1. Introduction
Since the spread of the COVID-19 pandemic in early 2020, a clear deficiency appeared in humans’ ability to reduce the spread of infection caused by this viral disease and to afford suitable medication for patients. Consequently, health organizations faced a challenging situation and health crises emerged. A few months later, China recorded a high death rate with a very low recovery rate. Since that, the knowledge about this novel virus has been developed, but is still not enough. An essential need for new diagnostics and medication to reduce or prevent virus transmission must be considered. Different vaccination types have been developed, in addition to applying the precautionary measures in many countries that resulted in increasing the recovery rate and reducing death cases, see [
1]. Biomaterials science has made a great contribution to the enhancement of new and personal protective tools, vaccines, and drug delivery which can be used to treat COVID-19 and its side effects. Furthermore, tissue engineers and cell-biology models can have a great influence on inspecting the disease, testing, and improving new vaccines, see [
2]. One of the main factors that indicate the improvement of the treatment process is the recovery rate; hence, modelling this factor is important and it will lead to many useful results and the development of biomedical research related to COVID-19 or any other viral disease. Modelling real-life phenomena from a statistical point of view have been widely considered in the literature. For example, one may refer to the book [
3], in which the author studied the factors affecting the degradation of an organic coating by forecasting the remaining lifetime of the coating. In many scientific fields, statistical models are required to describe the trend of the data under consideration. For the purpose of modeling real data, several new generators of distributions appeared in the literature. The most attractive ones are the beta and Kumaraswamy distributions, see [
4,
5]. However, since more data became available and spread quickly, there is a persistent need for new families of distributions. A unit distribution is a type of distribution that is usually used to model the extreme values in a population. It is characterized by a power-law relationship, where the probability of observing a value within a certain range decreases exponentially as the value increases. Consequently, more studies about unit modeling appeared in the literature. Generally, new unit distributions are formulated by transforming some familiar continuous distributions, which are more flexible than the original ones, without adding new parameters. For more examples of unit distributions, one may refer to the unit Weibull [
6,
7], the unit Gompertz [
8], the unit Birnbaum–Saunders [
9], the unit inverse Gaussian [
10], the unit generalized half normal [
11], the unit Rayleigh [
12], the unit Burr-XII [
13], the unit Chen [
14], and the unit power Weibull [
15] distributions.
In the literature, the Pareto (P) distribution is a well-known statistical continuous model that describes heavy-tailed data. Ref. [
16] explained the P distribution’s applications in modeling earthquakes, forest-fire areas, and oil- and gas-field sizes. Various generalizations appeared in the research work, which emphasizes the flexibility of the P model. For example, the Weibull-P distribution [
17], the Gamma-P distribution [
18], the beta-P distribution [
19], the beta generalized P distribution [
20], the Kumaraswamy exponentiated P distribution [
21] and a new Weibull-P distribution [
22]. One may refer to [
23,
24,
25,
26] for more generalized P distributions.
Some unit models were obtained using other transformation methods, see, for example, [
27,
28,
29,
30,
31,
32].
Ref. [
33] studied a new model based on the exponential and P distributions. The cumulative density function (CDF) and the probability density function (pdf) of the exponential P distribution (EPD) are given, respectively, as
and
where
,
is a positive shape parameter, and
and
are positive scale parameters. The EPD is a very flexible model and has many applications in different fields. However, in this paper, we introduce a new statistical model that has more flexibility than the EPD in many different fields and the application section recommended our claims. The newly suggested model is called the unit exponential Pareto distribution.
One of the main motivations for using the unit exponential Pareto distribution is its ability to capture the long-tailed nature of many real-world data sets. In addition to its ability to model long-tailed data, the unit exponential Pareto distribution is also useful for identifying patterns and trends in data. By analyzing the shape of the distribution, insight can be gained into the underlying factors that drive the data and informed decisions made about how to best allocate resources or make predictions about future observations.
The basic goal of this work is to introduce the new unit exponential Pareto distribution (UEPD), study its statistical properties, and explore the ability of UEPD by applying it to three real-life data examples. The stimulus for studying the UEPD are: (i) UEPD is a distinguished model since it is constructed on the bases of the unit interval [0,1] rather than the positive real numbers; (ii) it has remarkable flexibility with respect to tail features; hence, it is used in risk evaluation theory with relatively improved outputs; and (iii) the potency of the new mixture of the well-known exponential and Pareto distributions appears in modeling and fitting many real-life data with relatively fewer errors than other competitive models. This new model proves the suitability of fit to the COVID-19 recovery rate in two countries, Turkey and France, during the pandemic, in addition to the modeling production rate and failure times of components as an application in economic and engineering fields.
The rest of the article is presented as follows:
Section 2 investigates the UEPD and discusses the basic properties of this distribution. In
Section 3, some important distributional and statistical features of the UEPD are studied, such as the quantile function (
), moments (
s), moment-generating function (
), some entropy measures, order statistics (
), stress–strength (
), and stochastic ordering (
). Classical estimation methods are discussed in
Section 4, while the Bayesian approach is implemented in
Section 5.
Section 6 presents the results of examining three real-skewed data sets.
Section 7 investigates the performance of the suggested estimation techniques using Monte Carlo simulations. Finally,
Section 8 summarizes the relevant conclusions and remarks.
2. Unit Exponential Pareto Distribution
Assume
Y is a random variable (
) which follows the EPD with parameters
and
. Applying the transformation
then, the new unit distribution is created. The pdf and the CDF of the transformed variable
X are provided via
and
where
,
is the shape parameter and
and
are non-negative scale parameters.
Figure 1 represents the shape of the UEPD distribution, which is decreasing, upside down with uni-modal, skewed to the left, and symmetric for some selected parameter values.
The survival function (SF) is a function that provides the probability that a particular object will survive after a specific time. The term survival function is extensively used in human mortality to show the survival time of a patient beyond a specific time. Reliability analysis is used to show the performance of electric devices beyond a fixed time. The SF for the UEPD is provided as
The hazard function (HF) for the UEPD is obtained as
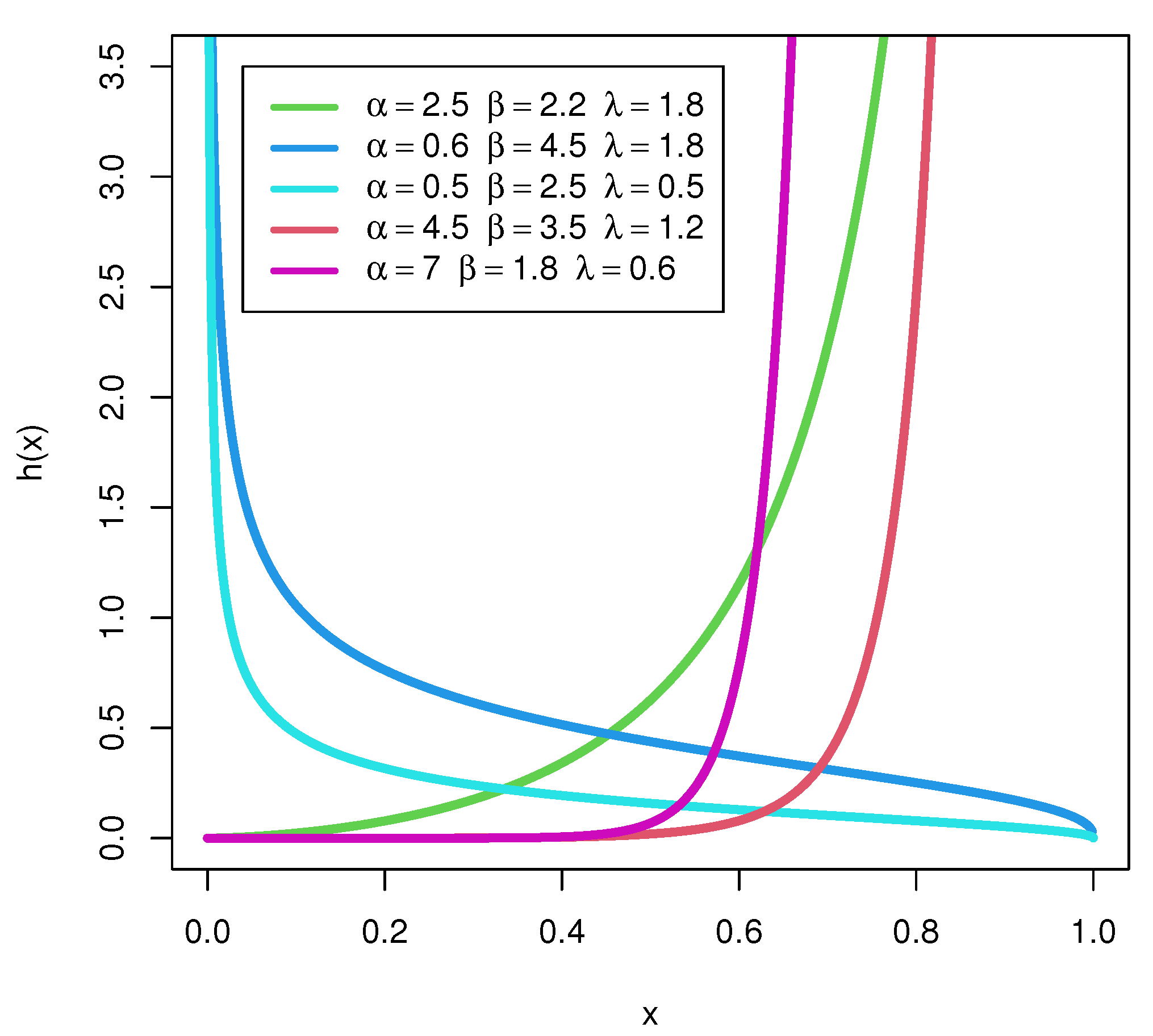
Figure 2 depicts the HFs of the UEPD distribution. It is noticed that the distribution has decreasing HFs if
, while it has increasing HFs for
, and is constant for
, with different values for the other two parameters. This shows the flexibility of the UEPD. The cumulative HF of the UEPD is obtained by
The reversed HF (RHF) is another important tool in reliability analysis and it is calculated as
3. Statistical Properties
Some important structural and statistical properties of the UEPD are investigated in this section.
3.1. Quantile Function
The
of the UEPD is a useful tool to perform a simulated sample and it can be calculated by solving
with respect to x, where
, that is,
. The simplified form of the
of the UEPD is provided by
The
th quartile function of the UEPD is defined as
For
, Equation
reveals the median of the UEPD
3.2. Moments
We derive the
rth Moment (
) for the UEPD. The first four
s are the most important to describe the shape and monotonicity of the distribution curve. Suppose
X via a
that follows
(
); then, the
rth
about the origin of the UEPD is provided via
By substituting Equation (
3) into Equation (
12), we obtain
where
Using the binomial expansion on
we obtained an expression for the moment as
It is important to note that the
s exist only when
In particular, the first four
s of UEPD are written as
and
As a result, the mean and variance are derived by
and
Table 1 shows some numerical values of the first four moments
,
,
,
; the variance (var); skewness (SK); kurtosis (KU); coefficient of variation (CV) and index of dispersion (ID).
3.3. The Moment-Generating Function
The
-generating function of the UEPD can be calculated from
As a result, using the Taylor series to extend
produces
3.4. Different Types of Entropy
Entropy measures are important measures in reliability and risk-analysis studies. It has been used in a wide range of applied biological, medical and physical fields.
3.4.1. Rényi Entropy
The essential shape of the distribution is measured using Rényi entropy (RE) [
34] and it is provided by
For UEPD, Rényi entropy is obtained as
where
Then
3.4.2. Havrda and Charvat Entropy
The Havrda and Charvat entropy (HCE) [
35] measure is provided via
For UEPD, Havrda and Charvat entropy is obtained as
where
Then
3.4.3. Tsallis Entropy
The Tsallis entropy (TE) [
36] measure is provided by
For UEPD, Tsallis entropy is obtained as
where
Then
3.4.4. Arimoto Entropy
The Arimoto entropy (AE) [
37] measure is provided by
For UEPD, Arimoto entropy is obtained as
where
Then
3.4.5. Mathai–Haubold Entropy
The Mathai–Haubold entropy (MHE) [
38] measure is provided by
For UEPD, Mathai–Haubold entropy is obtained as
where
Then
Table 2 and
Table 3 show some numerical values of the different types of entropy.
3.5. The Order Statistics
Assume (
) is a random sample of size n from the UEPD. Let
denote the corresponding
s. The pdf and the CDF of the
rth
,
,
, are given, respectively, as:
and
where
denote Beta function.
One can obtain the minimum and maximum ordered statistics by assuming and , respectively.
3.6. Stress–Strength
The
(R) is defined as
. In lifetime models, R depicts the lifetime of a component with random stress
X and a related random strength
Y. The component fails when the applied stress exceeds the component’s strength, and the component functions normally when
. Readers may find additional information in [
39].
Let
X and
Y be two independent
s, where
X follows the
(
) and
follows the
(
). Hence,
R is computed as below
where
Utilizing the exponential and the binomial expansions, we obtain
3.7. Stochastic Ordering
Assume the pdf, CDF, hrf, and the mean residual life function (MRL) of a non-negative continuous X are symbolized with and , respectively, and those of Y by and , respectively.
A X is lower than the Y with respect to the next definitions
The , () when , for all .
The HF order, () when , for all .
The MRL order, () when , for all .
The likelihood ratio order, () when is decreasing in x.
The following are some implications that are strongly supported:
()⇒() ⇒() and () ⇒ ().
Next, we show the for the UEPD. Let X have the () and have the ().
For
, the likelihood ratio function is
Consider log
and take the derivative with respect to
, we obtain
If , then which proves the ordered relation as: ()() () and ().
4. Maximum Likelihood Estimators
The maximum likelihood () estimation approach is the most popular classical inference in statistics. In this section, we estimate the parameters of the UEPD using this method. Let , ,…, represent a random sample with UEPD and the observed values , ,…, with = () representing the vector of parameters.
The log-likelihood function for the UEPD is computed as
The
estimates (
s) of
and
, say
and
, are then calculated by maximizing
regarding to the parameters. Therefore, the next nonlinear system of equations needs to be solved in terms of
, and
and
From Equation (
26), the
is obtained as function of
and
; therefore, it is given as
Fisher’s information matrix is utilized here to find the asymptotic CONIs for the UEPD’s parameters. The variance-covariance values are located in the entries of the inverse Fisher information matrix. Consequently, the information matrix that was seen is
where
and
The
is approximately normal and has a mean
and covariance matrix
; for more details, see [
40,
41,
42]. Then, for
and
the
CONIs can be computed as
where
is the
lower percentile of standard normal distribution.
5. Maximum Product Spacings
An alternative estimation method to the
is the maximum product spacings (
) method. Cheng and Amin [
43] proposed this method, and Ranneby [
44] independently developed it as an approximation to the Kullback–Leibler divergence measure. The
method relies on the assumption that the spacings between CDF values at successive points of data must be identically distributed. Assume the ordered observations of random variables are denoted by
,
,…,
and they follow the UEPD. The geometric mean of the differences is expressed as
where, the difference
is provided via
note that
and
The
estimation is achieved by maximizing the geometric mean (GM) of the differences; hence,
and
are the
estimators of the
and
parameters, respectively. Substituting the CDF of the UEPD in Equation (
29) and taking the logarithm resulted with
The
s are evaluated by solving the following nonlinear equations simultaneously
and
where
and
6. Bayesian Estimation
The Bayesian estimation method is a popular non-classical inference in statistics. In this method, the distribution parameters are considered as random variables and define uncertainties on the parameters using a joint prior distribution with some suggested loss functions, such as the squared error loss function (SEL). The parameters
,
, and
are considered to be independent and follow the gamma prior distributions.
The hyper-parameters and are assumed to be known non-negative values.
The joint posterior distribution of
and
can be produced using the following formula
The SEL function is considered as a symmetrical loss function that assumes equal loss to the over- and the underestimation. If
is the parameter to be estimated by an estimator
, then the SEL function is described as
Under the SEL function, the Bayes estimate of any function of
, and
, say
, can be measured as
where
To evaluate the triple integrals in Equation (
32), numerical techniques are used; with some algebraic simplifications of Equation (
32), the joint posterior distribution is provided by
The posterior conditional for
and
are
and
However, the conditional posteriors in Equations (36)–(38) are not in standard forms; hence, Gibbs sampling is not a good option. Therefore, the use of Metropolis–Hasting (M-H) sampling is required for the implementation of the MCMC technique. The steps for the M-H algorithm are written as
Start with initial values
Let
Use the M-H algorithm to generate and from
and
with the normal distributions
Generate a required from from and from
- (i)
Find the acceptance probabilities
- (ii)
From the uniform distribution, generate , and
- (iii)
If , accept the proposal and set ; else, set .
- (iv)
If , accept the proposal and set ; else, set .
- (v)
If , accept the proposal and set ; else, set
Set
Repeat steps N times and obtain and
To compute the CRs of as then the CRIs of is
To ensure convergence and remove the affection of initial value selection, the first
M simulated variants are discarded. The chosen samples are then
, for sufficiently large
N. The approximate Bayes estimates of
, depending on the SEL function, are provided by
7. Applications
In this section, the new UEPD is used to model different real data examples from various fields of science. When using the unit distribution, researchers should be aware of some limitations related to the features of this distribution, for example (i) Limited range of applicability: The UEPD is only applicable to data sets with a long tail of extreme values that follow a power law. If the data does not follow this pattern, the distribution may not be an appropriate model. (ii) Assumptions about the data: The UEPD assumes that the data follow a particular distributional form, and any deviations from this form may lead to inaccurate results.
Different distributions are suggested for comparison with UEPD, such as EPD by [
33], the unit-Weibull (UW) distribution by [
6], Kumaraswamy by [
44], beta by [
45], Kumaraswamy Kumaraswamy (KK) by [
46], Marshall-Olkin Kumaraswamy (MOK) by [
47], Marshall-Olkin extended Topp–Leone (MOETL) by [
48], unit-Gompertz (UG) by [
8], unit generalized log-Burr XII (UGLBXII) by [
49], Topp–Leone (TL) [
50], and unit gamma/ Gompertz (UGG) [
51].
Three measures of goodness of fit are used, the Kolmogorov-Smirnov (KS), Cramér–von Mises (CVM), and Anderson–Darling (AD) test. In addition, the p-value of the KS test (PVKS) is evaluated for reliable results.
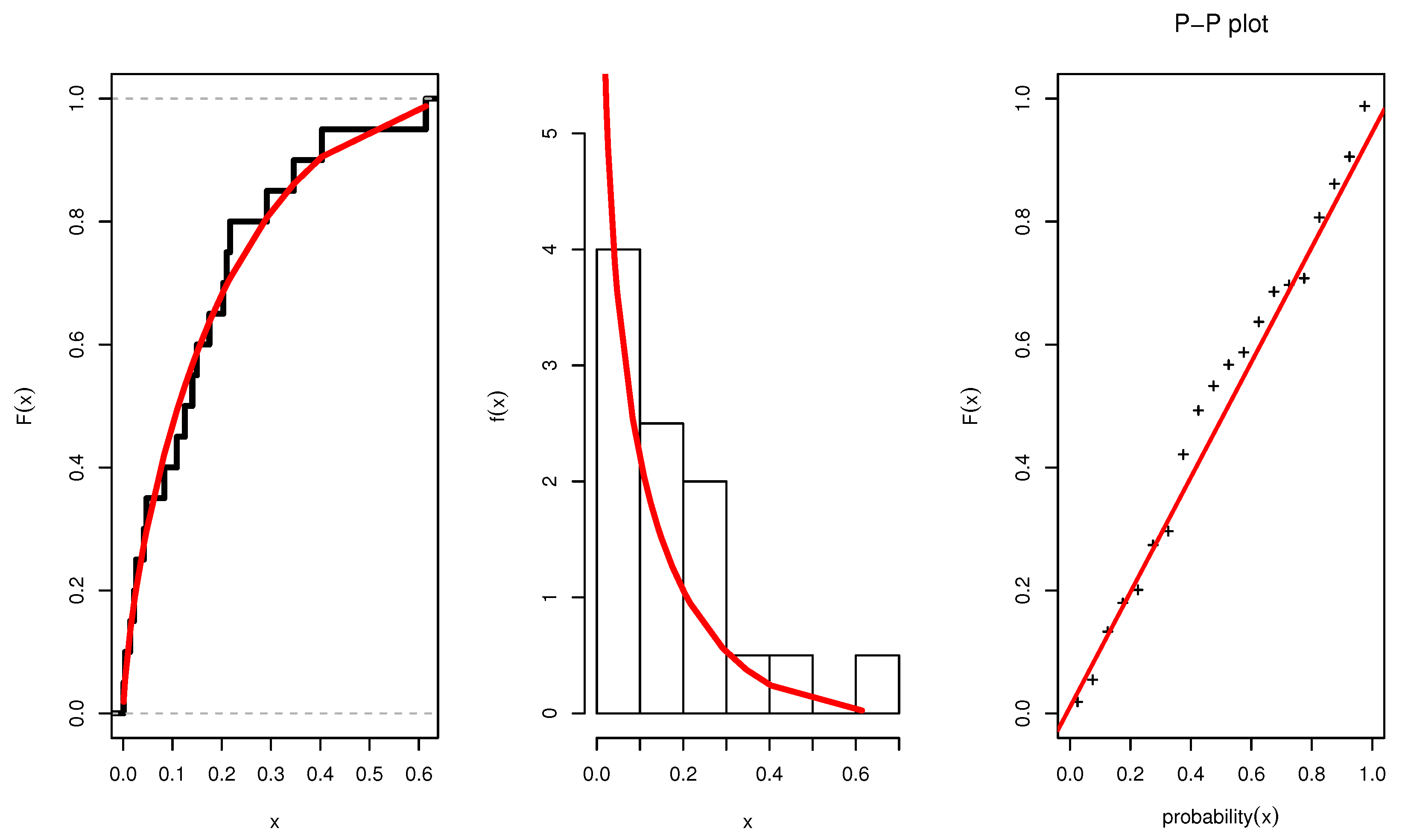
7.1. Recovery Rate of COVID-19 in Turkey
We are interested in studying the recovery rate of COVID-19 during the pandemic. To do so, recovery rates from Turkey were collected, see
Table 4. The World Health Organization (WHO) confirmed the first death from COVID-19 in Turkey occurred on 17 March 2020, with the first official recoveries from the pandemic virus occurring on 26 March 2020. On 20 April 2020, the Ministry of Health confirmed that the total number of cases had risen to 90,980, with 2140 deaths. As of 20 April, the total number of tests performed was 673,980. From 27 March to 20 April, 25 observations were made and the daily ratio of total recoveries calculated to the total number of confirmed cases in Turkey. Accessed by
https://en.wikipedia.org/wiki/COVID-19_pandemic_in_Turkey#Cumulative_cases,_recoveries,_and_deaths.
In
Table 5, eight different distributions are suggested for comparison with the UEPD. It is noticed that in all the measures of goodness of fit, the minimum value is attained for the UEPD, with a large value of PVKS (
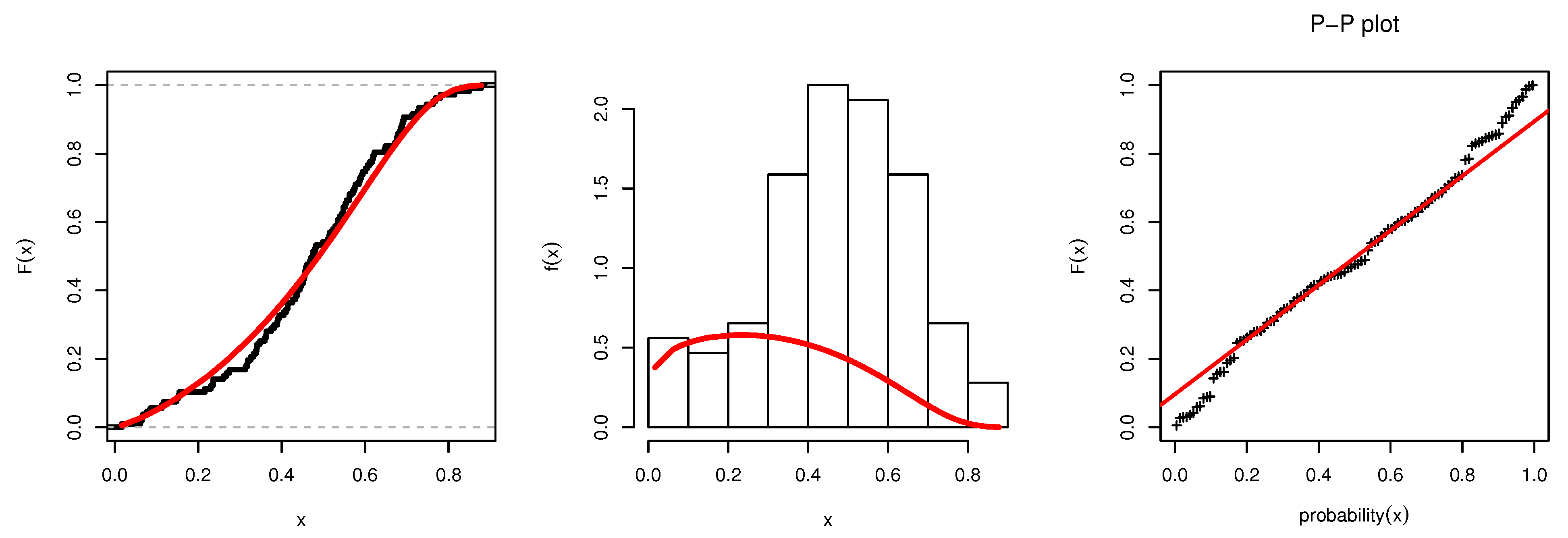
). This indicates its suitability and efficiency to model the recovery rate of COVID-19 data from Turkey, rather than using the other competitive distributions.
From
Table 6, it is noted that the values of estimators under the three methods of estimation are relatively close; in addition, the
asymptotic CONIs and the CRIs are computed for the parameters
,
and
. The lengths of the confidence intervals reveal that the shortest length is obtained when using the Bayesian credible interval.
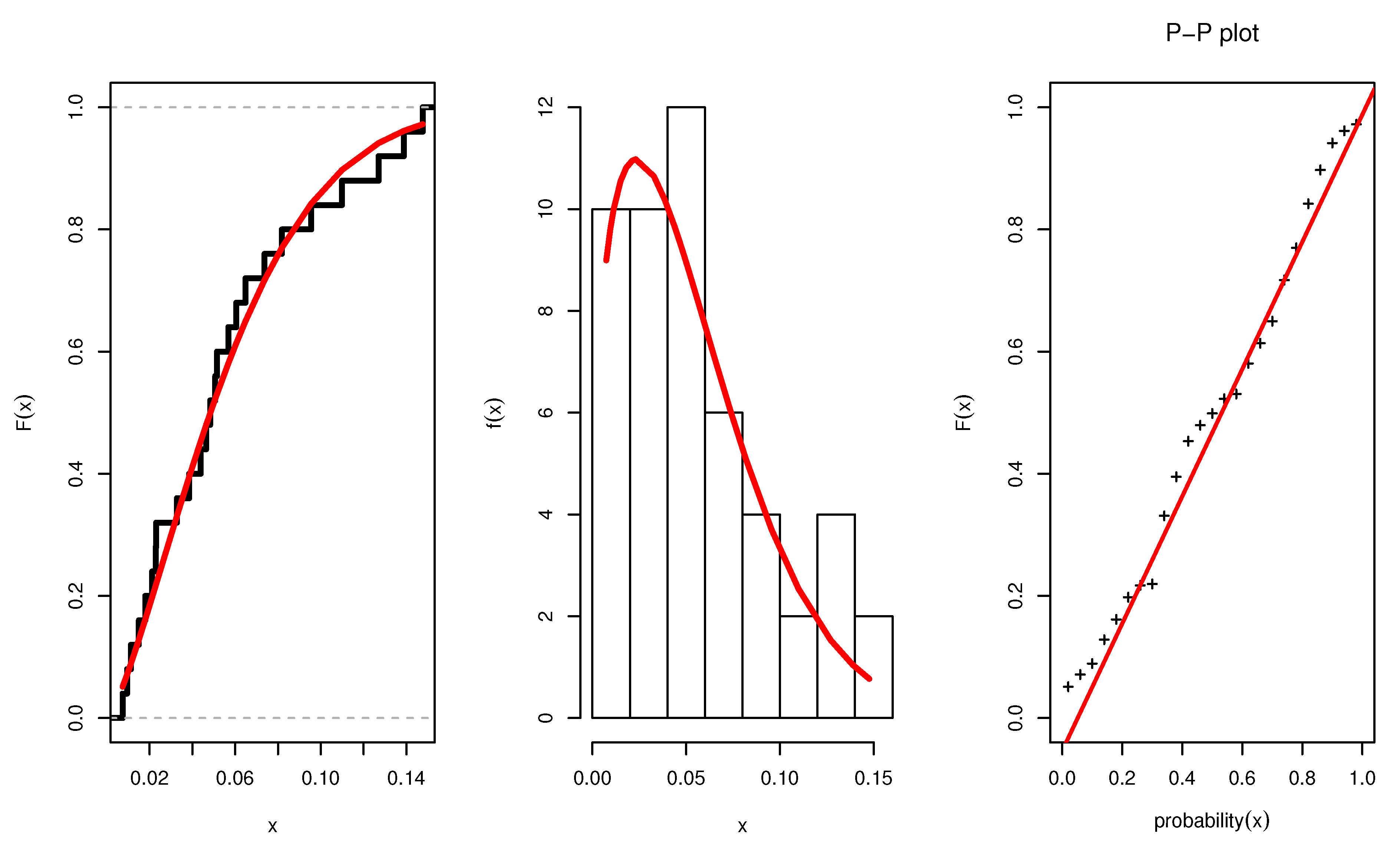
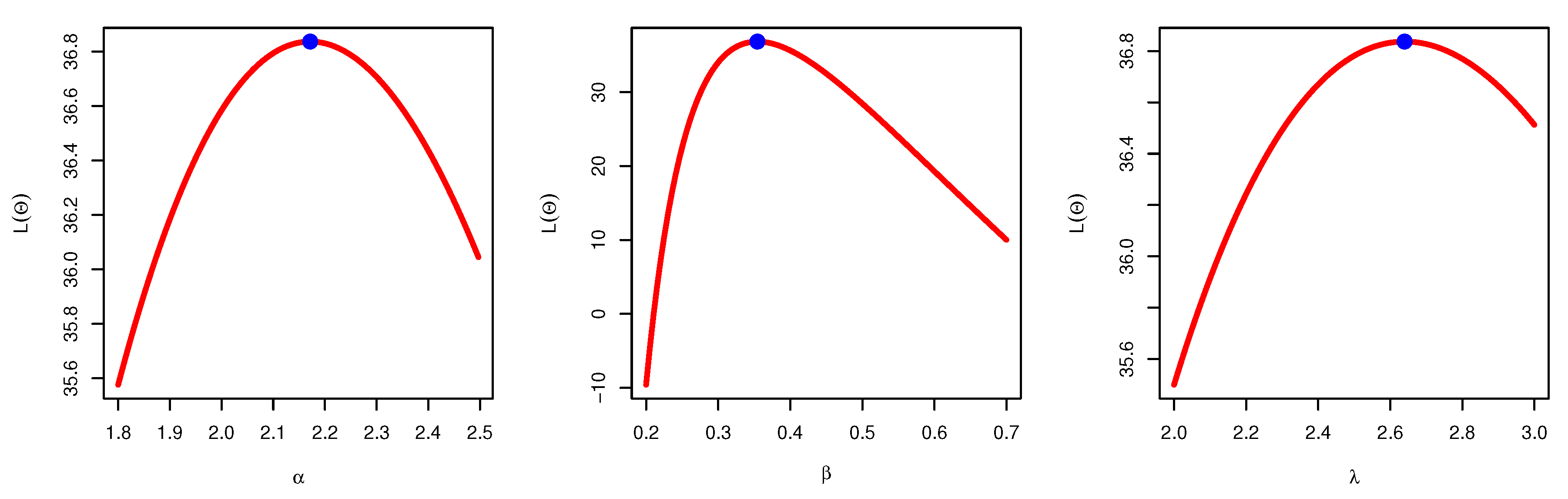
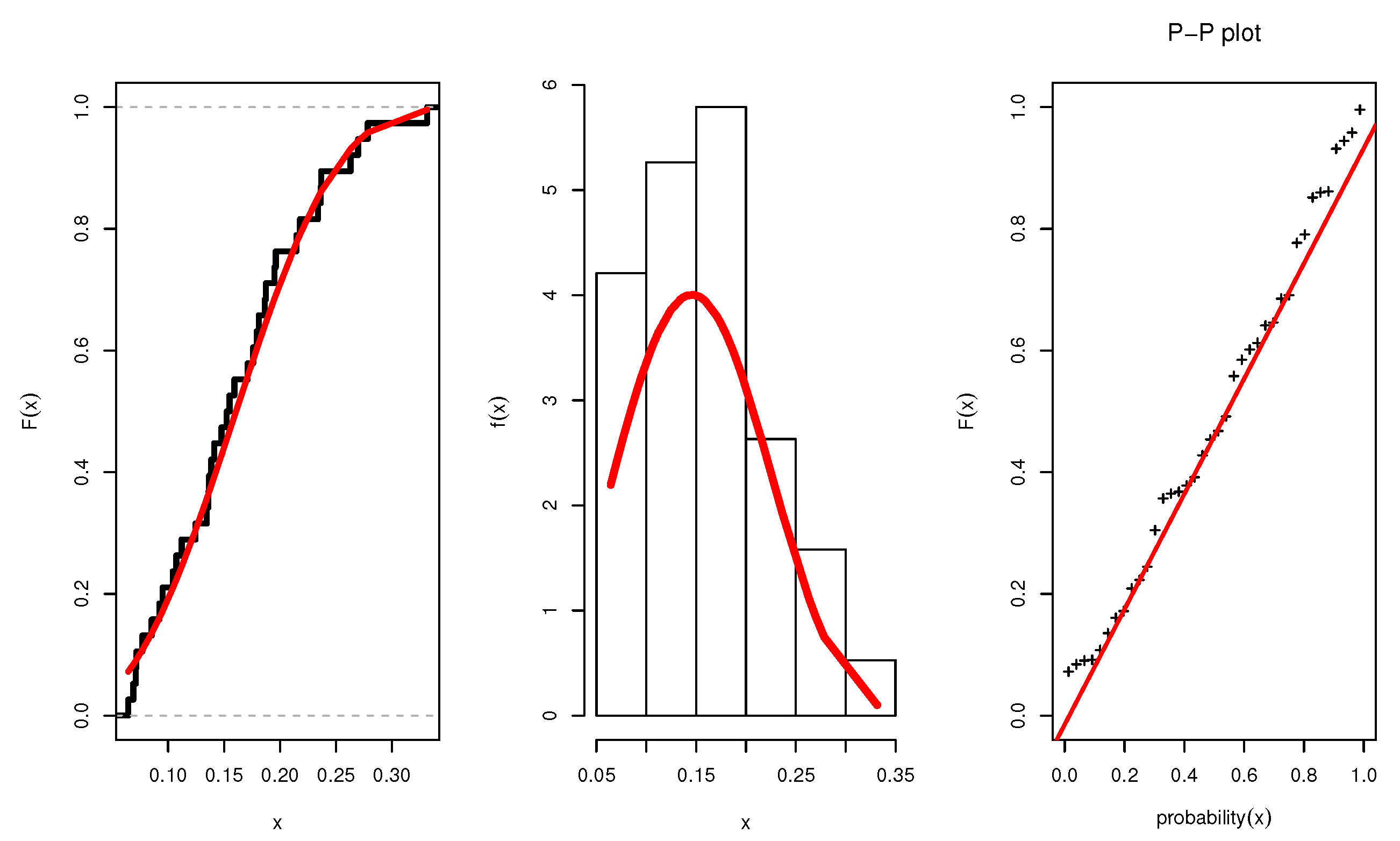
Figure 3 illustrates the plots of the estimated pdf, CDF, and P-P plots of UEPD for the recovery rate of COVID-19 data from Turkey. The profile likelihood appears in
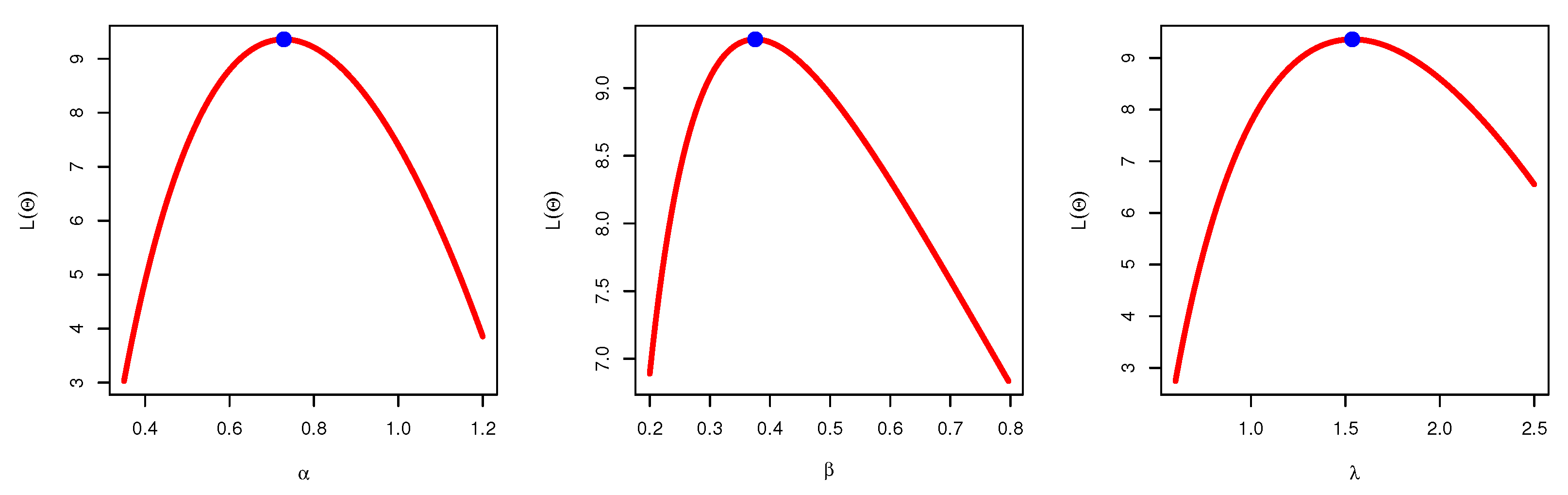
Figure 4, which indicates the uniqueness of the maximum values, and demonstrates the similarity of these maximum values to the MLEs which are obtained in
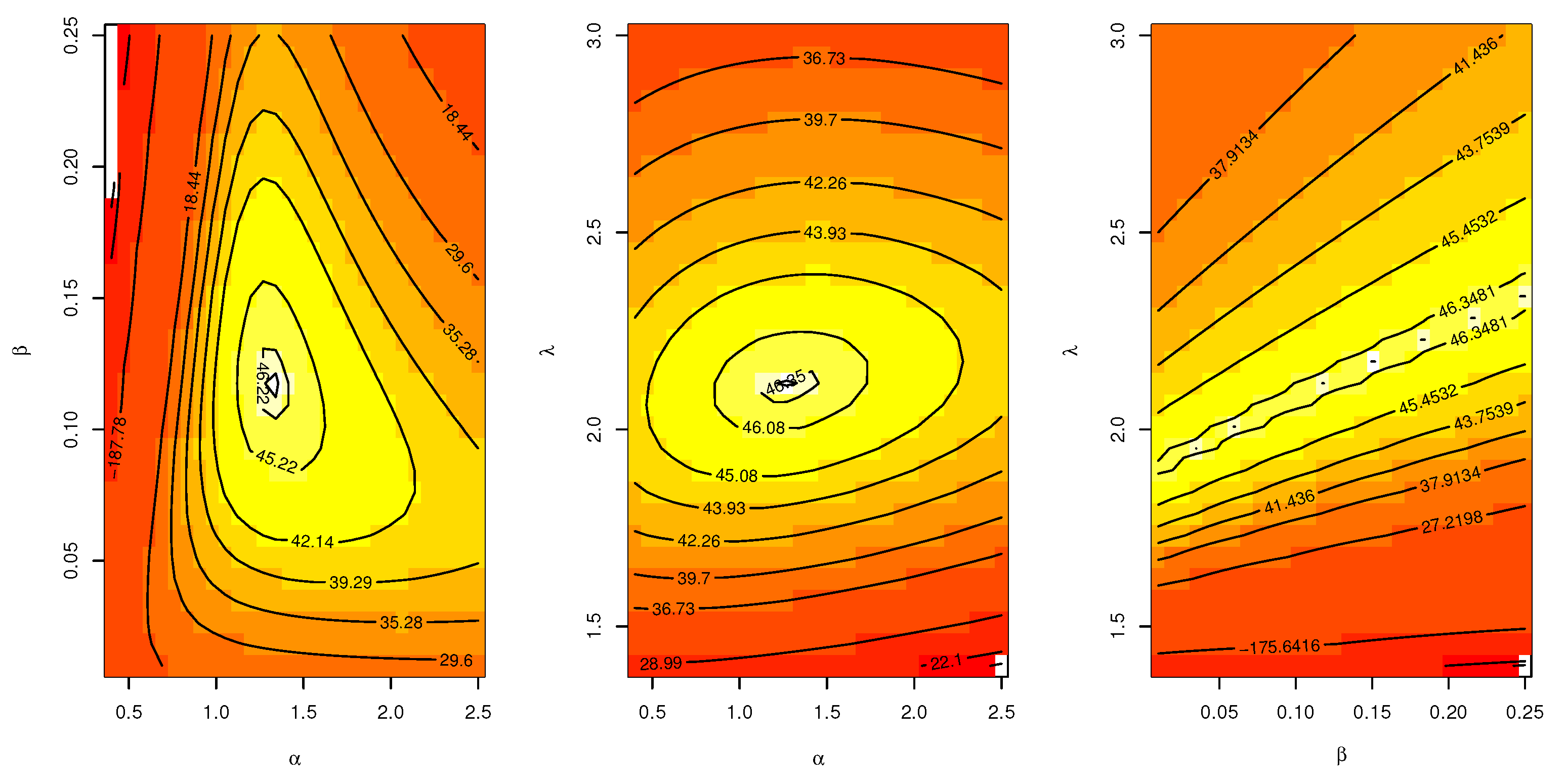
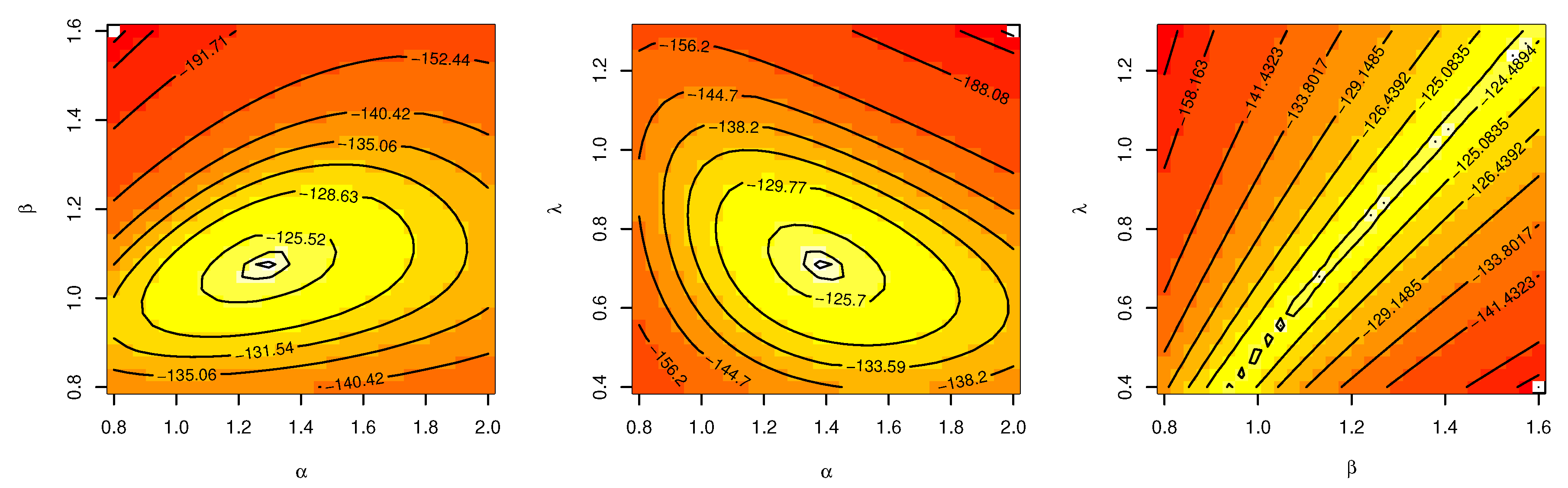
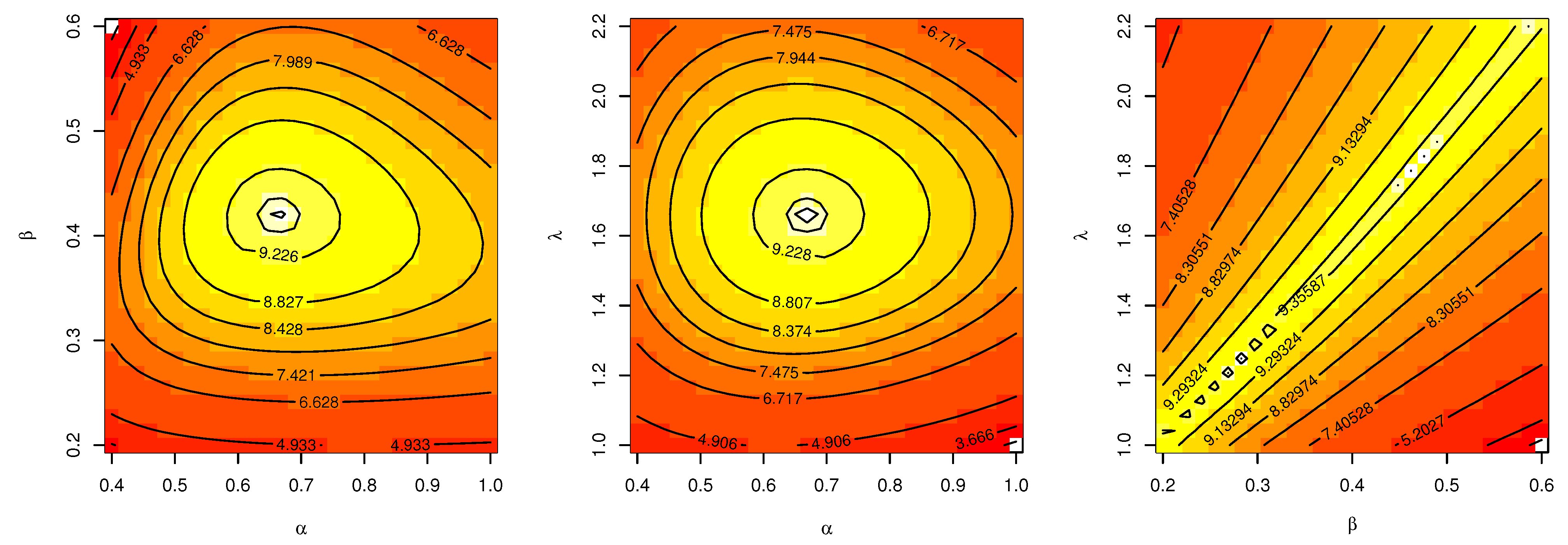
Table 6. The contour plot in
Figure 5 also shows that the
has unique values with maximum log-likelihood.
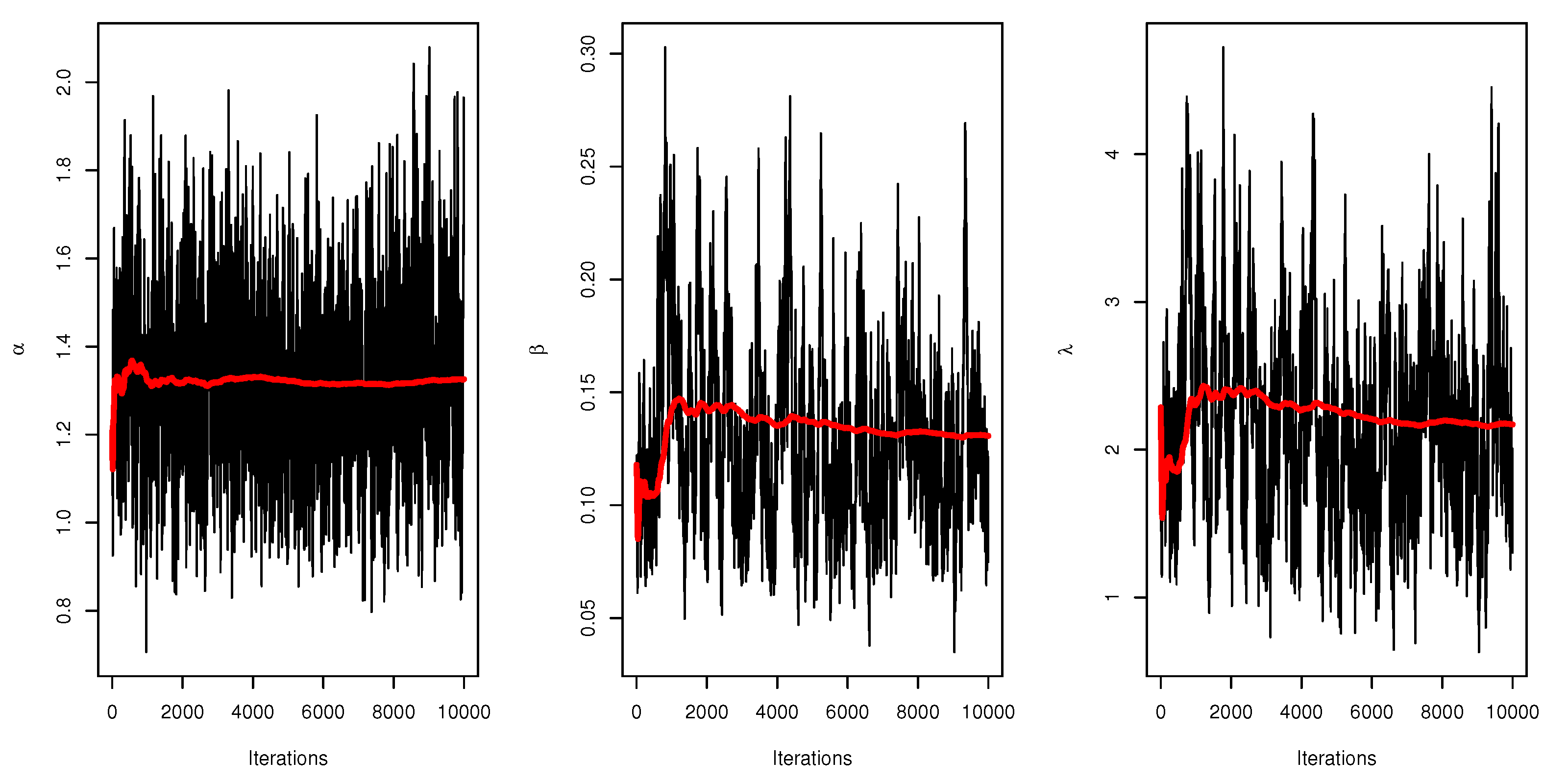
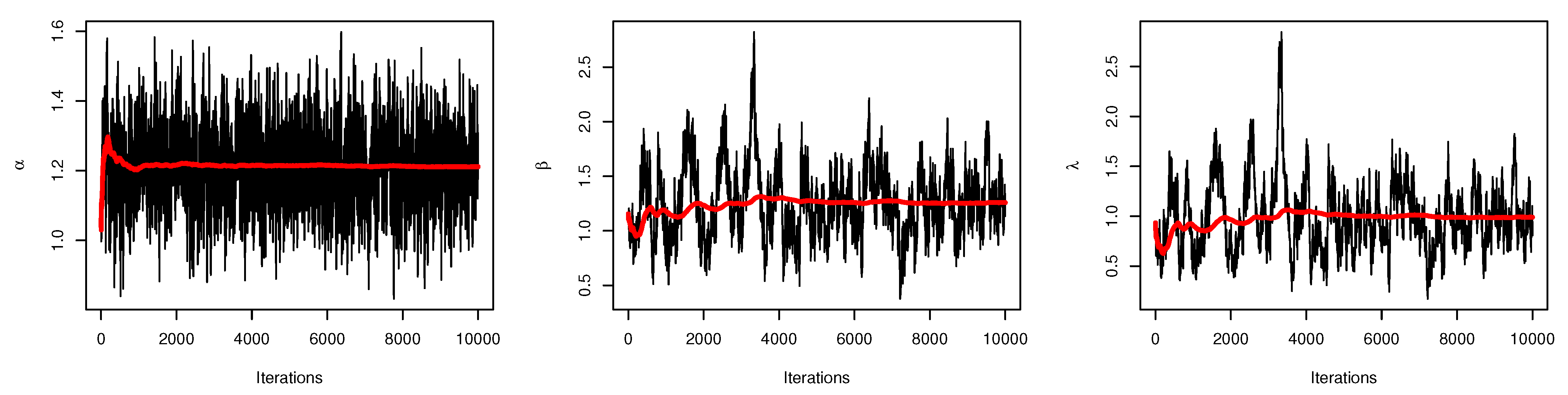
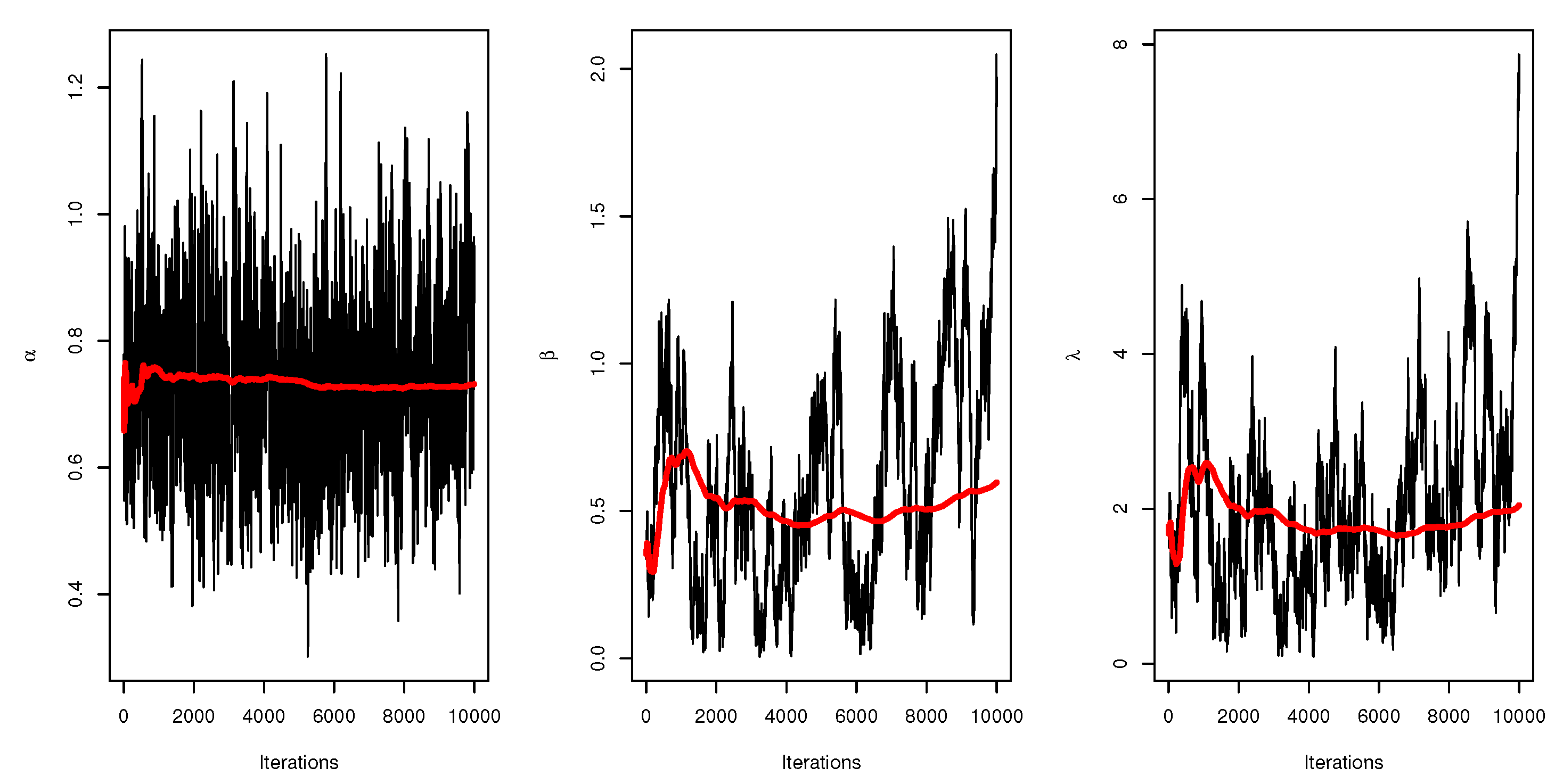
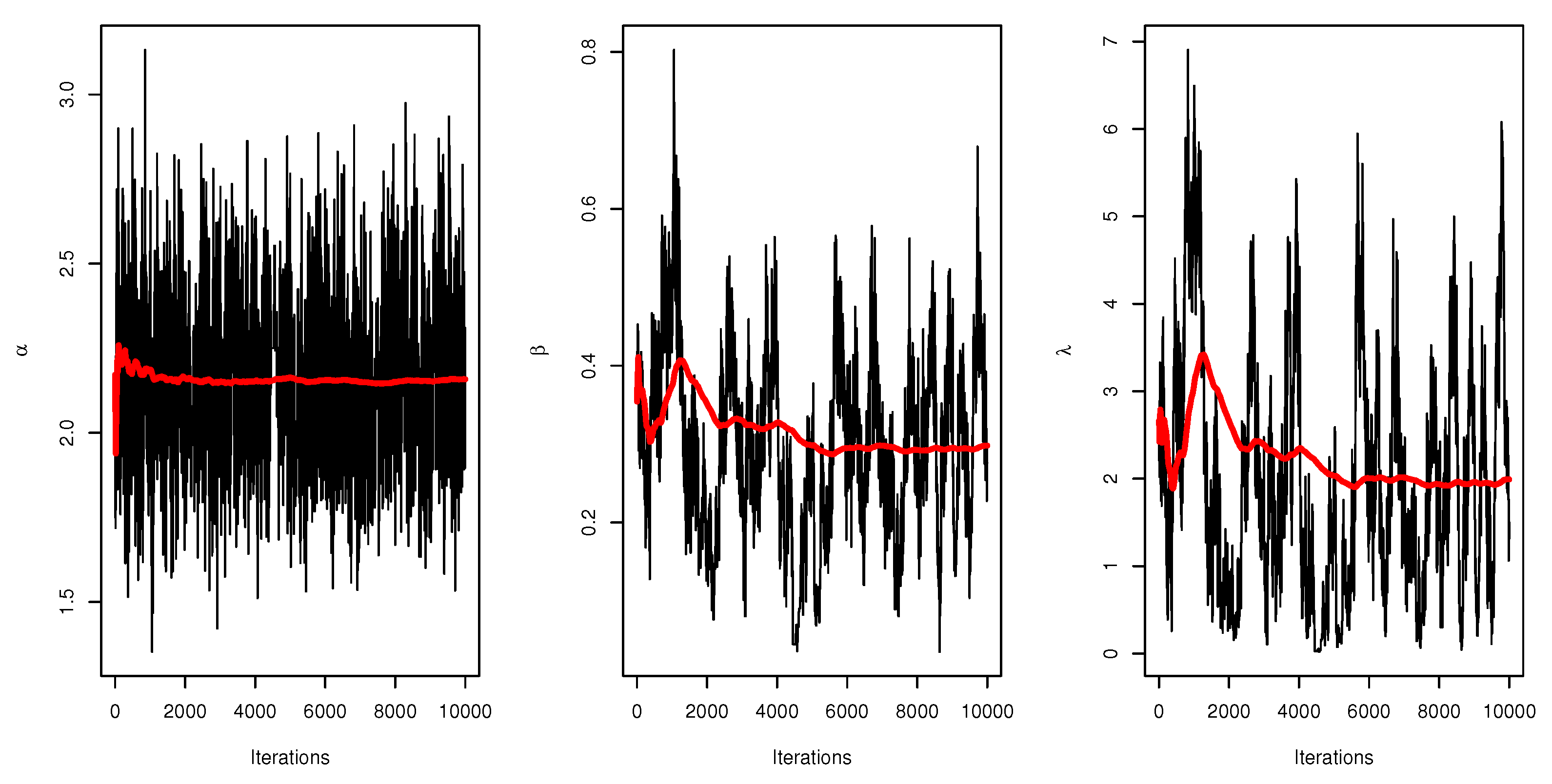
Figure 6 displays the trace plots of the MCMC results for the parameters of UEPD for the recovery rate of COVID-19 data from Turkey, which shows convergence results.
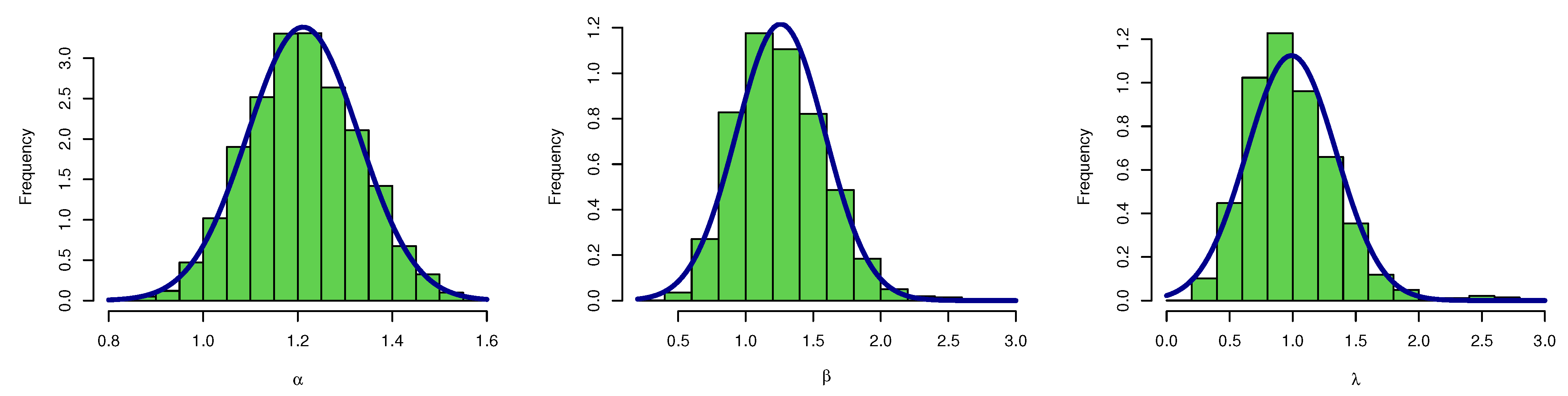
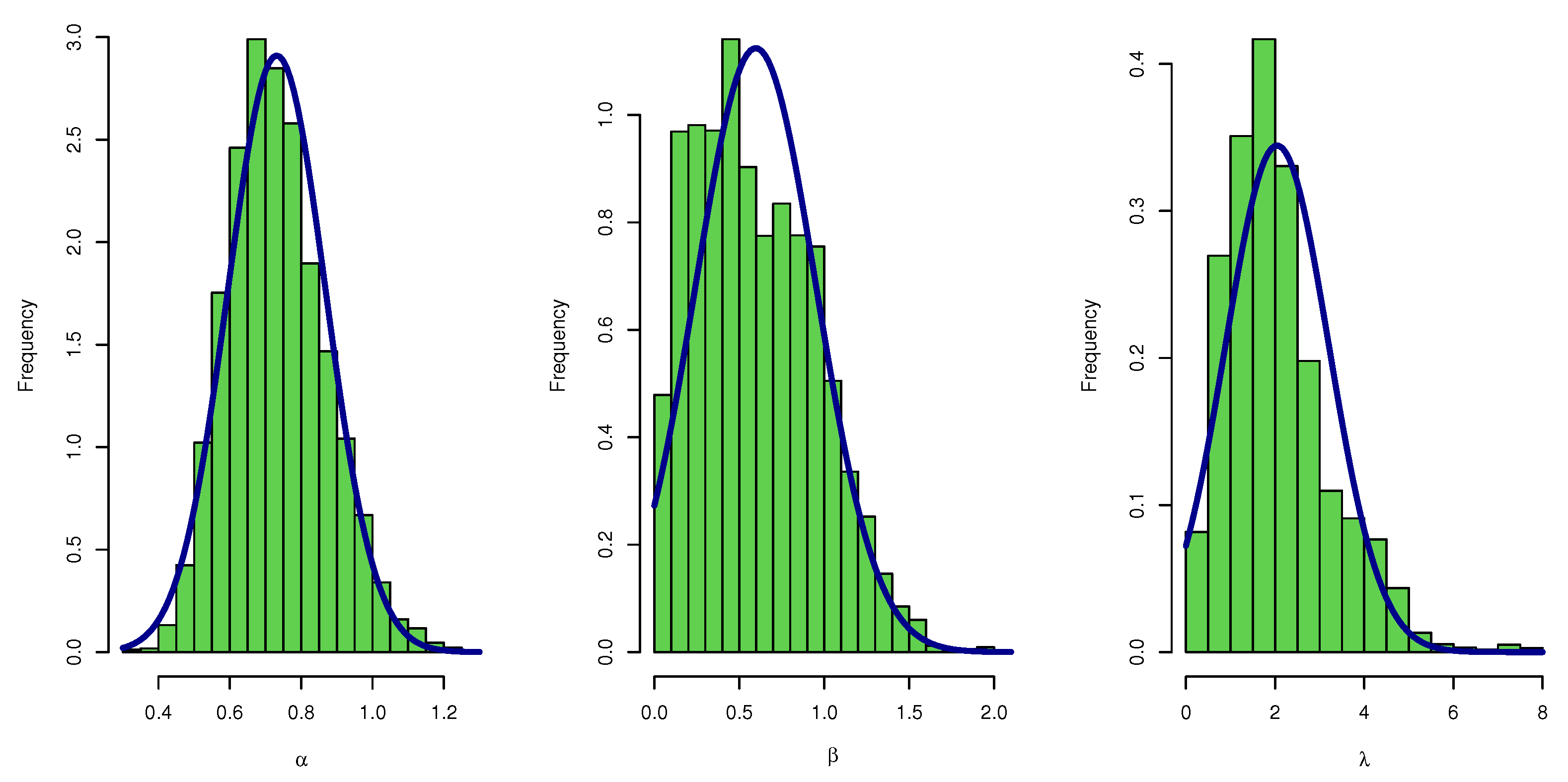
Figure 7 plots the posterior MCMC for the UEPD parameters to check the normality plot for these results.
7.2. Milk Production Data
This data set consists of the total milk output from the first calves of 107 SINDI race cows. The information is available in [
52]. In particular, the data set is included in
Table 7.
The
s estimation of the parameters and some measures of goodness of fit were obtained and are displayed in
Table 8. It is realized that the minimum values of the goodness-of-fit measures are achieved when using the UEPD, which indicates its superiority and efficiency for modeling the milk production data. The UEPD is compared with eight other suggested models.
Figure 8 displays the empirical and fitted CDF, histogram with fitted pdf, and P-P plot of the UEPD model. The profile log probability and the contour plot of the estimated parameters are displayed in
Figure 9 and
Figure 10. They demonstrate the existence and uniqueness of the MLE.
Table 9, summarizes the values of the
,
, and the Bayesian estimates for the UEPD’s parameters. It is noted that the estimated values for each parameter are relatively close when using any method of estimation. This reveals the consistency in our simulated analysis. The
asymptotic CONIs and CRIs for the parameters are also computed and the interval lengths show that its shortest length is when using the Bayesian CRI.
Figure 11 displays the trace plot of MCMC results for parameters of UEPD for the milk production data, which shows the convergence for these results.
Figure 12 displays the density plot of posterior MCMC results for parameters of UEPD for the milk production data, to check the normality plot for these results.
7.3. The Failure Components Data
In this subsection, a real data set is considered from Nigm et al. [
53] regarding the ordered failure of 20 components. The data is given in
Table 10.
The
s estimation of the parameters and the goodness-of-fit measures were obtained and are displayed in
Table 11. It is realized from
Table 11 that the minimum values of the goodness-of-fit measures are achieved when using the UEPD, which indicates its superiority and efficiency for modeling the failure time of components with respect to the six other suggested models. From
Table 12, note that the values of the
,
and Bayesian estimates are close together, which reveals a consistency in our simulated analysis; the
asymptotic CONIs and CRIs for the parameters were also computed.
Figure 13 displays the empirical and fitted CDF, histogram with fitted pdf, and P-P plot of the UEPD model. The profile log probability and the contour plot of the estimated parameters are displayed in
Figure 14 and
Figure 15. They demonstrate the existence and uniqueness of the MLE.
Figure 16 displays a trace plot of MCMC results for parameters of UEPD for the failure of 20 components data to check convergence for these results.
Figure 17 displays the density plot of posterior MCMC results for parameters of UEPD for the failures of 20 components data to check the normality plot for these results.
7.4. Recovery Rate of COVID-19 in France
The WHO confirmed that the first death from COVID-19 in France occurred on 17 March 2020, with the first official recoveries from the pandemic virus occurring on 26 March 2020. From 1 January to 7 February 2022, this data set contains 38 observations calculated as the daily ratio of total recoveries to the cumulative number of confirmed cases and different cumulative numbers of confirmed death cases in France, see
Table 13. Accessed by
https://en.wikipedia.org/wiki/COVID-19_pandemic_in_France).
From
Table 14, it is clear that the UEPD has the minimum goodness-of-fit measure and the maximum PVKS. This reveals the suitability of the UEPD to fit the recovery rate of COVID-19 in France.
Table 15 summarizes the values of estimators under MLE, MPS, and Bayesian methods. We can view the close values of estimators for all methods which prove consistency in this simulation. In addition, the
asymptotic CONIs and the CRIs for the parameters
,
and
are computed for the recovery rate of COVID-19 in France.
Figure 18 illustrates the plots of the estimated pdf, CDF, and P-P plots of UEPD for the recovery rate of COVID-19 in France. We plot the profile likelihood in
Figure 19, which indicates the uniqueness of the maximum. The contour plot in
Figure 20 also shows that the
has unique values with maximum log-likelihood.
Figure 21 displays the trace plots of the MCMC results for the above data, it shows convergence results. while
Figure 22 plots the posterior MCMC for the UEPD parameters to check the normality plot for these results.
8. Results of Simulation
Simulation research was carried out to evaluate the interpretation of the estimating techniques discussed previously. The following procedure was used to assess the efficiency of the estimators:
First step: Generate a random sample with size n from the UEPD.
Second step: The results of the first step are utilized to calculate the considering the , and Bayesian estimators.
Third step: Repeat the first and second steps N times.
Fourth step: Using and , calculate the bias and the average squared error (AVSE).
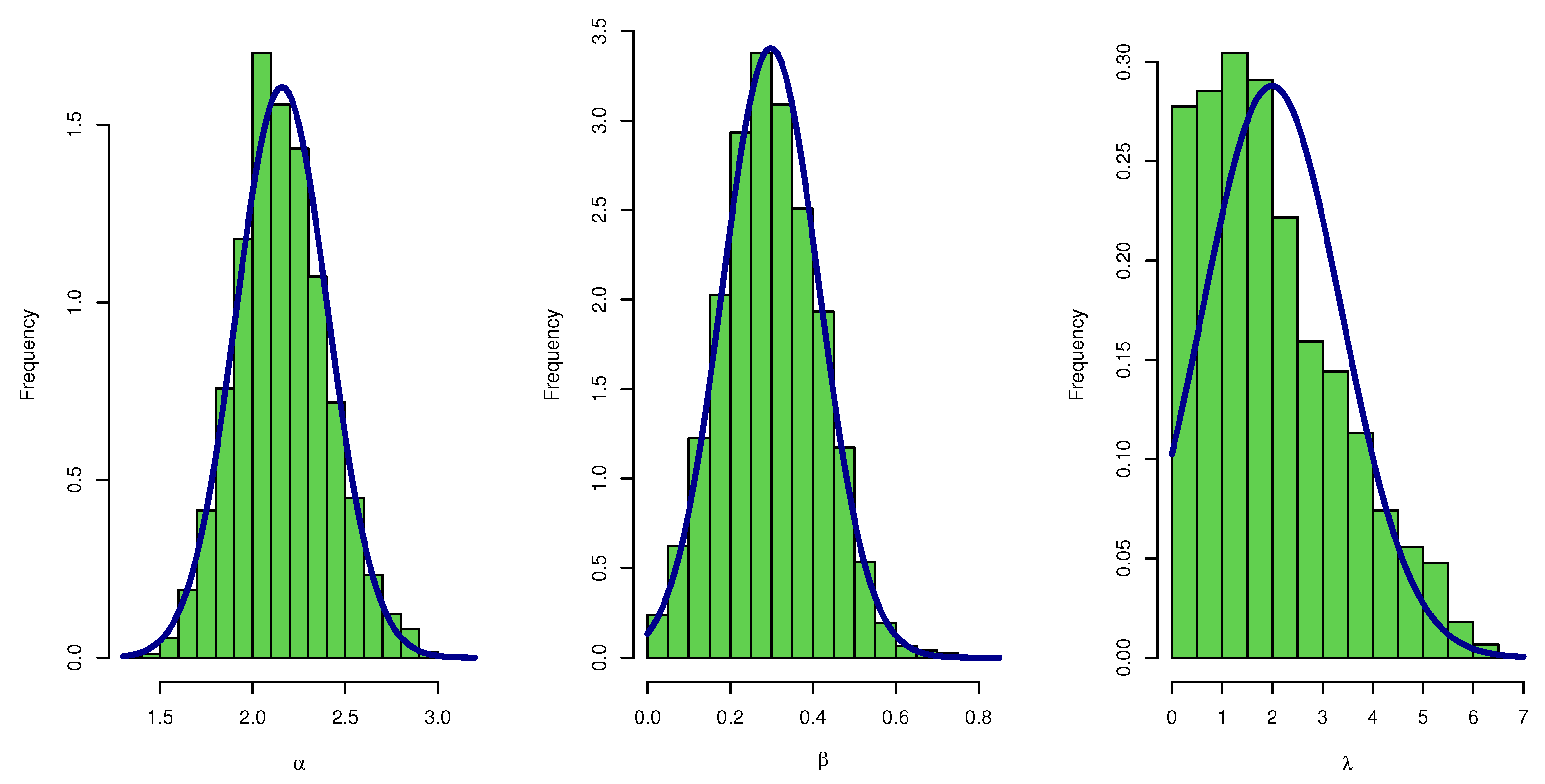
The numerical results were calculated using the function (in the stat package) and Nelder–Mead approach in R software. From the UEPD, 5000 samples were generated, where , and by choosing starting values , and , the comparisons of the different approaches of the estimators of and were considered regarding the AVSE, which is calculated for , as where is the number of simulated samples.
Another criterion was also applied for comparing the 95% CONIs; this is achieved using asymptotic distributions of the s and the with the idea of CRIs. They are compared in view of the average confidence interval lengths (ACLs). The hyper-parameters in the case of informative priors are chosen as follows: , and , while in case of non-informative priors, the values of , where , were chosen to be equal to zero.
All estimates illustrate the consistency property, which means the AVSEs decrease as the sample size increases for all parameter values. It is shown that the AVSEs illustrate that the estimation method outperforms all other estimators for all parameter combinations.
Table 16 and
Table 17 show the outcomes of the estimated parameters and their AVSE, as well as the asymptotic and credible intervals.
The findings of
Table 16 and
Table 17 show that the UEPD is stable because the range of biases, AVSEs, and CI lengths for all the parameters is quite narrow.
In some instances, we notice that as the sample size grows larger, the AVSEs for all estimations decrease.
This shows that various estimation techniques have good results for big sample sizes in terms of bias and AVSEs.
Estimation methods has better measures than the method.
The Bayesian estimation method is the best estimation method to estimate the parameter of UEPD.
The results of the simulation revealed empirical proof of the stability of our estimates.
9. Conclusions
In this paper, we introduced the unit exponential Pareto distribution to model real data examples from the recovery rate of COVID-19 during the pandemic, in addition to other applications from different fields. The unit exponential Pareto distribution is a powerful tool for modeling extreme values in a population and understanding the underlying patterns and trends in the data. It can provide valuable insights into the characteristics of the data and help to make good decisions. Consequently, many important characteristics were studied, such as the shape of the density and hazard rate functions; the , s, ; five different types of entropy such as RE, HCE, TE, AE and MHE; and , and were obtained. The unknown parameters were estimated using the method, the , and the Bayesian estimations, and their performances were compared using simulation experiments. In general, it was observed that the s estimation outperforms the other methods; hence, for small sample sizes, the estimators have a lower mean squared error than the others. We used three goodness-of-fit criteria to prove the benefit of the proposed UEPD for modeling real skewed data sets. A simulation was carried out to examine the parameters’ bias, average square error, and confidence interval lengths. Based on observed results, one can use this model to describe other features related to biomedical, mechanical, and economical fields.







_Masoom_Ali.png)







