Early Forecasting of Rice Blast Disease Using Long Short-Term Memory Recurrent Neural Networks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
2.1.1. Rice Blast Disease Score Data
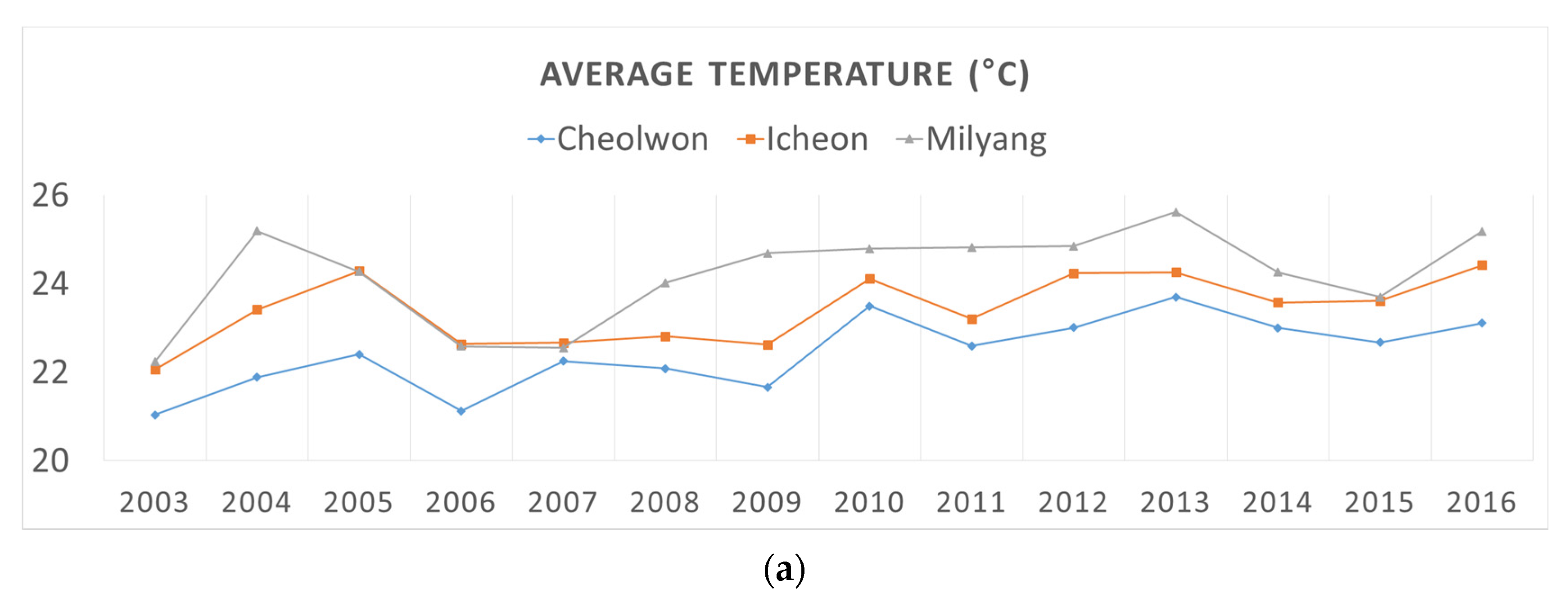
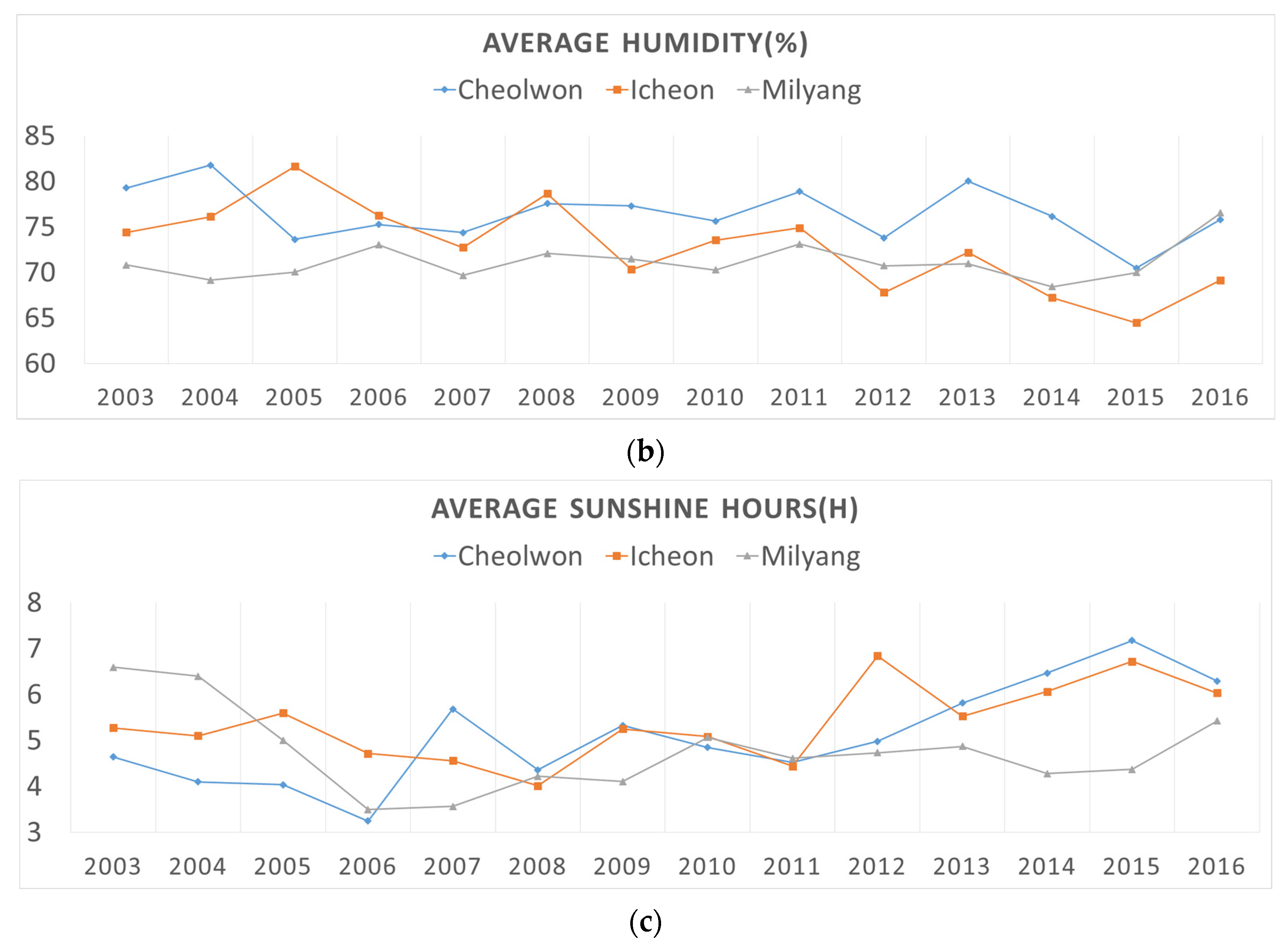
2.1.2. Historical Climatic Data
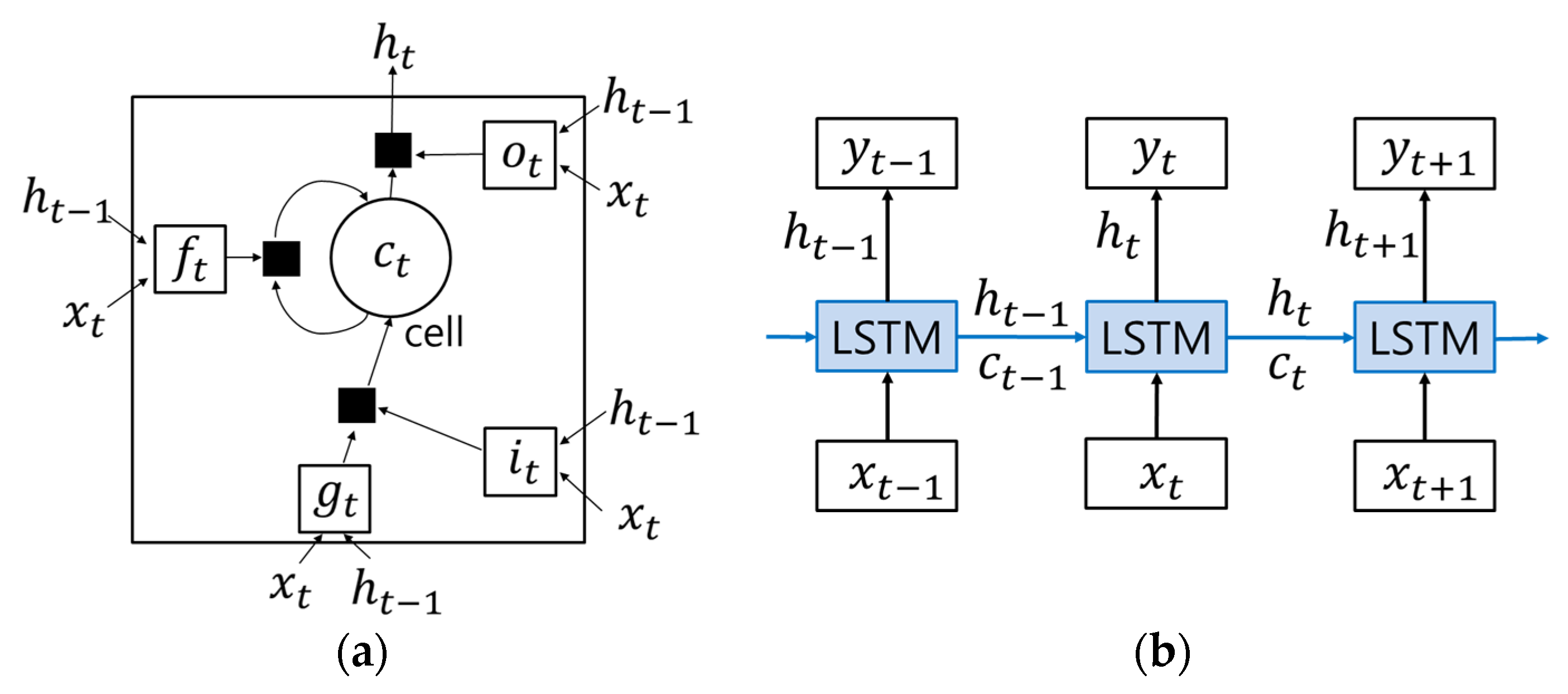
2.2. Long Short-Term Memory (LSTM) Recurrent Neural Networks (RNNs)
3. Experiments
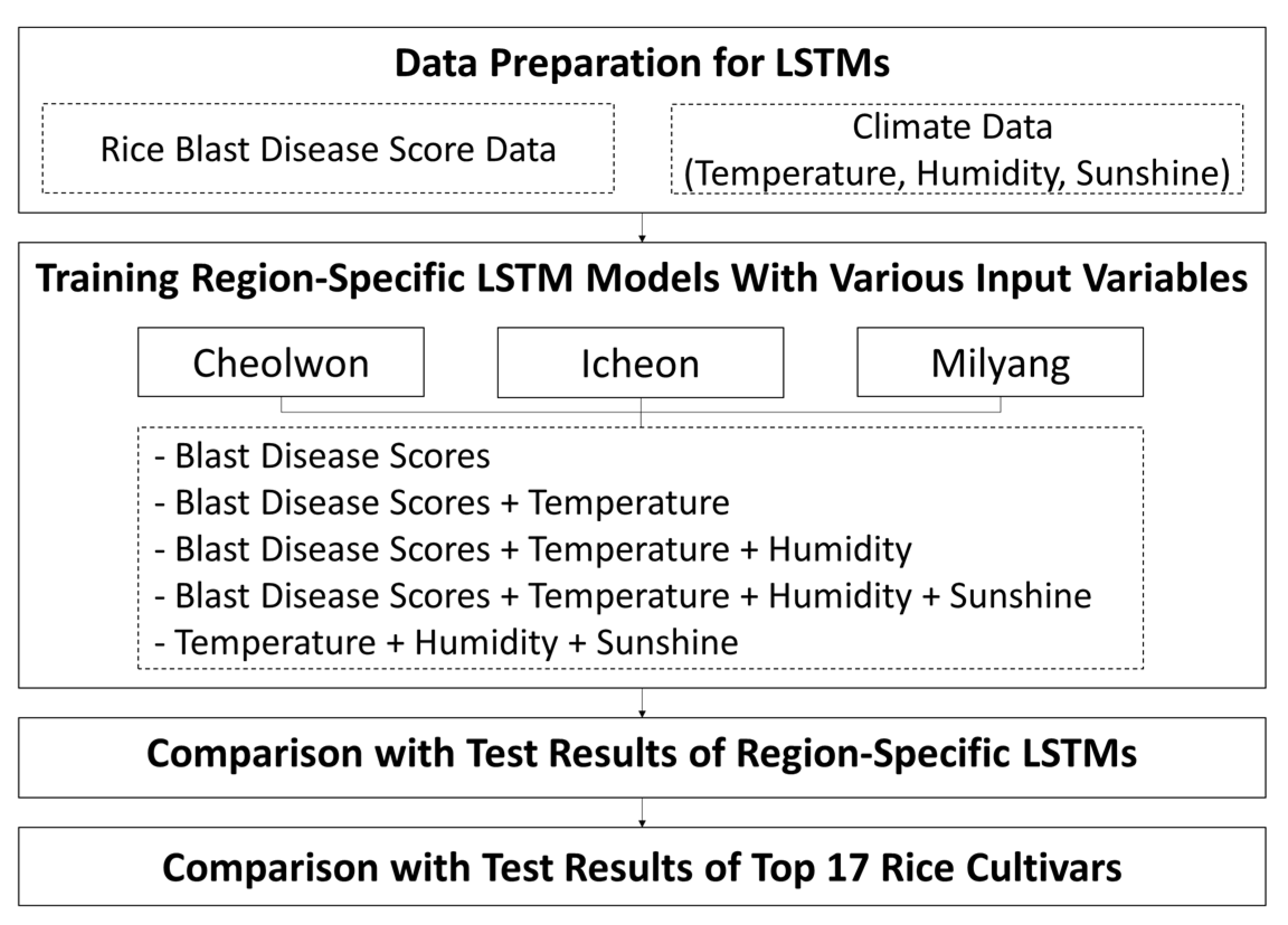
3.1. Data Preparation for LSTMs
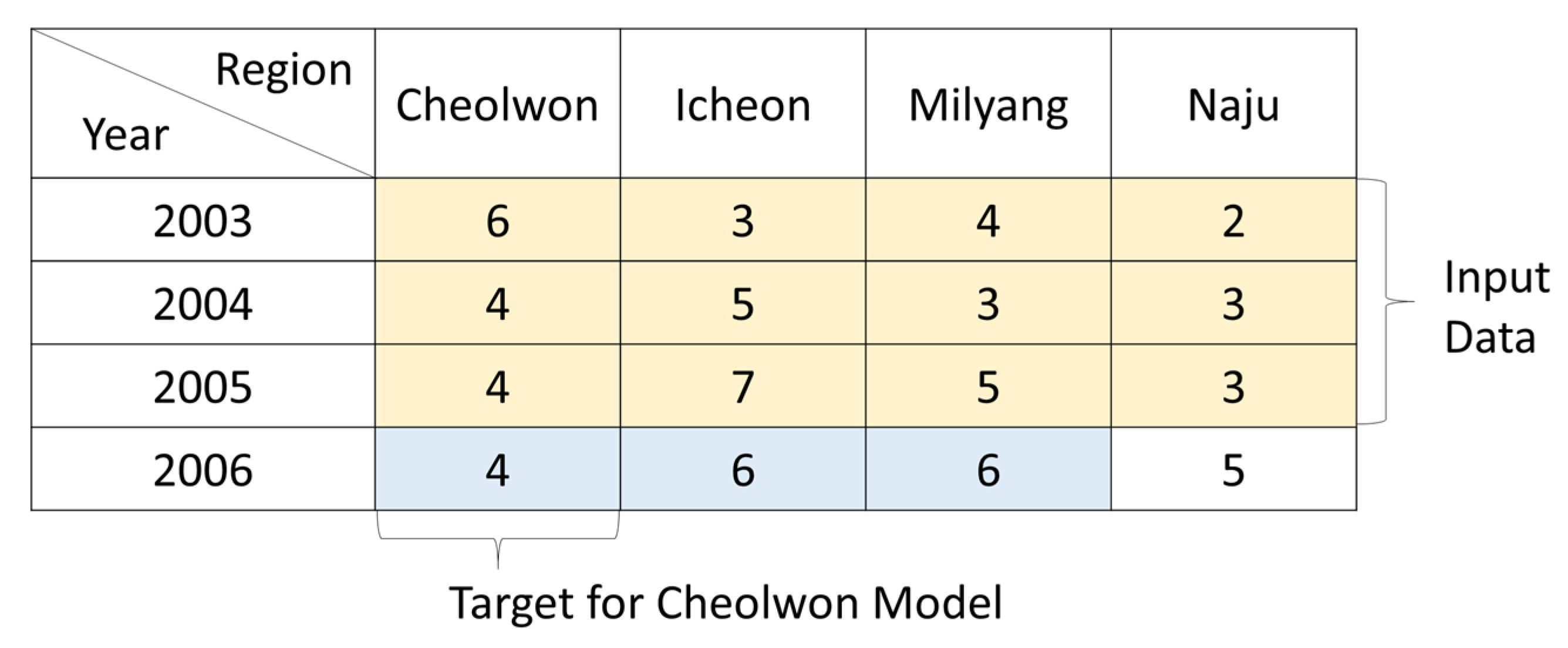
3.1.1. Blast Disease Score Data Preparation
3.1.2. Climate Data Preparation
3.2. Model Design
4. Results and Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, G.-L.; Valent, B. Durable resistance to rice blast. Science 2017, 355, 906–907. [Google Scholar] [CrossRef] [PubMed]
- Kato, H. Rice blast disease. Pestic. Outlook 2001, 12, 23–25. [Google Scholar] [CrossRef]
- Li, Y.B.; Wu, C.J.; Jiang, G.H.; Wang, L.Q.; He, Y.Q. Dynamic analyses of rice blast resistance for the assessment of genetic and environmental effects. Plant Breed. 2007, 126, 541–547. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Ra, D.-S.; Yeh, W.-H.; Choi, H.-W.; Myung, I.-S.; Lee, S.-W.; Lee, Y.-H.; Han, S.-S.; Shim, H.-S. Survey of major disease incidence of rice in Korea during 1999–2008. Res. Plant Dis. 2010, 16, 183–190. [Google Scholar] [CrossRef]
- Cho, Y.C.; Kwon, S.W.; Choi, I.S.; Lee, S.K.; Jeon, J.S.; Oh, M.G.; Roh, J.H.; Hwang, H.G.; Kim, Y.G. Identification of major blast resistance genes in Korean rice varieties (Oryza sativa L.) using molecular markers. J. Crop Sci. Biotechnol. 2007, 10, 265–276. [Google Scholar]
- Han, S.S.; Ryu, J.D.; Shim, H.S.; Lee, S.W.; Hong, Y.K.; Cha, K.H. Breakdown of resistant cultivars by new race KI-1117a and race distribution of rice blast fungus during 1999–2000 in Korea. Res. Plant Dis. 2001, 7, 86–92. [Google Scholar]
- Huhn-Pal, M. Genetic diversity of high-quality rice cultivars based on SSR markers linked to blast resistance genes. Korean J. Crop Sci. 2004, 49, 251–255. [Google Scholar]
- Ishiguro, K.; Hashimoto, A. Recent advances in forecasting of rice blast epidemics using computers in Japan. In Tropical Agriculture Research Series, Proceedings of the 23rd International Symposium on Tropical Agriculture Research, Tsu, Japan, 20–22 September 1989; The Agriculture, Forestry and Fisheries Research Information Technology Center: Ibaraki, Japan, 1989. [Google Scholar]
- Teng, P.S.; Klein-Gebbinck, H.W.; Pinnschmidt, H. An analysis of the blast pathosystem to guide modeling and forecasting. In Rice Blast Modeling and Forecasting, Proceedings of the International Rice Research Conference, Seoul, Korea, 27–31 August 1990; International Rice Research Institute (IRRI): Los Baños, Philippines, 1991. [Google Scholar]
- Kim, C.-K.; Choong, H.K. The rice leaf blast simulation model EPIBLAST. In Systems Approaches for Agricultural Development; Penning de Vries, F., Teng, P., Metselaar, K., Eds.; Springer: Dordrecht, The Netherlands, 1993; pp. 309–321. [Google Scholar]
- Calvero, S.B., Jr.; Coakley, S.M.; Teng, P.S. Development of empirical forecasting models for rice blast based on weather factors. Plant Pathol. 1996, 45, 667–678. [Google Scholar] [CrossRef]
- Katsantonis, D.; Kadoglidou, K.; Dramalis, C.; Puigdollers, P. Rice blast forecasting models and their practical value: A review. Phytopathol. Mediterr. 2017, 56, 187–216. [Google Scholar] [CrossRef]
- Kaundal, R.; Kapoor, A.S.; Raghava, G.P.S. Machine learning techniques in disease forecasting: A case study on rice blast prediction. BMC Bioinform. 2006, 7, 485. [Google Scholar] [CrossRef] [PubMed]
- Malicdem, A.R.; Fernandez, P.L. Rice blast disease forecasting for northern Philippines. WSEAS Trans. Inf. Sci. Appl. 2015, 12, 120–129. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016. [Google Scholar] [CrossRef]
- Taigman, Y.; Yang, M.; Ranzato, M.A.; Wolf, L. Deepface: Closing the gap to human-level performance in face verification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; IEEE: Piscataway, NJ, USA, 2014. [Google Scholar] [CrossRef]
- Graves, A.; Mohamed, A.; Hinton, G. Speech recognition with deep recurrent neural networks. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Vancouver, BC, Canada, 26–31 May 2013; IEEE: Piscataway, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Dauphin, Y.N.; Bengio, Y. Big neural networks waste capacity. arXiv, 2013; arXiv:1301.3583. [Google Scholar]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. In Proceedings of the 27th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; MIT Press: Cambridge, MA, USA, 2014; Volume 2, pp. 3104–3112. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Sak, H.; Senior, A.; Beaufays, F. Long short-term memory recurrent neural network architectures for large scale acoustic modeling. In Proceedings of the Fifteenth Annual Conference of the International Speech Communication Association, Singapore, 14–18 September 2014. [Google Scholar]
- Greff, K.; Srivastava, R.K.; Koutník, J.; Steunebrink, B.R.; Schmidhuber, J. LSTM: A search space odyssey. IEEE Trans. Neural Netw. Learn. Syst. 2017, 28, 2222–2232. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Yue, J.; Rao, Y. A deep learning framework for financial time series using stacked autoencoders and long-short term memory. PLoS ONE 2017, 12, e0180944. [Google Scholar] [CrossRef]
- International Rice Research Institute (IRRI). Standard Evaluation System for Rice (SES); International Rice Research Institute: Los Banos, Philippines, 1988. [Google Scholar]
- Asuyama, H. Morphology, taxonomy, host range, and life cycle of Pyricularia oryzae. In The Rice Blast Disease, Proceedings of a Symposium at the International Rice Research Institute, Los Banos, Philippines, 4–8 February 1963; IRRI, Ed.; Johns Hopkins Press: Baltimore, ML, USA, 1965; pp. 9–22. [Google Scholar]
- Ou, S.H. Rice Diseases; International Rice Research Institute: Los Baños, Philippines, 1985. [Google Scholar]
- Alizadeh, A.; Mousanejad, S.; Safaie, N. Effect of weather factors on sporulation of rice blast disease causal agent in Guilan Province. J. Water Soil Sci. 2009, 13, 315–326. Available online: http://jstnar.iut.ac.ir/article-1-1009-en.html (accessed on 23 December 2017).
- Chetri, D.K.; Daiho, L.; Upadhyay, D.N. Tentative identification of critical weather factors to circumvent leaf blast with altered dates of sowing of rice in the foot-hills of Nagaland, India. Int. J. Bio-Res. Stress Manag. 2011, 2, 298–301. [Google Scholar]
- Bengio, Y.; Simard, P.; Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Netw. 1994, 5, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Fausett, L. Fundamentals of Neural Networks: Architectures, Algorithms, and Applications; Prentice-Hall: Upper Saddle River, NJ, USA, 1994. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Werbos, P.J. Backpropagation through time: What it does and how to do it. Proc. IEEE 1990, 78, 1550–1560. [Google Scholar] [CrossRef]
- Yu, L.; Wang, S.; Lai, K.K. An integrated data preparation scheme for neural network data analysis. IEEE Trans. Knowl. Data Eng. 2006, 18, 217–230. [Google Scholar] [CrossRef]
- Priddy, K.L.; Keller, P.E. Artificial Neural Networks: An Introduction; SPIE Press: Bellingham, WA, USA, 2005; Volume 68. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. arXiv, 2014; arXiv:1412.6980. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv, 2016; arXiv:1603.04467. [Google Scholar]
- Bohnert, H.U.; Fudal, I.; Dioh, W.; Tharreau, D.; Notteghem, J.L.; Lebrun, M.H. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 2004, 16, 2499–2513. [Google Scholar] [CrossRef] [PubMed]
- Farman, M.L.; Eto, Y.; Nakao, T.; Tosa, Y.; Nakayashi, H.; Mayama, S.; Leong, S.A. Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol. Plant-Microbe Interact. 2002, 15, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; McAdams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct interaction of resistance gene and avirulene gene products confers rice blast resistance. EMBO J. 2000, 37, 554–565. [Google Scholar] [CrossRef]
- Kang, S.; Sweigard, J.A.; Valent, B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 1995, 8, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, B.; Wu, J.; Lu, G.; Hu, Y.; Zhang, X.; Zhang, Z.; Zhao, Q.; Feng, Q.; Zhang, H.; et al. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 2009, 22, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Orbah, M.J.; Farrall, L.; Sweigard, J.A.; Chrmley, F.G.; Valent, B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2019–2032. [Google Scholar] [CrossRef]
- Park, S.Y.; Milgroom, M.G.; Han, S.S.; Kang, S.; Lee, Y.H. Genetic differentiation of Magnaporthe oryzae populations from scouting plots and commercial rice fields in Korea. Phytopathology 2008, 98, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Go, J.; Kang, I.J.; Shim, H.-W.; Shin, D.B.; Heu, S.; Roh, J.-H. Distribution of rice blast disease and pathotype analysis in 2014 and 2015 in Korea. Res. Plant Dis. 2016, 22, 264–268. [Google Scholar] [CrossRef]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pest. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef] [PubMed]





| Cultivar | Icheon | Suwon | Cheolwon | … | Milyang | Sangju | Naju |
|---|---|---|---|---|---|---|---|
| Nampyung | 9 | 1 | 3 | … | 3 | 7 | 2 |
| Ilpum | 3 | 1 | 5 | … | 7 | 8 | 0 |
| Dongang | 0 | 1 | 3 | … | 5 | NA | 2 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| Date (YYYY-MM-DD) | Average Temperature (°C) | Relative Humidity (%) | Sunshine Hours (h) |
|---|---|---|---|
| 2003-06-01 | 20.4 | 57 | 11.9 |
| 2003-06-02 | 19.7 | 46 | 12 |
| ⋮ | ⋮ | ⋮ | ⋮ |
| 2016-07-31 | 26.1 | 89 | 5.7 |
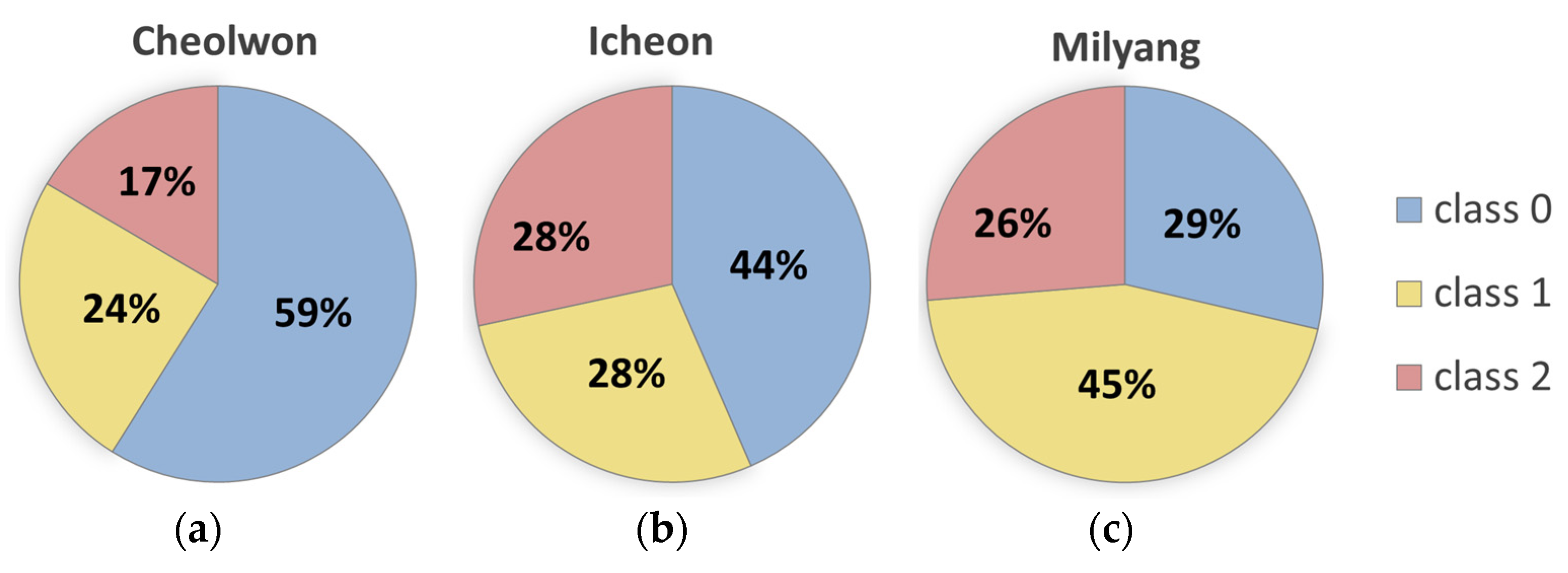
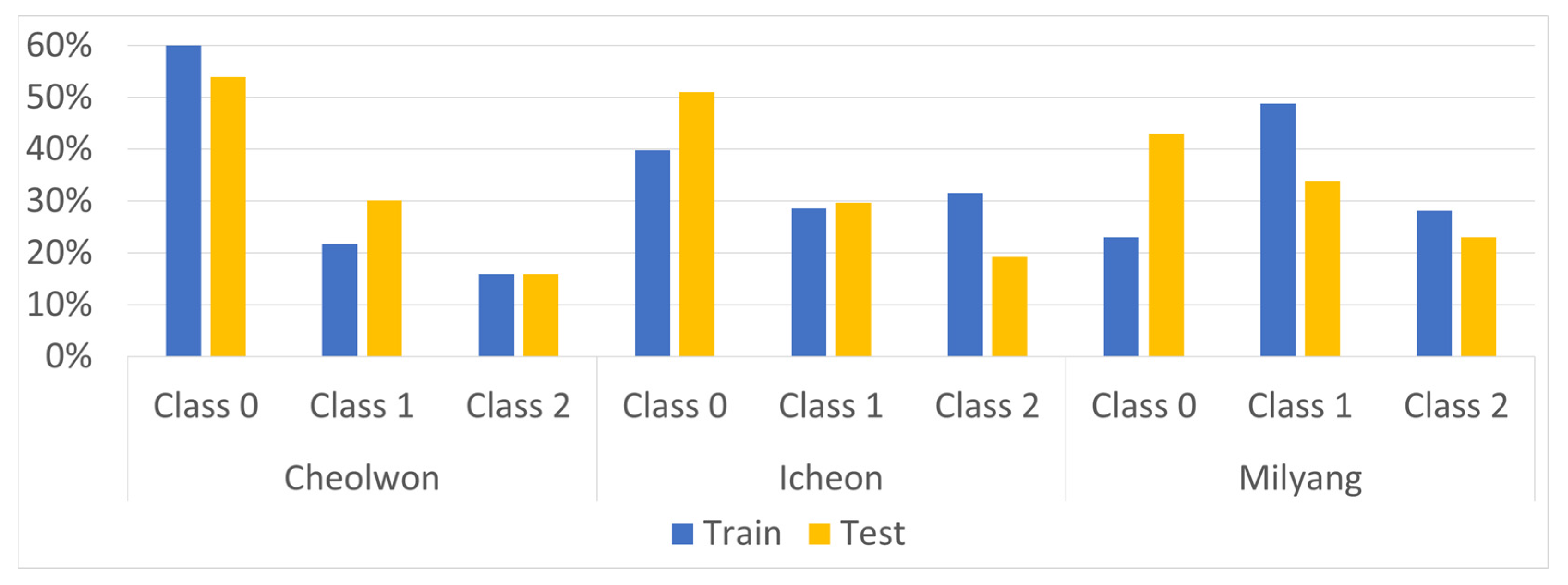
| Region | Class | Train | Validation | Test |
|---|---|---|---|---|
| Cheolwon | Class 0 | 518 (62%) | 55 (46%) | 129 (54%) |
| Class 1 | 182 (22%) | 38 (32%) | 72 (30%) | |
| Class 2 | 133 (16%) | 26 (22%) | 38 (16%) | |
| Icheon | Class 0 | 332 (40%) | 64 (54%) | 122 (51%) |
| Class 1 | 238 (29%) | 26 (22%) | 71 (30%) | |
| Class 2 | 263 (32%) | 29 (24%) | 46 (19%) | |
| Milyang | Class 0 | 192 (23%) | 46 (39%) | 103 (43%) |
| Class 1 | 407 (49%) | 49 (41%) | 81 (34%) | |
| Class 2 | 234 (28%) | 24 (20%) | 55 (23%) |
| Model Variation | Input Variables (size) | Input Size per Time Step |
|---|---|---|
| Blast_LSTM | Blast disease scores for four regions (4) | 4 |
| BlastT_LSTM | Blast disease scores for four regions (4) + Target region temperature (4) | 8 |
| BlastTH_LSTM | Blast disease scores for four regions (4) + Target region temperature (4) + Target region humidity(4) | 12 |
| BlastTHS_LSTM | Blast disease scores for four regions (4) + Target region temperature (4) + Target region humidity (4) + Target region sunshine hours (4) | 16 |
| Climate_LSTM | Target region temperature (4) + Target region humidity (4) + Target region sunshine hours (4) | 12 |
| Model Name | Cheolwon | Icheon | Milyang | |||
|---|---|---|---|---|---|---|
| Accuracy | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | |
| Blast_LSTM | 62.3% | 59.6% | 61.5% | 59.8% | 46.9% | 44.9% |
| BlastT_LSTM | 65.7% | 61.0% | 62.8% | 60.7% | 51.0% | 51.1% |
| BlastTH_LSTM | 66.9% | 63.4% | 62.8% | 61.4% | 52.3% | 52.6% |
| BlastTHS_LSTM | 67.4% | 63.6% | 63.2% | 62.1% | 54.4% | 53.4% |
| Climate_LSTM | 55.2% | 49.7% | 52.7% | 46.1% | 44.4% | 38.0% |
| Model | Cheolwon | Icheon | Milyang |
|---|---|---|---|
| Blast_LSTM | 69.1% | 60.3% | 44.4% |
| BlastT_LSTM | 72.1% | 60.3% | 48.1% |
| BlastTH_LSTM | 77.9% | 61.8% | 48.1% |
| BlastTHS_LSTM | 79.4% | 64.7% | 55.6% |
| Climate_LSTM | 45.6% | 54.4% | 40.7% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Roh, J.-H.; Kim, H.Y. Early Forecasting of Rice Blast Disease Using Long Short-Term Memory Recurrent Neural Networks. Sustainability 2018, 10, 34. https://doi.org/10.3390/su10010034
Kim Y, Roh J-H, Kim HY. Early Forecasting of Rice Blast Disease Using Long Short-Term Memory Recurrent Neural Networks. Sustainability. 2018; 10(1):34. https://doi.org/10.3390/su10010034
Chicago/Turabian StyleKim, Yangseon, Jae-Hwan Roh, and Ha Young Kim. 2018. "Early Forecasting of Rice Blast Disease Using Long Short-Term Memory Recurrent Neural Networks" Sustainability 10, no. 1: 34. https://doi.org/10.3390/su10010034
APA StyleKim, Y., Roh, J.-H., & Kim, H. Y. (2018). Early Forecasting of Rice Blast Disease Using Long Short-Term Memory Recurrent Neural Networks. Sustainability, 10(1), 34. https://doi.org/10.3390/su10010034




