Text-Mining to Identify Gene Sets Involved in Biocorrosion by Sulfate-Reducing Bacteria: A Semi-Automated Workflow
Abstract
:1. Introduction
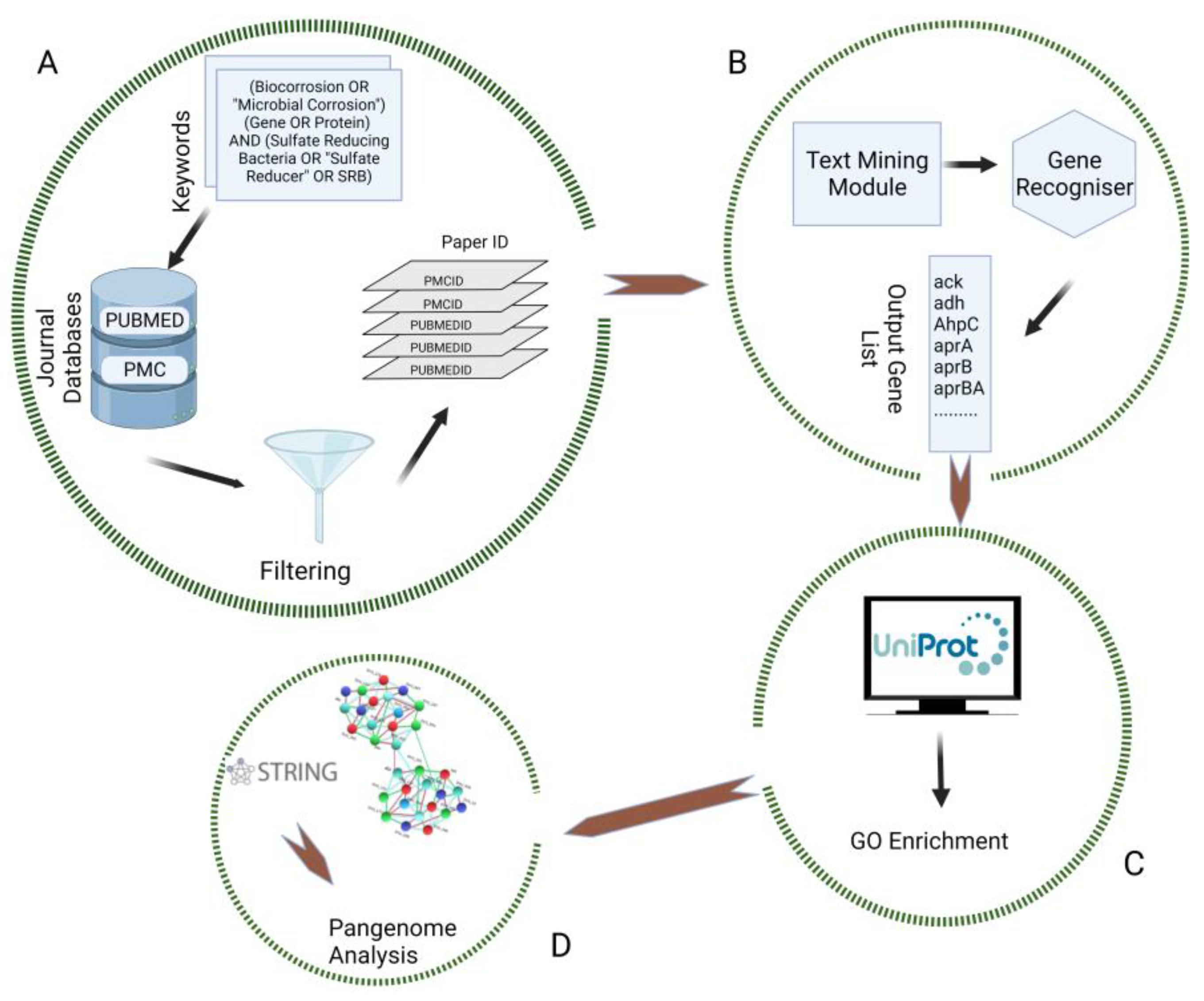
2. Materials and Methods
2.1. Data Retrieval
2.2. Data Selection and Preprocessing
2.3. GO Enrichment and Biological Network Building
2.4. Gene Set Distribution Analysis across the SRB Genome
3. Results and Discussions
3.1. Data Mining and Preprocessing
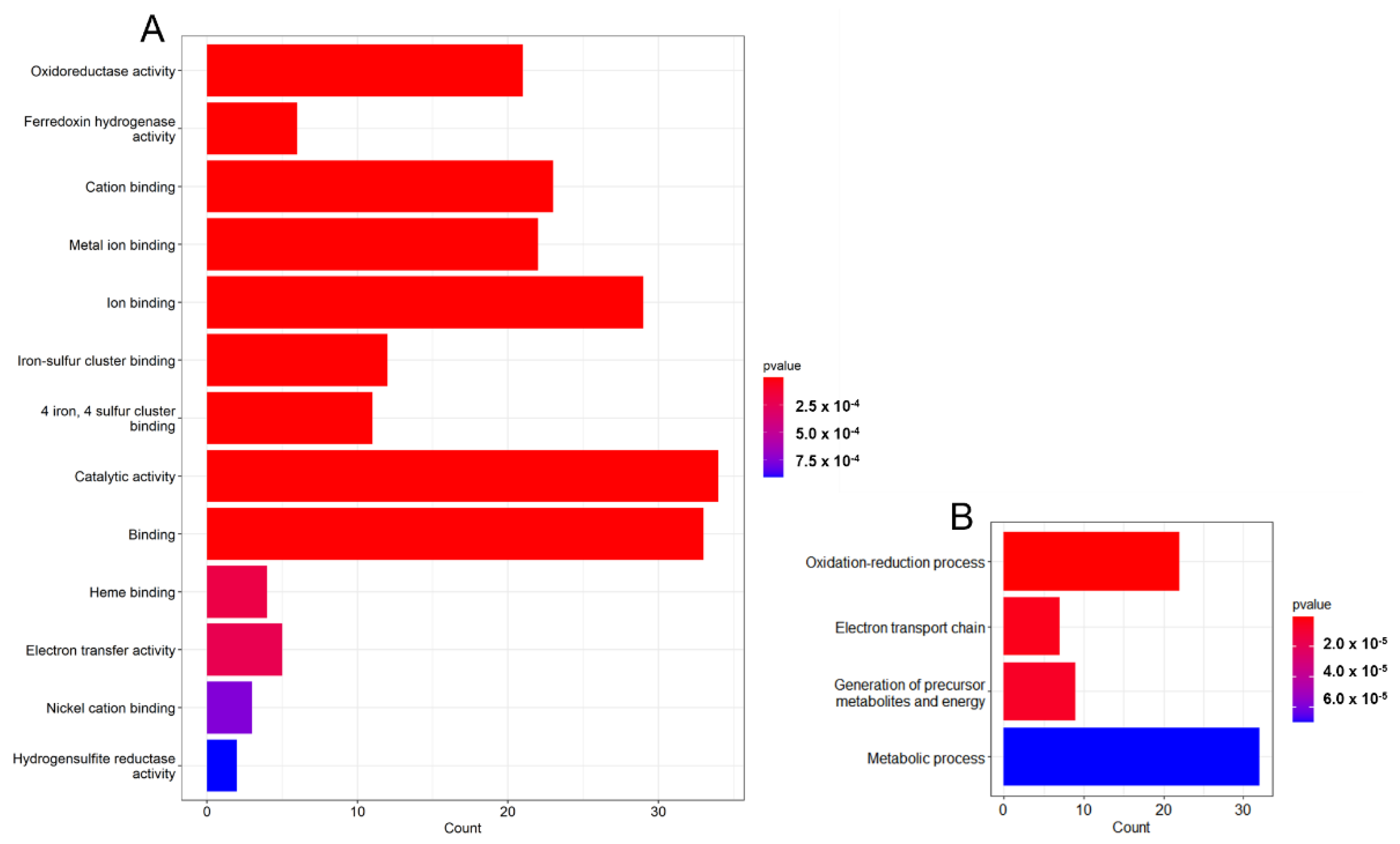
3.2. Gene Analysis Based on GO Terms
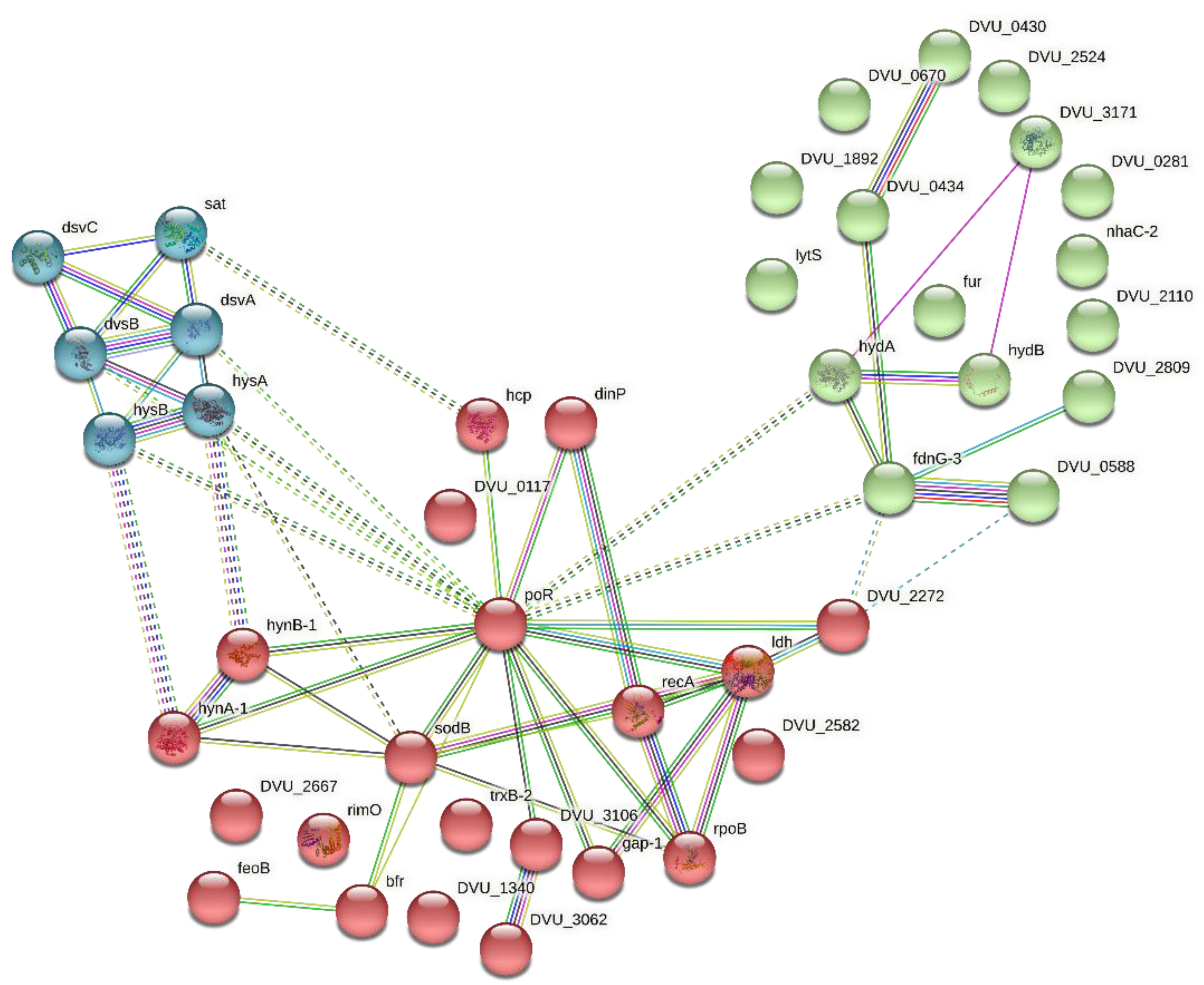
3.3. Network Building and Cluster Analysis
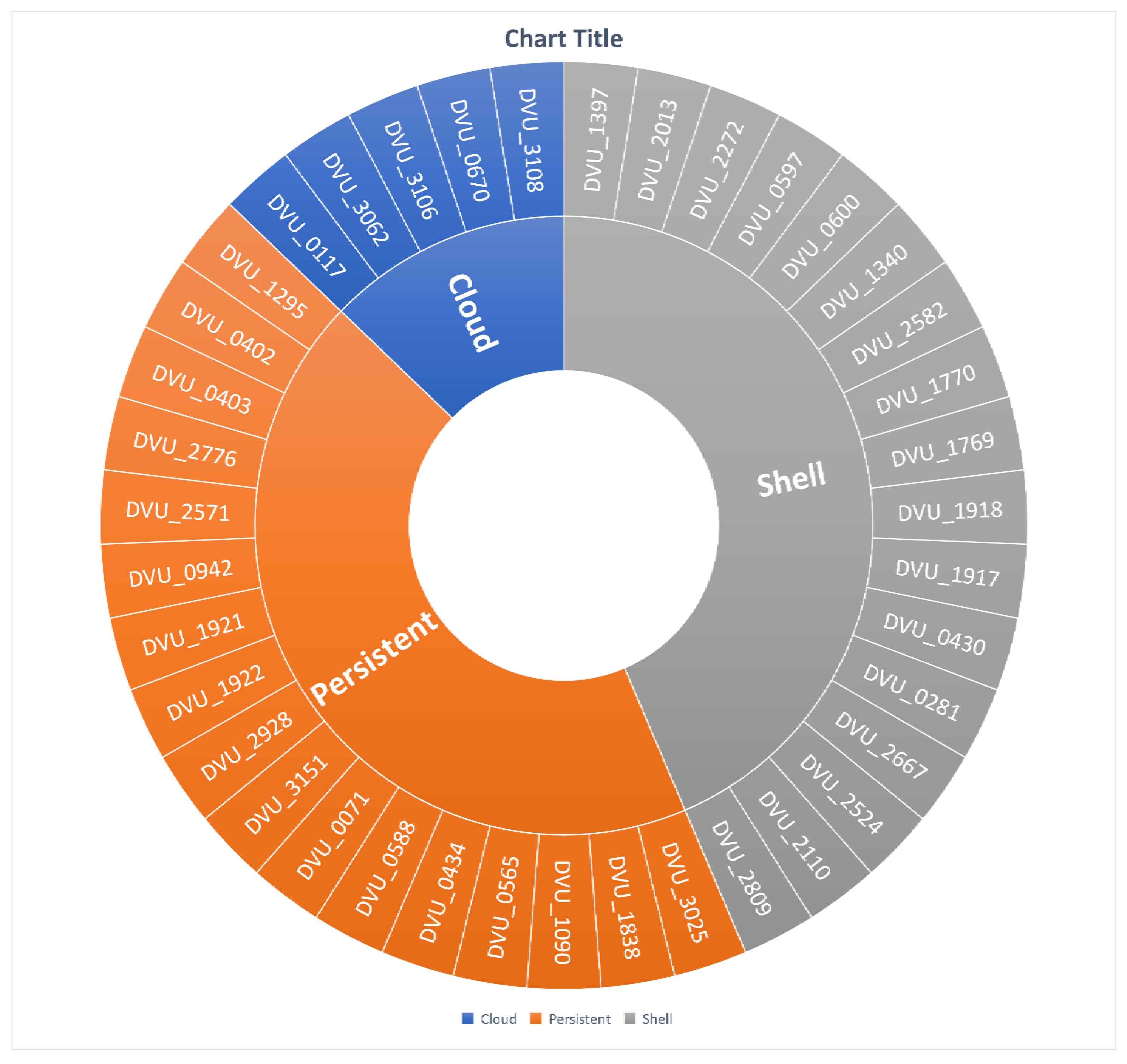
3.4. Gene Segregation Based on SRB-Pangenome Analysis
3.4.1. Persistent Gene Family
3.4.2. Shell Gene Family
3.4.3. Cloud Gene Family
3.5. Metabolic Pathway Analysis
- Enzyme secretion (e.g., hydrogenases) or secretion of active molecules (e.g., cytochrome, flavins).
- Acid production (e.g., acetic acid and sulfuric acid) and other corrosive compounds such as ammonia and sulfide.
- Formation of biofilm and EPS production that hold sites for metal ion binding [44].
3.5.1. Energy Metabolism
3.5.2. Electron Transport
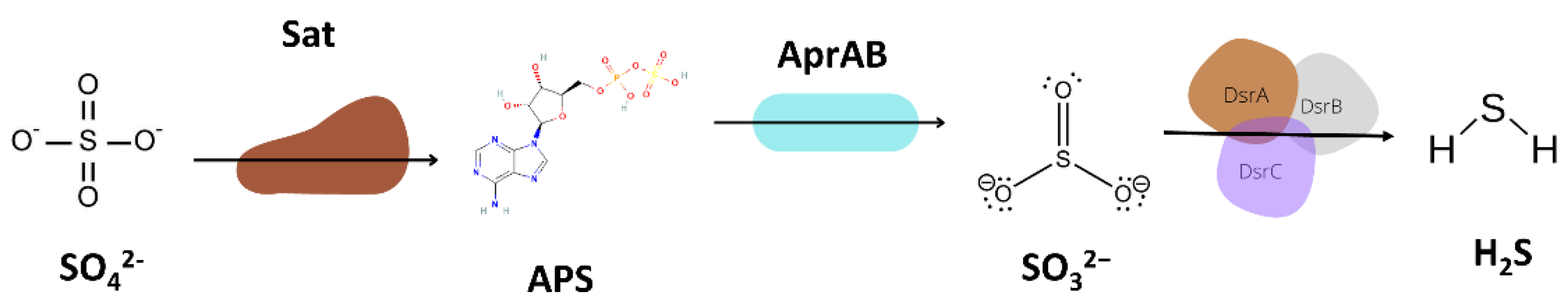
3.5.3. Sulfur Metabolism
3.5.4. Biofilm Formation
3.5.5. Stress Response
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kip, N.; van Veen, J.A. The dual role of microbes in corrosion. ISME J. 2015, 9, 542–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beech, I.B.; Sunner, J. Biocorrosion: Towards understanding interactions between biofilms and metals. Curr. Opin. Biotechnol. 2004, 15, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.K.; Thakur, P.; Saxena, P.; Rauniyar, S.; Gopalakrishnan, V.; Singh, R.N.; Gadhamshetty, V.; Gnimpieba, E.Z.; Jasthi, B.K.; Sani, R.K. Gene Sets and Mechanisms of Sulfate-Reducing Bacteria Biofilm Formation and Quorum Sensing with Impact on Corrosion. Front. Microbiol. 2021, 12, 3120. [Google Scholar] [CrossRef] [PubMed]
- Enning, D.; Garrelfs, J. Corrosion of Iron by Sulfate-Reducing Bacteria: New Views of an Old Problem. Appl. Environ. Microbiol. 2014, 80, 1226–1236. [Google Scholar] [CrossRef] [Green Version]
- Machuca Suarez, L. Understanding and addressing microbiologically influenced corrosion (MIC). Corros. Mater. 2019, 44, 88–96. [Google Scholar]
- Dou, W.; Xu, D.; Gu, T. Biocorrosion caused by microbial biofilms is ubiquitous around us. Microb. Biotechnol. 2021, 14, 803–805. [Google Scholar] [CrossRef]
- Elmouaden, K.; Jodeh, S.; Chaouay, A.; Oukhrib, R.; Salghi, R.; Bazzi, L.; Hilali, M. Sulfate-Reducing Bacteria Impact on Copper Corrosion Behavior in Natural Seawater Environment. J. Surf. Eng. Mater. Adv. Technol. 2016, 06, 36–46. [Google Scholar] [CrossRef] [Green Version]
- Blackwood, D.J. An Electrochemist Perspective of Microbiologically Influenced Corrosion. Corros. Mater. Degrad. 2020, 1, 59–76. [Google Scholar] [CrossRef] [Green Version]
- Kakooei, S.; Ismail, M.C.; Ariwahjoedi, B. Mechanisms of microbiologically influenced corrosion: A review. World Appl. Sci. J. 2012, 17, 524. [Google Scholar]
- Winnenburg, R.; Wächter, T.; Plake, C.; Doms, A.; Schroeder, M. Facts from text: Can text mining help to scale-up high-quality manual curation of gene products with ontologies? Brief. Bioinform. 2008, 9, 466–478. [Google Scholar] [CrossRef] [Green Version]
- Pham, B.; Jovanovic, J.; Bagheri, E.; Antony, J.; Ashoor, H.; Nguyen, T.T.; Rios, P.; Robson, R.; Thomas, S.M.; Watt, J.; et al. Text mining to support abstract screening for knowledge syntheses: A semi-automated workflow. Syst. Rev. 2021, 10, 156. [Google Scholar] [CrossRef] [PubMed]
- Fotseu, E.B.F.; Nembot, T.K.; Sani, R.K.; Gadhamshetty, V.; Gnimpieba, Z.E.; Bomgni, A.B. GenNER-A highly scalable and optimal NER method for text-based gene and protein recognition. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 3562–3569. [Google Scholar] [CrossRef]
- Gautreau, G.; Bazin, A.; Gachet, M.; Planel, R.; Burlot, L.; Dubois, M.; Perrin, A.; Médigue, C.; Calteau, A.; Cruveiller, S.; et al. PPanGGOLiN: Depicting microbial diversity via a partitioned pangenome graph. PLoS Comput. Biol. 2020, 16, e10077322020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidelberg, J.F.; Seshadri, R.; Haveman, S.A.; Hemme, C.L.; Paulsen, I.T.; Kolonay, J.F.; Eisen, J.A.; Ward, N.; Methe, B.; Brinkac, L.M.; et al. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hilden-borough. Nat. Biotechnol. 2004, 22, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Ueki, T.; Lovley, D.R. Desulfovibrio vulgaris as a model microbe for the study of corrosion under sulfate-reducing con-ditions. mLife 2022, 1, 13–20. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Geesey, G. What is biocorrosion? In Biofouling and Biocorrosion in Industrial Water Systems; Springer: Berlin/Heidelberg, Germany, 1991; pp. 155–164. [Google Scholar]
- Salgar-Chaparro, S.J.; Lepkova, K.; Pojtanabuntoeng, T.; Darwin, A.; Machuca, L.L. Nutrient Level Determines Biofilm Characteristics and Subsequent Impact on Microbial Corrosion and Biocide Effectiveness. Appl. Environ. Microbiol. 2020, 86, e02885-19. [Google Scholar] [CrossRef] [Green Version]
- Guala, D.; Ogris, C.; Müller, N.; Sonnhammer, E.L.L. Genome-wide functional association networks: Background, data & state-of-the-art resources. Brief. Bioinform. 2020, 21, 1224–1237. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Enright, A.J.; Ouzounis, C.A. Functional associations of proteins in entire genomes by means of exhaustive detection of gene fusions. Genome Biol. 2001, 2, research0034.1. [Google Scholar] [CrossRef]
- Karn, S.K.; Bhambri, A.; Jenkinson, I.R.; Duan, J.; Kumar, A. The roles of biomolecules in corrosion induction and inhibition of corrosion: A possible insight. Corros. Rev. 2020, 38, 403–421. [Google Scholar] [CrossRef]
- Hamilton, W.A. Microbially Influenced Corrosion as a Model System for the Study of Metal Microbe Interactions: A Unifying Electron Transfer Hypothesis. Biofouling 2003, 19, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, S.; Basseguy, R.; Bergel, A. Electron transfer between hydrogenase and 316L stainless steel: Identification of a hydrogenase-catalyzed cathodic reaction in anaerobic mic. J. Electroanal. Chem. 2004, 561, 93–102. [Google Scholar] [CrossRef]
- Lovley, D.R.; Phillips, E.J.P. Novel Processes for Anaerobic Sulfate Production from Elemental Sulfur by Sulfate-Reducing Bacteria. Appl. Environ. Microbiol. 1994, 60, 2394–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cottis, R. Shreir’s Corrosion; Elsevier: Amsterdam, The Netherlands, 2010; Volume 2. [Google Scholar]
- Beech, I.B.; Sunner, J.A. Sulphate-reducing bacteria and their role in. In Sulphate-Reducing Bacteria: Environmental and Engineered Systems; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Gumiere, T.; Meyer, K.M.; Burns, A.R.; Gumiere, S.J.; Bohannan, B.J.; Andreote, F.D. A probabilistic model to identify the core microbial community. bioRxiv 2018, 491183. [Google Scholar] [CrossRef]
- Snipen, L.; Almøy, T.; Ussery, D.W. Microbial comparative pan-genomics using binomial mixture models. BMC Genom. 2009, 10, 385. [Google Scholar] [CrossRef] [Green Version]
- Snipen, L.-G.; Liland, K.H. micropan: An R-package for microbial pan-genomics. BMC Bioinform. 2015, 16, 79. [Google Scholar] [CrossRef]
- Venceslau, S.S.; Stockdreher, Y.; Dahl, C.; Pereira, I.A.C. The “bacterial heterodisulfide” DsrC is a key protein in dissimilatory sulfur metabolism. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 1148–1164. [Google Scholar] [CrossRef] [Green Version]
- Caffrey, S.M.; Park, H.S.; Voordouw, J.K.; He, Z.; Zhou, J.; Voordouw, G. Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hil-denborough. J. Bacteriol. 2007, 189, 6159–6167. [Google Scholar] [CrossRef] [Green Version]
- Bender, K.S.; Yen, H.-C.B.; Hemme, C.L.; Yang, Z.; He, Z.; He, Q.; Zhou, J.; Huang, K.H.; Alm, E.J.; Hazen, T.C.; et al. Analysis of a Ferric Uptake Regulator (Fur) Mutant of Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 2007, 73, 5389–5400. [Google Scholar] [CrossRef] [Green Version]
- Arragain, S.; Garcia-Serres, R.; Blondin, G.; Douki, T.; Clemancey, M.; Latour, J.-M.; Forouhar, F.; Neely, H.; Montelione, G.T.; Hunt, J.F.; et al. Post-translational Modification of Ribosomal Proteins: Structural and functional characterization of rimo from thermotoga maritima, a radical s-adenosylmethionine methylthiotransferase. J. Biol. Chem. 2010, 285, 5792–5801. [Google Scholar] [CrossRef] [Green Version]
- Rhee, H.J.; Kim, E.-J.; Lee, J.K. Physiological polyamines: Simple primordial stress molecules. J. Cell. Mol. Med. 2007, 11, 685–703. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Pimentel, C.; Valente, F.M.A.; Rodrigues-Pousada, C.; Pereira, I.A.C. Tungsten and Molybdenum Regulation of Formate Dehydrogenase Expression in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 2011, 193, 2909–2916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valette, O.; Tran, T.T.; Cavazza, C.; Caudeville, E.; Brasseur, G.; Dolla, A.; Talla, E.; Pieulle, L. Biochemical function, molecular structure and evolution of an atypical thioredoxin reductase from Desul-fovibrio vulgaris. Front. Microbiol. 2017, 8, 1855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.R.; Lee, H.J.; Lee, H.K.; Kim, Y.K.; Oh, Y.S.; Choi, S.C. Involvement of organic acid during corrosion of iron coupon by Desulfovibrio desulfuricans. J. Microbiol. Biotechnol. 2003, 13, 937–941. [Google Scholar]
- Gu, T. Theoretical Modeling of the Possibility of Acid Producing Bacteria Causing Fast Pitting Biocorrosion. J. Microb. Biochem. Technol. 2014, 6, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Heggendorn, F.L.; Fraga, A.G.M.; Ferreira, D.D.C.; Gonçalves, L.S.; Lione, V.D.O.F.; Lutterbach, M.T.S. Sulfate-Reducing Bacteria: Biofilm Formation and Corrosive Activity in Endodontic Files. Int. J. Dent. 2018, 2018, 8303450. [Google Scholar] [CrossRef] [Green Version]
- Borenstein, S.W. Microbiologically Influenced Corrosion Handbook; Industrial Press Inc.: New York, NY, USA, 1994. [Google Scholar] [CrossRef]
- Gadd, G.-M.; Burford, E.-P.; Fomina, M. Biogeochemical activities of microorganisms in mineral transformations: Con-sequences for metal and nutrient mobility. J. Microbiol. Biotechnol. 2003, 13, 323–331. [Google Scholar]
- Wang, D.; Liu, J.; Jia, R.; Dou, W.; Kumseranee, S.; Punpruk, S.; Li, X.; Gu, T. Distinguishing two different microbiologically influenced corrosion (MIC) mechanisms using an electron mediator and hydrogen evolution detection. Corros. Sci. 2020, 177, 108993. [Google Scholar] [CrossRef]
- Dall’Agnol, L.T. Deeper Insights into SRB-Driven Biocorrosion Mechanisms; Universidade NOVA de Lisboa (Portugal): Lisbon, Portugal, 2013. [Google Scholar]
- Morais-Silva, F.O.; Santos, C.I.; Rodrigues, R.; Pereira, I.A.; Rodrigues-Pousada, C. Roles of HynAB and Ech, the only two hydrogenases found in the model sulfate reducer Desulfovibrio gigas. J. Bacteriol. 2013, 195, 4753–4760. [Google Scholar] [CrossRef] [Green Version]
- Valente, F.M.A.; Oliveira, A.S.F.; Gnadt, N.; Pacheco, I.; Coelho, A.; Xavier, A.V.; Teixeira, M.; Soares, C.; Pereira, I.A.C. Hydrogenases in Desulfovibrio vulgaris Hildenborough: Structural and physiologic characterisation of the membrane-bound [NiFeSe] hydrogenase. JBIC J. Biol. Inorg. Chem. 2005, 10, 667–682. [Google Scholar] [CrossRef]
- Pereira, P.; He, Q.; Valente, F.M.A.; Xavier, A.V.; Zhou, J.; Pereira, I.A.C.; Louro, R.O. Energy metabolism in Desulfovibrio vulgaris Hildenborough: Insights from transcriptome analysis. Antonie Van Leeuwenhoek 2008, 93, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Cordas, C.M.; Moura, I.; Moura, J.J. Direct electrochemical study of the multiple redox centers of hydrogenase from Desulfovibrio gigas. Bioelectrochemistry 2008, 74, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Marshall, M.J.; Plymale, A.E.; Kennedy, D.W.; Shi, L.; Wang, Z.; Reed, S.B.; Dohnalkova, A.C.; Simonson, C.J.; Liu, C.; Saffarini, D.A.; et al. Hydrogenase- and outer membrane c-type cytochrome-facilitated reduction of technetium(VII) by Shewanella oneidensis MR-1. Environ. Microbiol. 2008, 10, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Goenka, A.; Voordouw, J.K.; Lubitz, W.; Gaertner, W.; Voordouw, G. Construction of a [NiFe]-hydrogenase deletion mutant of Desulfovibrio vulgaris Hildenborough. Bio-Chem. Soc. Trans. 2005, 33 Pt 1, 59–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohorelic, B.K.J.; Voordouw, J.K.; Lojou, E.; Dolla, A.; Harder, J.; Voordouw, G. Effects of Deletion of Genes Encoding Fe-Only Hydrogenase of Desulfovibrio vulgaris Hildenborough on Hydrogen and Lactate Metabolism. J. Bacteriol. 2002, 184, 679–686. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-W.; Helmann, J.D. Functional specialization within the Fur family of metalloregulators. BioMetals 2007, 20, 485–499. [Google Scholar] [CrossRef]
- Zhang, W.; Culley, D.E.; Nie, L.; Scholten, J.C.M. Comparative transcriptome analysis of Desulfovibrio vulgaris grown in planktonic culture and mature biofilm on a steel surface. Appl. Microbiol. Biotechnol. 2007, 76, 447–457. [Google Scholar] [CrossRef]
- Matias, P.; Pereira, I.A.C.; Soares, C.; Carrondo, M.A. Sulphate respiration from hydrogen in Desulfovibrio bacteria: A structural biology overview. Prog. Biophys. Mol. Biol. 2005, 89, 292–329. [Google Scholar] [CrossRef]
- Aubert, C.; Brugna, M.; Dolla, A.; Bruschi, M.; Giudici-Orticoni, M.-T. A sequential electron transfer from hydrogenases to cytochromes in sulfate-reducing bacteria. Biochim. Et Biophys. Acta (BBA)-Protein Struct. Mol. Enzym. 2000, 1476, 85–92. [Google Scholar] [CrossRef]
- Zhou, J.; He, Q.; Hemme, C.L.; Mukhopadhyay, A.; Hillesland, K.; Zhou, A.; He, Z.; Van Nostrand, J.D.; Hazen, T.C.; Stahl, D.A.; et al. How sulphate-reducing microorganisms cope with stress: Lessons from systems biology. Nat. Rev. Genet. 2011, 9, 452–466. [Google Scholar] [CrossRef] [Green Version]
- Rosenbaum, M.; Aulenta, F.; Villano, M.; Angenent, L.T. Cathodes as electron donors for microbial metabolism: Which extracellular electron transfer mecha-nisms are involved? Bioresour. Technol. 2011, 102, 324–333. [Google Scholar] [CrossRef]
- Kloeke, F.V.O.; Bryant, R.D.; Laishley, E.J. Localization of Cytochromes in the Outer Membrane of Desulfovibrio vulgaris (Hildenborough) and their Role in Anaerobic Biocorrosion. Anaerobe 1995, 1, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Carepo, M.; Baptista, J.F.; Pamplona, A.; Fauque, G.; Moura, J.J.G.; Reis, M.A.M. Hydrogen metabolism in Desulfovibrio desulfuricans strain New Jersey (NCIMB 8313)—Comparative study with D. vulgaris and D. gigas species. Anaerobe 2002, 8, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, W.D.; Venceslau, S.S.; Waldbauer, J.; Smith, D.A.; Pereira, I.A.C.; Bradley, A.S. Proteomic and Isotopic Response of Desulfovibrio vulgaris to DsrC Perturbation. Front. Microbiol. 2019, 10, 658. [Google Scholar] [CrossRef] [Green Version]
- Carbonero, F.; Benefiel, A.C.; Alizadeh-Ghamsari, A.H.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448. [Google Scholar] [CrossRef] [Green Version]
- Duncan, K.E.; Davidova, I.A.; Nunn, H.S.; Stamps, B.W.; Stevenson, B.S.; Souquet, P.J.; Suflita, J.M. Design features of offshore oil production platforms influence their susceptibility to biocorrosion. Appl. Microbiol. Biotechnol. 2017, 101, 6517–6529. [Google Scholar] [CrossRef] [PubMed]
- Basera, P.; Lavania, M.; Lal, B. Potential of dynamic bacterial communities in the bio-corrosion process: A proof study with surface morphology of metal coupons. RSC Adv. 2019, 9, 17040–17050. [Google Scholar] [CrossRef] [PubMed]
- Little, B.J.; Ray, R.I.; Pope, R.K. Relationship Between Corrosion and the Biological Sulfur Cycle: A Review. Corrosion 2000, 56, 433–443. [Google Scholar] [CrossRef]
- Zhang, W.; Culley, D.E.; Wu, G.; Brockman, F.J. Two-Component Signal Transduction Systems of Desulfovibrio vulgaris: Structural and Phylogenetic Analysis and Deduction of Putative Cognate Pairs. J. Mol. Evol. 2006, 62, 473–487. [Google Scholar] [CrossRef]
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular Mechanisms of Two-Component Signal Transduction. J. Mol. Biol. 2016, 428, 3752–3775. [Google Scholar] [CrossRef] [Green Version]
- Lewandowskiy, Z.; Dickinsony, W.; Leey, W. Electrochemical interactions of biofilms with metal surfaces. Water Sci. Technol. 1997, 36, 295–302. [Google Scholar] [CrossRef]
- Hori, K.; Matsumoto, S. Bacterial adhesion: From mechanism to control. Biochem. Eng. J. 2010, 48, 424–434. [Google Scholar] [CrossRef]
- Caffrey, S.M.; Park, H.S.; Been, J.; Gordon, P.; Sensen, C.W.; Voordouw, G. Gene Expression by the Sulfate-Reducing Bacterium Desulfovibrio vulgaris Hildenborough Grown on an Iron Electrode under Cathodic Protection Conditions. Appl. Environ. Microbiol. 2008, 74, 2404–2413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Poosarla, V.G.; Song, S.; Wood, T.L.; Miller, D.S.; Yin, B.; Wood, T.K. Glycoside hydrolase DisH from Desulfovibrio vulgaris degrades the N-acetylgalactosamine component of diverse biofilms. Environ. Microbiol. 2018, 20, 2026–2037. [Google Scholar] [CrossRef]
- Beech, I.; Cheung, C. Interactions of exopolymers produced by sulphate-reducing bacteria with metal ions. Int. Biodeterior. Biodegrad. 1995, 35, 59–72. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Bautista, B.E.T.; Wikieł, A.J.; Datsenko, I.; Vera, M.; Sand, W.; Seyeux, A.; Zanna, S.; Frateur, I.; Marcus, P. Influence of extracellular polymeric substances (EPS) from Pseudomonas NCIMB 2021 on the corrosion behaviour of 70Cu–30Ni alloy in seawater. J. Electroanal. Chem. 2015, 737, 184–197. [Google Scholar] [CrossRef] [Green Version]
- Černoušek, T.; Ševců, A.; Shrestha, R.; Steinová, J.; Kokinda, J.; Vizelková, K. Microbially influenced corrosion of container material. In The Microbiology of Nuclear Waste Disposal; Elsevier: Amsterdam, The Netherlands, 2021; pp. 119–136. [Google Scholar]
- Chan, K.-Y.; Xu, L.-C.; Fang, H.H.P. Anaerobic Electrochemical Corrosion of Mild Steel in the Presence of Extracellular Polymeric Substances Produced by a Culture Enriched in Sulfate-Reducing Bacteria. Environ. Sci. Technol. 2002, 36, 1720–1727. [Google Scholar] [CrossRef]
- Clark, M.E.; He, Z.; Redding, A.M.; Joachimiak, M.P.; Keasling, J.D.; Zhou, J.Z.; Arkin, A.P.; Mukhopadhyay, A.; Fields, M.W. Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: Carbon and energy flow con-tribute to the distinct biofilm growth state. BMC Genom. 2012, 13, 138. [Google Scholar] [CrossRef] [Green Version]
- Beese-Vasbender, P.F.; Nayak, S.; Erbe, A.; Stratmann, M.; Mayrhofer, K.J. Electrochemical characterization of direct electron uptake in electrical microbially influenced corrosion of iron by the lithoautotrophic SRB Desulfopila corrodens strain IS4. Electrochim. Acta 2015, 167, 321–329. [Google Scholar] [CrossRef]
- Tran, T.T.T.; Kannoorpatti, K.; Padovan, A.; Thennadil, S. A study of bacteria adhesion and microbial corrosion on different stainless steels in environment containing Desulfovibrio vulgaris. R. Soc. Open Sci. 2021, 8, 201577. [Google Scholar] [CrossRef] [PubMed]
- Lopes, F.; Morin, P.; Oliveira, R.; Melo, L. The influence of nickel on the adhesion ability of Desulfovibrio desulfuricans. Colloids Surf. B Biointerfaces 2005, 46, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Scarascia, G.; Lehmann, R.; Machuca, L.L.; Morris, C.; Cheng, K.Y.; Kaksonen, A.; Hong, P.-Y. Effect of Quorum Sensing on the Ability of Desulfovibrio vulgaris To Form Biofilms and To Biocorrode Carbon Steel in Saline Conditions. Appl. Environ. Microbiol. 2019, 86, e01664-19. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012, 67, 2069–2089. [Google Scholar] [CrossRef] [Green Version]
- Barton, L.L.; Hamilton, W.A. Sulphate-Reducing Bacteria: Environmental and Engineered Systems; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Timóteo, C.G.; Guilherme, M.; Penas, D.; Folgosa, F.; Tavares, P.; Pereira, A.S. Desulfovibrio vulgaris bacterioferritin uses H2O2 as a co-substrate for iron oxidation and reveals DPS-like DNA protection and binding activities. Biochem. J. 2012, 446, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, M.C.; Lobo, S.; Carita, J.N.; Nobre, L.S.; Saraiva, L.M. Bacterioferritin protects the anaerobe Desulfovibrio vulgaris Hildenborough against oxygen. Anaerobe 2012, 18, 454–458. [Google Scholar] [CrossRef]
- Carrondo, M.A. New EMBO member’s review: Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 2003, 22, 1959–1968. [Google Scholar] [CrossRef]
- Soldano, A.; Yao, H.; Chandler, J.R.; Rivera, M. Inhibiting Iron Mobilization from Bacterioferritin in Pseudomonas aeruginosa Impairs Biofilm Formation Irrespective of Environmental Iron Availability. ACS Infect. Dis. 2020, 6, 447–458. [Google Scholar] [CrossRef] [Green Version]
- Punchi Hewage, A.N.; Fontenot, L.; Guidry, J.; Weldeghiorghis, T.; Mehta, A.K.; Donnarumma, F.; Rivera, M. Mobilization of iron stored in bacterioferritin is required for metabolic homeostasis in Pseu-domonas aeruginosa. Pathogens 2020, 9, 980. [Google Scholar] [CrossRef]
- Qi, Z.; Chen, L.; Zhang, W. Comparison of Transcriptional Heterogeneity of Eight Genes between Batch Desulfovibrio vulgaris Biofilm and Planktonic Culture at a Single-Cell Level. Front. Microbiol. 2016, 7, 597. [Google Scholar] [CrossRef] [Green Version]
- Pieulle, L.; Stocker, P.; Vinay, M.; Nouailler, M.; Vita, N.; Brasseur, G.; Garcin, E.B.; Sebban-Kreuzer, C.; Dolla, A. Study of the Thiol/Disulfide Redox Systems of the Anaerobe Desulfovibrio vulgaris Points Out Pyruvate:Ferredoxin Oxidoreductase as a New Target for Thioredoxin 1. J. Biol. Chem. 2011, 286, 7812–7821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, E.R.; Tzianabos, A.O.; Smith, C.J. Thioredoxin Reductase Is Essential for Thiol/Disulfide Redox Control and Oxidative Stress Survival of the Anaerobe Bacteroides fragilis. J. Bacteriol. 2007, 189, 8015–8023. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Kim, J.-C.; Jeon, B. Stimulation of biofilm formation by oxidative stress in Campylobacter jejuni under aerobic conditions. Virulence 2016, 7, 846–851. [Google Scholar] [CrossRef] [Green Version]
- Bade, K.; Manz, W.; Szewzyk, U. Behavior of sulfate reducing bacteria under oligotrophic conditions and oxygen stress in particle-free systems related to drinking water. FEMS Microbiol. Ecol. 2000, 32, 215–223. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakur, P.; Alaba, M.O.; Rauniyar, S.; Singh, R.N.; Saxena, P.; Bomgni, A.; Gnimpieba, E.Z.; Lushbough, C.; Goh, K.M.; Sani, R.K. Text-Mining to Identify Gene Sets Involved in Biocorrosion by Sulfate-Reducing Bacteria: A Semi-Automated Workflow. Microorganisms 2023, 11, 119. https://doi.org/10.3390/microorganisms11010119
Thakur P, Alaba MO, Rauniyar S, Singh RN, Saxena P, Bomgni A, Gnimpieba EZ, Lushbough C, Goh KM, Sani RK. Text-Mining to Identify Gene Sets Involved in Biocorrosion by Sulfate-Reducing Bacteria: A Semi-Automated Workflow. Microorganisms. 2023; 11(1):119. https://doi.org/10.3390/microorganisms11010119
Chicago/Turabian StyleThakur, Payal, Mathew O. Alaba, Shailabh Rauniyar, Ram Nageena Singh, Priya Saxena, Alain Bomgni, Etienne Z. Gnimpieba, Carol Lushbough, Kian Mau Goh, and Rajesh Kumar Sani. 2023. "Text-Mining to Identify Gene Sets Involved in Biocorrosion by Sulfate-Reducing Bacteria: A Semi-Automated Workflow" Microorganisms 11, no. 1: 119. https://doi.org/10.3390/microorganisms11010119
APA StyleThakur, P., Alaba, M. O., Rauniyar, S., Singh, R. N., Saxena, P., Bomgni, A., Gnimpieba, E. Z., Lushbough, C., Goh, K. M., & Sani, R. K. (2023). Text-Mining to Identify Gene Sets Involved in Biocorrosion by Sulfate-Reducing Bacteria: A Semi-Automated Workflow. Microorganisms, 11(1), 119. https://doi.org/10.3390/microorganisms11010119







