CrossRef]
- Lim, J.; Jee, S.; Lee, D.; Roh, E.; Jung, K.; Oh, C.; Heu, S. Biocontrol of Pectobacterium carotovorum subsp. carotovorum using bacteriophage PP1. J. Microbiol. Biotechnol. 2013, 23, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Navarro, R.; Takeshita, K.; Tago, K.; Hayatsu, M.; Hori, T.; Kikuchi, Y. Bacterial population succession and adaptation affected by insecticide application and soil spraying history. Front. Microbiol. 2014, 5, 457. [Google Scholar] [CrossRef] [Green Version]
- Hamm, A.C.; Tenuta, M.; Krause, D.O.; Ominski, K.H.; Tkachuk, V.L.; Flaten, D.N. Bacterial communities of an agricultural soil amended with solid pig and dairy manures, and urea fertilizer. Appl. Soil Ecol. 2016, 103, 61–71. [Google Scholar] [CrossRef]
- Ye, J.; Perez, P.G.; Zhang, R.; Nielsen, S.; Huang, D.; Thomas, T. Effects of different C/N ratios on bacterial compositions and processes in an organically managed soil. Biol. Fertil. Soils 2017, 54, 137–147. [Google Scholar] [CrossRef]
- Degrune, F.; Theodorakopoulos, N.; Dufrêne, M.; Colinet, G.; Bodson, B.; Hiel, M.P.; Taminiau, B.; Nezer, C.; Daube, G.; Vandenbol, M. No favorable effect of reduced tillage on microbial community diversity in a silty loam soil (Belgium). Agric. Ecosyst. Environ. 2016, 224, 12–21. [Google Scholar] [CrossRef]
- Tu, C.; Ristaino, J.B.; Hu, S. Soil microbial biomass and activity in organic tomato farming systems: Effects of organic inputs and straw mulching. Soil Biol. Biochem. 2006, 38, 247–255. [Google Scholar] [CrossRef]
- Venter, Z.S.; Jacobs, K.; Hawkins, H.J. The impact of crop rotation on soil microbial diversity: A meta–analysis. Pedobiologia 2016, 59, 215. [Google Scholar] [CrossRef]
- Lenc, L.; Kwasna, H.; Sadowski, C.; Grabowski, A. Microbiota in wheat roots, rhizosphere and soil in crops grown in organic and other production systems. J. Phytopathol. 2015, 163, 245–263. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, Y.; Lu, S.; Xiang, Q.; Yu, X.; Zhao, K.; Zou, L.; Chen, Q.; Tu, S.; Zhang, X. Long–term Fertilization Structures Bacterial and Archaeal Communities along Soil Depth Gradient in a Paddy Soil. Front. Microbiol. 2017, 8, 1516. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.P.; Babujia, L.C.; Franchini, J.C.; Ralisch, R.; Hungria, M.; Guimaraes, M.D. Soil structure and its influence on microbial biomass in different soil and crop management systems. Soil Tillage Res. 2014, 142, 42–53. [Google Scholar] [CrossRef]
- FAO (Food Agriculture Organization). FAO Soil Map of the World; FAO: Paris, France, 1974. [Google Scholar]
- Ma, B.; Li, X.; Chang, S.X. Capping material type affects rhizosphere bacteria community structure in the cover soil in oil sands reclamation. J. Soil Sediment 2017, 17, 2516–2523. [Google Scholar] [CrossRef]
- Blake, G. Bulk density. In Methods of Soil Analysis. Part 1. Physical and Mineralogical Properties, Including Statistics of Measurement and Sampling; American Society of Agronomy: Madison, WI, USA, 1965. [Google Scholar]
- Bremner, J. Total nitrogen. In Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties; American Society of Agronomy: Madison, WI, USA, 1965. [Google Scholar]
- Carroll, J.A.; Caporn, S.J.; Johnson, D.; Morecroft, M.; Lee, J.A. The interactions between plant growth, vegetation structure and soil processes in semi–natural acidic and calcareous grasslands receiving long–term inputs of simulated pollutant nitrogen deposition. Environ. Pollut. 2003, 121, 363–376. [Google Scholar] [CrossRef]
- Kandeler, E.; Gerber, H. Short–term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils 1988, 6, 68–72. [Google Scholar] [CrossRef]
- Abdelmagid, H.; Tabatabai, M. Nitrate reductase activity of soils. Soil Biol. Biochem. 1987, 19, 421–427. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pẽa, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high–throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera–checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microb. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Vick–Majors, T.J.; Priscu, J.C.; Amaral–Zettler, L.A. Modular community structure suggests metabolic plasticity during the transition to polar night in ice–covered Antarctic lakes. ISME J. 2014, 8, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Guo, D.; Zhang, Y.; Sun, X.; Jiang, S.; Guo, Z.; Huang, H.; Liang, W.; Li, R.; Zhang, Z. Using bamboo biochar with compost for the stabilization and phytotoxicity reduction of heavy metals in mine–contaminated soils of China. Sci. Rep. 2017, 7, 2690. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.L.; Sun, T.H.; He, W.X.; Zhou, Q.X.; Chen, S. Single and joint effects of pesticides and mercury on soil urease. J. Environ. Sci. 2007, 19, 210–216. [Google Scholar] [CrossRef]
- Degrune, F.; Dufrêne, M.; Colinet, G.; Massart, S.; Taminiau, B.; Bodson, B.; Hiel, M.P.; Daube, G.; Nezer, C.; Vandenbol, M. A novel sub–phylum method discriminates better the impact of crop management on soil microbial community. Agron. Sustain. Dev. 2015, 35, 1157–1166. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, S.; Rong, Y.; Zhu, X.; Zhao, X.; Cai, Z. Pyrosequencing reveals bacterial communities and enzyme activities differences after application of novel chiral insecticide Paichongding in aerobic soils. Appl. Soil Ecol. 2017, 112, 18–27. [Google Scholar] [CrossRef]
- Guo, W.; Li, D.; Tao, Y.; Gao, P.; Hu, J. Isolation and Description of a Stable Carbazole–Degrading Microbial Consortium Consisting of Chryseobacterium sp. NCY and Achromobacter sp. NCW. Curr. Microbiol. 2008, 57, 251. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, C.S.; Hjelmsø, M.H. Agricultural soils, pesticides and microbial diversity. Curr. Opin. Biotechnol. 2014, 27, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.E.; Gross, D.C. Drippy pod of white lupine: A new bacterial disease caused by a pathovar of Brenneriaquercina. Plant Dis. 2010, 94, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Toth, I.K.; Pritchard, L.; Birch, P.R.J. Comparative Genomics Reveals What Makes An Enterobacterial Plant Pathogen. Annu. Rev.Phytopathol. 2006, 44, 305. [Google Scholar] [CrossRef] [PubMed]
- Van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottovā, D.; Krištůfek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xun, W.; Zhao, J.; Xue, C.; Zhang, G.; Ran, W.; Wang, B.; Shen, Q.; Zhang, R. Significant alteration of soil bacterial communities and organic carbon decomposition by different long-term fertilization management conditions of extremely low-productivity arable soil in South China. Environ. Microbiol. 2016, 18, 1907–1917. [Google Scholar] [CrossRef]
- Wang, X.; Sharp, C.E.; Jones, G.M.; Grasby, S.E.; Brady, A.L.; Dunfield, P.F. Stable–Isotope probing identifies uncultured planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl. Environ. Microb. 2015, 81, 4607–4615. [Google Scholar] [CrossRef] [PubMed]
- Mcgenity, T.J.; Folwell, B.D.; Mckew, B.A.; Sanni, G.O. Marine crude–oil biodegradation: A central role for interspecies interactions. Aquat. Biosyst. 2012, 8, 1–19. [Google Scholar] [CrossRef]
- Ivanova, A.; Philippov, D.; Kulichevskaya, I.; Dedysh, S. Distinct diversity patterns of Planctomycetes associated with the freshwater macrophyte Nuphar lutea (L.) Smith. Antonie Van Leeuwenhoek 2018, 111, 811–823. [Google Scholar] [CrossRef]
- Lee, S.; Strand, S.; Stensel, H.; Herwig, R. Pseudonocardiachloroethenivorans sp. nov., a chloroethene–degrading actinomycete. Int. J. Syst. Evol. Microbiol. 2004, 54, 131–139. [Google Scholar] [CrossRef]
- Rawat, S.; Männistö, M.; Bromberg, Y.; Häggblom, M. Comparative genomic and physiological analysis provides insights into the role of Acidobacteria in organic carbon utilization in Arctic tundra soils. FEMS Microbiol. Ecol. 2012, 82, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Tahon, G.; Willems, A. Isolation and characterization of aerobic anoxygenic phototrophs from exposed soils from the SørRondane Mountains, East Antarctica. Syst. Appl. Microbiol. 2017, 40, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, L.; Hassan, M.; Xie, B. Succession of the functional microbial communities and the metabolic functions in maize straw composting process. Bioresour. Technol. 2018, 256, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Embarcadero-Jiménez, S.; Rivera-Orduña, F.N.; Wang, E.T. Bacterial communities estimated by pyrosequencing in the soils of chinampa, a traditional sustainable agro–ecosystem in Mexico. J. Soil Sediment 2016, 16, 1001–1011. [Google Scholar] [CrossRef]
- Sánchez–Marañón, M.; Miralles, I.; Aguirre–Garrido, J.; Anguita–Maeso, M.; Millán, V.; Ortega, R.; García–Salcedo, J.; Martínez–Abarca, F.; Soriano, M. Changes in the soil bacterial community along a pedogenic gradient. Sci. Rep. 2017, 7, 14593. [Google Scholar] [CrossRef]
- Hamedi, J.; Mohammadipanah, F. Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J. Ind. Microbiol. Biotechnol. 2015, 42, 157–171. [Google Scholar] [CrossRef]
- Mendes, L.W.; Raaijmakers, J.M.; de Hollander, M.; Mendes, R.; Tsai, S.M. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018, 12, 212–224. [Google Scholar] [CrossRef]
- De Mandal, S.; Chatterjee, R.; Kumar, N.S. Dominant bacterial phyla in caves and their predicted functional roles in C and N cycle. BMC Microbiol. 2017, 17, 90. [Google Scholar] [CrossRef]
- Wang, H.; Zeng, Y.; Guo, C.; Bao, Y.; Lu, G.; Reinfelder, J.R.; Dang, Z. Bacterial, archaeal, and fungal community responses to acid mine drainage–laden pollution in a rice paddy soil ecosystem. Sci. Total Environ. 2018, 616, 107–116. [Google Scholar] [CrossRef]
- Bates, S.T.; Berg–Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011, 5, 908–917. [Google Scholar] [CrossRef]
- Khalaf, E.M.; Raizada, M.N. Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens including powdery mildew. Front. Microbiol. 2018, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Matthias, A.; Benigno, P.; Ignasi, B.; Anna, T. Consequences of plant invasions on compartmentalization and species’ roles in plant–pollinator networks. Proc. Biol. Sci. 2014, 281, 20140773. [Google Scholar]
| Practice | CP | CF | NF |
|---|---|---|---|
| Fertilizer type | Chemical fertilizer | Chemical fertilizer | Bioorganic fertilizer |
| Inputs (kg ha−1 y−1) | |||
| N | 21.2 | 21.2 | 7.2 |
| P | 12.6 | 12.6 | 6.84 |
| K | 8.8 | 8.8 | 1.8 |
| Tillage | Ploughing with 15 cm depth | Ploughing with 15 cm depth | Reduced-tillage with 5 cm depth |
| Pest and disease control | Agrochemicals and bioagent | None | None |
| Rotation system | Cabbage | Cabbage | Cabbage-Rye |
| Residue mulch | None | None | Rye straw |
| Depth | 0–10 cm | 10–20 cm | ||||
|---|---|---|---|---|---|---|
| Cultivation System | CP | CF | NF | CP | CF | NF |
| Water content (%) | 34.38 ± 0.18 e | 33.55 ± 0.17 f | 37.87 ± 0.33 a | 35.43 ± 0.16 c | 34.50 ± 0.15 d | 36.68 ± 0.15 b |
| Bulk density (g cm−3) | 1.01 ± 0.03 b | 1.10 ± 0.03 a | 0.96 ± 0.02 bc | 0.93 ± 0.01 cd | 0.96 ± 0.02 bc | 0.87 ± 0.03 d |
| pH | 6.00 ± 0.04 e | 5.94 ± 0.06 e | 6.92 ± 0.03 b | 6.38 ± 0.01 d | 6.50 ± 0.02 c | 7.05 ± 0.01 a |
| EC (dSm−1) | 0.14 ± 0.03 a | 0.10 ± 0.01 ab | 0.08 ± 0.01 bc | 0.09 ± 0.00 bc | 0.08 ± 0.00 bc | 0.05 ± 0.00 c |
| NH4+-N (mg kg−1) | 6.70 ± 1.15 ab | 5.56 ± 1.73 ab | 8.78 ± 3.81 a | 1.52 ± 0.53 b | 2.54 ± 0.78 b | 3.72 ± 0.87 ab |
| NO3−-N (mg kg−1) | 33.24 ± 10.39 a | 31.90 ± 5.92 a | 12.78 ± 1.69 b | 16.50 ± 3.19 b | 12.32 ± 0.91 b | 8.80 ± 0.80 b |
| Urease activity 1 | 45.27 ± 8.26 b | 22.27 ± 5.71 c | 69.05 ± 7.96 a | 18.26 ± 3.37 c | 17.01 ± 4.54 c | 21.48 ± 2.56 c |
| Nitrate reductase activity 2 | 9.79 ± 1.84 c | 3.18 ± 0.40 c | 101.92 ± 6.35 a | 9.21 ± 2.15 c | 7.09 ± 1.37 c | 49.26 ± 3.48 b |
| TC (%) | 4.21 ± 0.05 c | 4.55 ± 0.02 b | 4.94 ± 0.11 a | 4.00 ± 0.09 d | 4.48 ± 0.07 b | 4.25 ± 0.03 c |
| TN (%) | 0.38 ± 0.01 b | 0.39 ± 0.01 b | 0.43 ± 0.01 a | 0.33 ± 0.01 d | 0.35 ± 0.00 c | 0.34 ± 0.01 cd |
| C/N | 10.99 ± 0.25 b | 11.55 ± 0.15 b | 11.38 ± 0.08 b | 12.27 ± 0.20 a | 12.73 ± 0.22 a | 12.37 ± 0.15 a |
| Depth | Cultivation System | Reads | OTUs | Coverage (%) | ACE | Chao1 | Shannon |
|---|---|---|---|---|---|---|---|
| 0–10 cm | CP | 35283 | 4680 | 0.97 | 1229 ± 89 c | 1140 ± 75 c | 5.62 ± 0.32 bc |
| CF | 32882 | 5357 | 0.95 | 1756 ± 137 a | 1675 ± 175 a | 6.91 ± 0.98 a | |
| NF | 31859 | 4969 | 0.95 | 1578 ± 131 ab | 1475 ± 100 ab | 6.39 ± 1.20 ab | |
| 10–20 cm | CP | 32966 | 3986 | 0.97 | 1197 ± 239 c | 1082 ± 207 c | 5.30 ± 0.29 c |
| CF | 31229 | 4232 | 0.96 | 1364 ± 330 bc | 1238 ± 292 bc | 5.41 ± 0.53 c | |
| NF | 32889 | 3979 | 0.97 | 1245 ± 271 c | 1127 ± 228 c | 5.34 ± 0.25 c |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, J.; Xu, Q.; Xu, H.; Huang, D. Natural Farming Improves Soil Quality and Alters Microbial Diversity in a Cabbage Field in Japan. Sustainability 2019, 11, 3131. https://doi.org/10.3390/su11113131
Liao J, Xu Q, Xu H, Huang D. Natural Farming Improves Soil Quality and Alters Microbial Diversity in a Cabbage Field in Japan. Sustainability. 2019; 11(11):3131. https://doi.org/10.3390/su11113131
Chicago/Turabian StyleLiao, Jianli, Qicong Xu, Huilian Xu, and Danfeng Huang. 2019. "Natural Farming Improves Soil Quality and Alters Microbial Diversity in a Cabbage Field in Japan" Sustainability 11, no. 11: 3131. https://doi.org/10.3390/su11113131
APA StyleLiao, J., Xu, Q., Xu, H., & Huang, D. (2019). Natural Farming Improves Soil Quality and Alters Microbial Diversity in a Cabbage Field in Japan. Sustainability, 11(11), 3131. https://doi.org/10.3390/su11113131




