Abstract
When oil spill occurs, it poses serious toxic hazards to all forms of life. Mixed culture of Burkholderia cepacia and Corynebacterium sp isolated from refinery sludge using selective enrichment technique was used for bioremediation of refinery wastewater in a laboratory-scale bioreactor. Physicochemical parameters of both raw and treated water were as determined and compared with Federal Environmental Protection Agency (FEPA-limit, Abuja, Nigeria) to asses the efficiency of the bioremediation process. Each of the bacterium was screened for the presence of plasmid DNA and for the involvement or otherwise of plasmid in the bioremediation of wastewater. The immobilised cells showed percentage decrease in chemical oxygen demand (97%), biochemical oxygen demand (94%), phenol (98%), total petroleum hydrocarbon (79%), oil and grease (90%) of the refinery waste water after 20 days of treatment while their transconjugants showed the multiplicative effect by achieving the same percentage after 10 days of treatment. Therefore, the findings revealed that bioaugmentation of wastewater using transmissible catabolic plasmid will enhance efficiency of the bioremediation by spreading the plasmid among indigenous microbial community either through horizontal gene transfer or transformation.
Introduction
Wastewater released by petrochemical industries are characterized by the presence of large quantity of polycyclic and aromatic hydrocarbons, phenols, metal derivatives, surface active substances, sulphides, naphethy lenic acids and other chemicals [1,2].
The process of refining crude oil consumes large amounts of water. Consequently, significant volumes of wastewater are generated [3]. Coelho et al. [3] reported that the volume of refinery effluent generated during processing is 0.4-1.6 times the amount of the crude oil processed. Thus, based on the current yield of 84 million barrels per day (mbpd) of crude oil, a total of 33.6 mbpd of effluent is generated globally [4].
Mobile genetic elements (MGEs) that encode catabolic genes are considered to play a major role in the adaptation of microbial populations to xenobiotic organic compounds, through either by spreading the genes in a community and thereby increasing the diversity of organisms able to metabolise these compounds, or by rearranging and combining pre-existing genes or gene fragments from different microorganisms to constitute a new pathway dealing with a new compound. Such catabolic mobile elements include plasmids, transposable elements and elements using phage-like integrases [5,6].
Effluents from the oil industry will continually be produced and discharged into the world’s main water bodies. However, these pollutants pose serious toxic hazards to the environment. Petroleum refinery effluent can vary greatly depending on the type of oil being processed, the plant configuration, and operation procedures [7]. They enter into waterways and have negatives impact on the aquatic and terrestrial eco-systems.
Bioremediation has long been applied as a treatment technology that is cost-effective, ecologically friendly and efficient for the decontamination of hydrocarbon polluted soils [8,9,10,11,12,13,14,15].
There is a need for the survival of inoculum and to remain physiologically stable for a very long time in a pollutant, the inoculation of strains with MGEs that encode the relevant genetic information and their subsequent genetic horizontal transfer to one or various well established and competitive indigenous bacterial populations of an ecosystem is an alternative strategy. In this case, the survival of the introduced donor strain is no longer needed once the catabolic genes are transferred and expressed in the indigenous bacteria within the ecosystem [16].
The immense potential of such microorganisms to degrade petroleum hydrocarbon does not depend solely upon the wealth of catabolic enzymes that these organisms possess, but also upon their capacity for adaptive change [17]. The present work identified mixed culture of crude oil degrading Burkholderia cepacia and Corynebacterium sp isolated from refinery oil sludge and their ability to transfer plasmid to the indigenous bacteria in the ecosystem for the preparation of bacterial blend that could be used in bioremediation of refinery waste water.
Materials and Methods
Collection of the wastewater Samples
The wastewater samples from different concrete wastewater reservoirs in Kaduna Refinery & Petrochemical Company (KRPC; Kaduna, Nigeria) were collected into four litres plastic bottles and stored in an ice block cooler and transferred to the laboratory immediately for analysis [18,19]. All the collected samples were preserved in accordance with guidelines and International Standards. The samples collected were analysed for pH, biological oxygen demand (BOD5), chemical oxygen demand (COD), total dissolved solids, phenol, oil and grease, sulphide and some selected heavy metals. All other quality assurance and quality control procedure relevant to samples collection, custody and analyses were strictly adhered to for the determination of physicochemical parameters [20,21,22,23].
Immobilization of cells
Mixed culture of Burkholderia cepacia and Corynebacterium sp that showed degradative potential during mineralization of Bonny Light crude oil were immobilised on agar-agar as follows: agar-agar solution and inoculi for immobilisation were prepared separately as thus; Fifty mL of each of the inoculi was inoculated into nutrient broth and incubated for 24 h. A solution containing 3% bacteriological agar was dissolve in 0.9% sodium chloride (100 mL) in a 250 mL Erlenmeyer flask, sterilized and cooled to 40-45°C. The inoculi with optical density (OD)600nm=1.0 which correspond to 109 cfu/mL was added to the prepared molten agaragar maintained at 40°C, shaken well for few seconds (without forming foam), poured into sterile flat bottom 4-inch-diameter petriplates, solidification occurred after 10 min. The solidified agar block was cut into equal size cubes, then added to sterile 0.1 M phosphate buffer (pH 7.0), and kept in the refrigerator (1 h) for curing. Phosphate buffer was decanted and the cubes were washed with sterile distilled water 3 to 4 times before used [24].
Bioremediation using immobilized mixed culture
Initial physicochemical characterization of the refinery wastewater was done.
Two litres of the refinery wastewater supplemented with minimum basal medium in g/L [NaCl (0.8); MgSO4.7H2O (0.001); KH2P04 (2); NaN03 (2); CaCl2.2H20 (0.5); NaHP04.12H20 (2)] was sterilized in an autoclave at 121°C for 15 min. One litre of the sterilized wastewater was dispensed into 5 L bioreactor fitted with air sparger aseptically. Thirty grams each of the immobilized bacterial cells was added [25]. Physicochemical parameters were determined in triplicate at every five days intervals to monitor the progress of bioremediation for 20 days.
Preparation of crude oil degrading transconjugant culture
Method of Bathe and Hausner (2010) [26] was modified and used for the preparation of crude oil degrading transconjugants. This was done to transfer catabolic plasmid from oil degrading host strains to a mixture of non-oil degrading indigenous strains in the oil sludge. Overnight donor cell of mixed culture of Burkholderia cepacia and Corynebacterium sp grown in Luria-Bertani (LB) medium supplemented with 1% crude oil. Also ten grams of oil sludge was added to 90 mL of Bushnell-Haas (BH) medium and incubated for 48 h to obtain oil sludge suspension. Two mL of donor strains was mixed with 2 mL of pre-grown suspension of oil sludge. Drop of 1 mL of this mixture was plated on LB agar plates and incubated at 37°C for 24 h for the conjugation to occur.
The mating spot was scrapped of the agar surface and resuspended the cells in 1 mL of phosphate buffered saline (PBS), centrifuged and discarded the supernatant. The pellet mixtures were spread on oil agar plates (BH medium supplemented with 2% crude oil) to select transconjugants.
Prior to mixing of donor and recipient indigenous suspension was unable to grow in oil agar. Transconjugant was selected, purified in PBS and later used for bioremediation process. The colonies on the oil agar were removed and transferred into 1 mL of PBS, vortex vigorously, centrifuged for 10 min at 5000 rpm. Pellet was re-suspended again in PBS. The cell pellet was washed twice and resuspended with BH medium until OD600 was equivalent to 1.0.
Screening of transconjugant bioremediation capabilities
Ten grams of wood chips was added to five litres bioreactor fitted with air sparger to serve as biofilm carrier material. One litre of refinery wastewater supplemented with minimum basal medium was transferred to bioreactor. The 10% of cells of crude oil degrading transconjugant with OD600nm=1.0 which correspond to 109 cfu/mL was added to the bioreactor control sample contained only the sludge inoculums without culture. Set up was left for 10 days. The following parameter was used for assessment, e.g. COD, oil and grease, phenol, % oil loss and sulphide.
Plasmid profiling and agar gel electrophoresis
Plasmid extraction was carried out using the method described by Ojo and Oso (2009) [27] with slight modification. Pure isolates were inoculated on tars broth and incubated. The grown cells were harvested and suspended in 200 µL of solution A [100 mm glucose-50 mm tris hydrochloride (pH 8)-10 mm ethylenediaminetetraacetic acid (EDTA)] containing 10 mg tanolysin and incubated for 30 min at 37°C in a dodecyl sulphate in 0.2 µL NaOH was added and the samples were mixed by inverting tubes. Three hundred uL of a 30% potassium acetate solution (pH 4.8) was added and the samples were mixed by vortexing. The supernatant was removed and extracted once with a phenol – chloroform mixture (1:1) and precipitated with an equal volume of isopropanol. The plasmid DNA was then dissolved in 100 µL of tris-EDTA buffer. Electrophoresis of the DNA was carried out on a 0.8% agarose gel in a 0.5×concentration of tris-borate-EDTA buffer. After boiling, the solution was allowed to cool 10 µL of ethidium bromide was added to the cooled agarose solution. This was poured into a casting tray with a comb placed across its rim to form wells. The gel was allowed to set for 30 min and the comb was removed. Twenty µL of the plasmid DNA samples were then loaded into the wells after mixing with 2 µL of bromophenol blue. A DNA molecular weight marker was also loaded into one of the well. The gels were thereafter electropholesed in a horizontal tank at a constant voltage of 60v for about 1 h and 30 min. After electrophoresis, plasmid DNA bands were viewed by fluorescence of bound ethidium bromide under a short wave ultraviolet light transilluminator and the photograph were taken using a digital camera.
Curing of plasmid
Curing of plasmid was done to identify the catabolic plasmid encoded in the isolated bacteria. The modified methods [28,29] were used. A stock solution of 10% sodium dodecyl sulfate (SDS) was prepared by the addition of 10 g SDS to 100 mL of nutrient broth. The pH was adjusted to 7.8-8.0. The mixture was steamed for 1 h and kept aside as stock. The overnight cultures of selected isolates in LB were diluted 100-fold and 0.5 mL volume of each of the isolates was transferred aseptically into fresh 30 mL volume of nutrient broth. The cultures were incubated with shaking for 3 h and SDS stock solution was added to give the required final concentration 1% (w/v). The cultures were incubated at room temperature with mild agitation for 72 h. The cells were then tested for crude oil degradation: after transferring 0.5 mL sample of each of the isolates into nutrient broth supplemented with crude oil 2% (w/v), the plasmid content was determined by carrying out the plasmid isolation procedure described above.
Results and Discussion
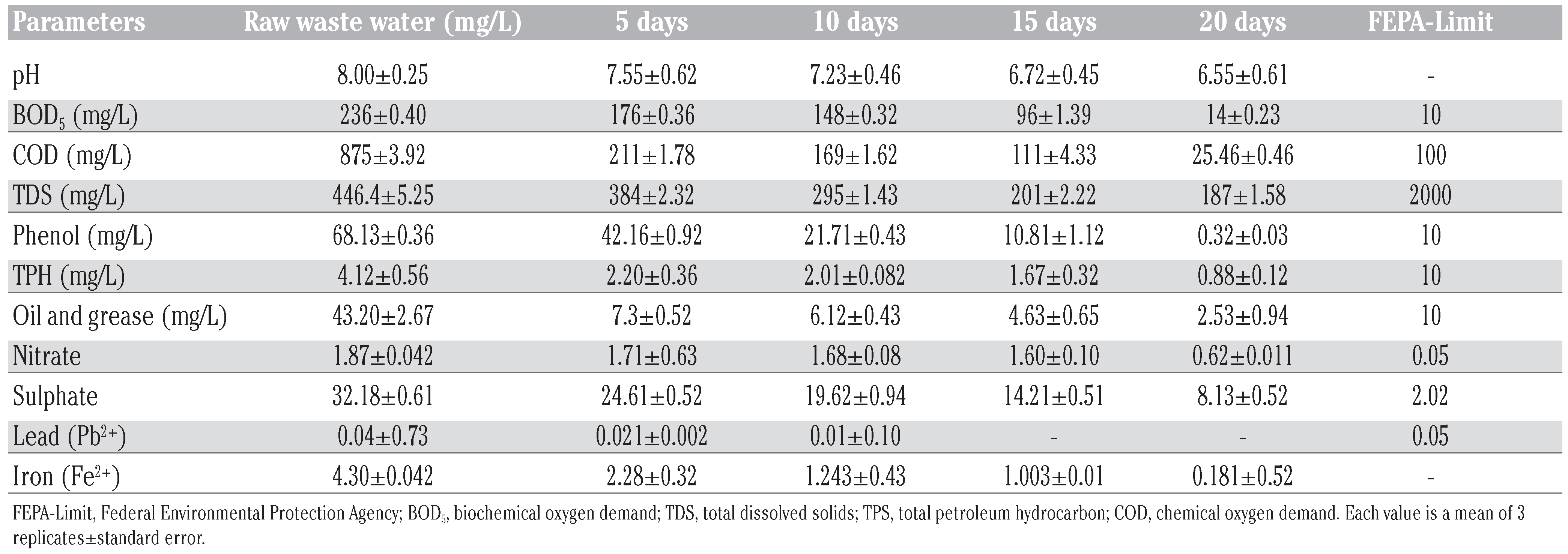
The result of both raw and treated kaduna refinery wastewater using immobilised mixed culture of Burkholderia cepacia and Corynebacterium sp. are presented in Table 1 while the result of treated wastewater using transconjugants are shown in Table 2. The pH values varies between (8.0-6.55) while COD, phenol and oil and grease in raw waste water are 875 mg/L, 68.13 mg/L and 43.20 mg/L respectively while final level obtained in both immobilised cell treated wastewater and transconjugant treated wastewater are: 25.46 mg/L and 68 mg/L; (0.32 mg/L and 1.06 mg/L; 4.27 mg/L and 2.53 mg/L, respectively. Result revealed an overall reduction in all the physicochemical parameters below FEPA-Limit. Lead (Pb2+) was completely biosorptioned, while nitrate and sulphate were significantly reduced. BOD5 test was found to reduce to (14 mg/mL). Both BOD and COD are useful parameters in determining the relative waste loading and higher degree therefore indices the presence of large amount of organic pollutant and relatively higher level of microbial activities with consequent depletion of oxygen content [30]. However, nitrate and phosphate are essential nutrients to plants life, but when found in excess quantities, stimulates excessive plant growth leading to potentially toxic algal blooms [31].

Table 1.
Physicochemical characterization of raw and treated wastewater of Kaduna Refining & Petrochemical Company (KRPC), Kaduna, Nigeria, compared with Federal Environmental Protection Agency (FEPA, Abuja, Nigeria)-Limit.

Table 2.
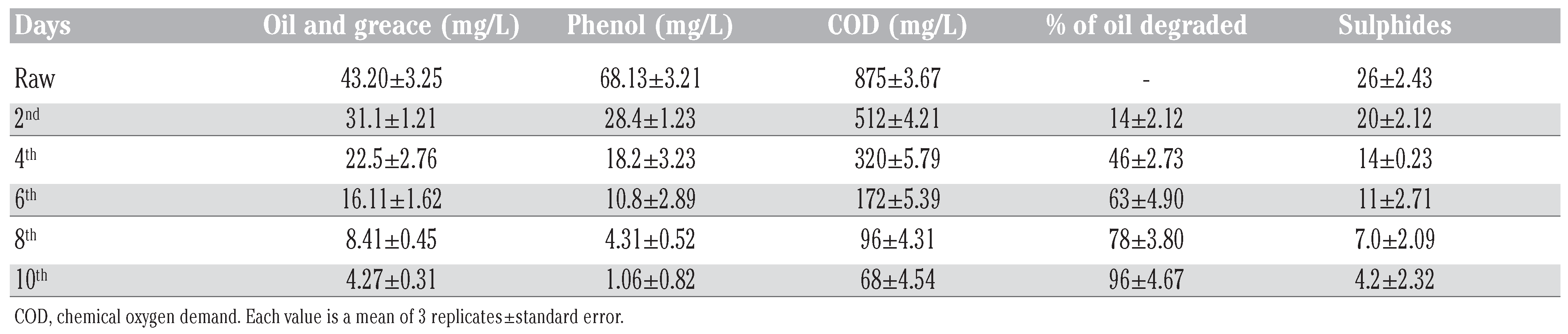
Bioremediation of refinery wastewater using transconjugant.
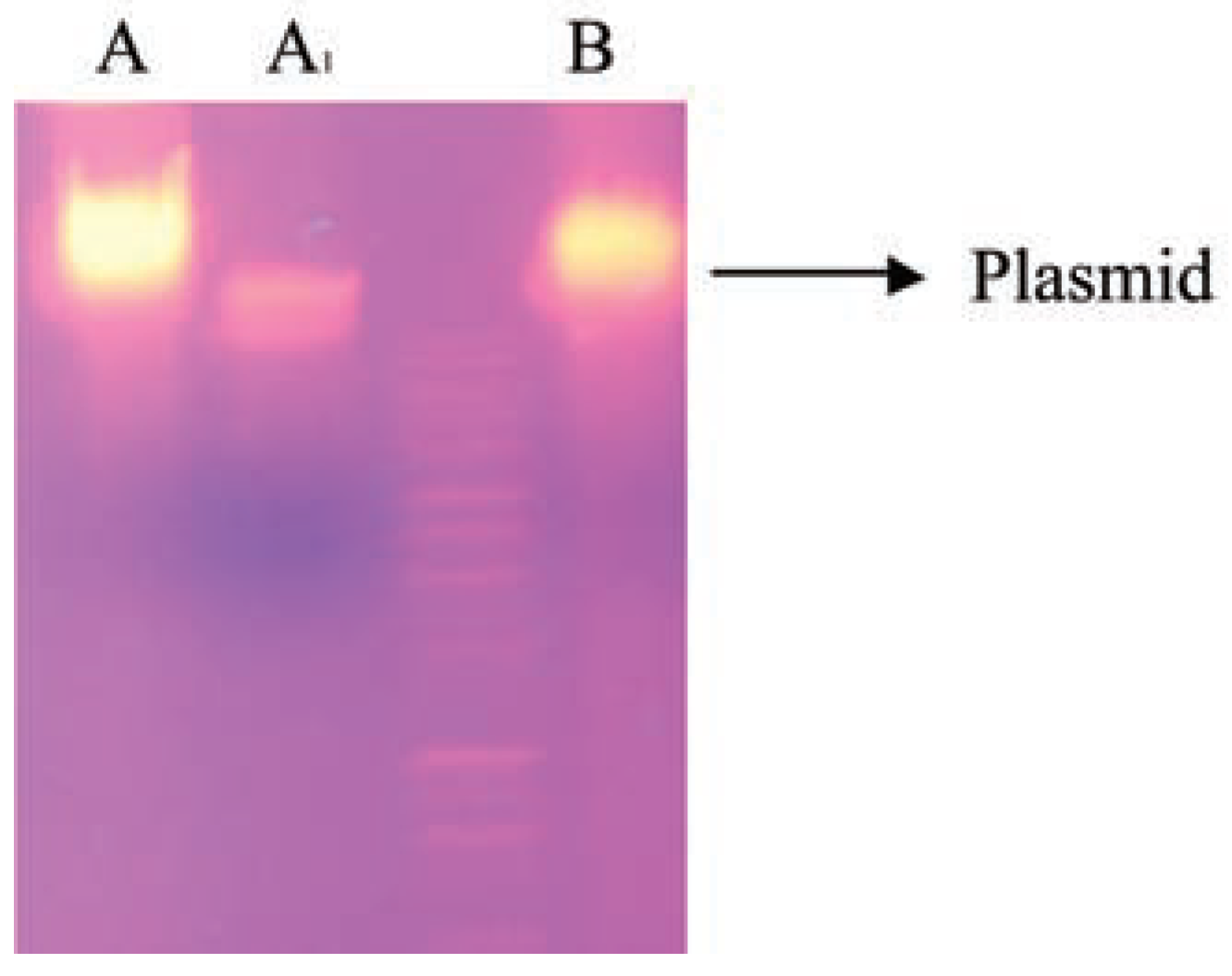
The results obtained in the plasmid profiling in Figure 1 showed that both isolates contained plasmid. Plasmids that encode enzymes involved in the transformation of environmental pollutants are known as catabolic plasmids [32]. The incidence of such plasmids in oil degrading bacteria had been reported by many researchers [33], reported that the catabolic gene which encode degradation routes of different aromatic and aliphatic hydrocarbons are frequently located on plasmids, although degradative genes can be located on either chromosome or plasmid [34]. Whyte and colleagues [35] extracted plasmids from Burkholderia and Corynebacterium strains isolated from oil-sludge conferred ability to degrade crude oil in order to confirm whether the observed biodegradation potential in the isolates was plasmid or chromosomal mediated, the cured colony was unable to grow on Bushnell-Haas medium agar with crude oil as sole carbon source, therefore, the total loss of biodegradation ability after plasmid curing shows the involvement of plasmid in catabolic activity of hydrocarbon biodegradation, this indicates that the gene(s) responsible for crude oil degradation might have been cured, thus not allowing the colony to grow in oil agar [36].
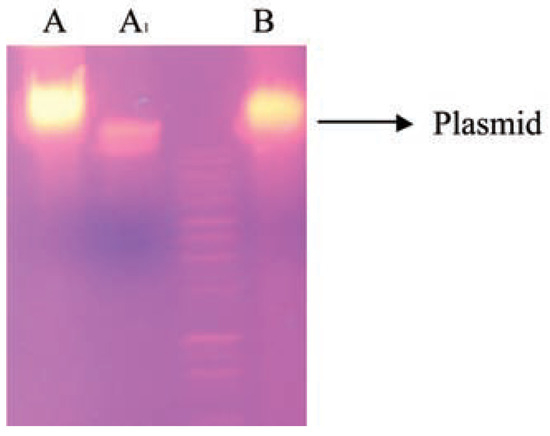
Figure 1.
Electrophoretic separation profiles of plasmids from cured and non-cured strains. Lane A: Corynebacterium sp; Lane A1: cured Corynebacterium sp; Lane B: Burkholderia cepacia.
Catabolic pathways, which encoded different aromatic hydrocarbon degradation routes, are frequently located on plasmids, although genes involved in degradation can be located on either chromosomal DNA or plasmid [37]. Our findings show that the capacity to mineralize crude oil by Burkholderia and Corynebacterium species are plasmid-encoded. Agarose gel electrophoretic separation profiles (Figure 1) of plasmidic DNA isolated from non-cured cultures showed that Burkholderia and Corynebacterium species harboured single plasmids with high molecular weight. To determine the plasmid harbouring the catabolic genes involved in crude oil degradation, electrophoretic profiles of plasmids isolated from the cured strains were compared with those of the non-cured one. The findings showed that the introduction of the transmissible catabolic plasmid into the indigenous bacteria strains in the oil sludge increase their catabolic ability. Result shown in Table 2 with 96% of oil degradation (Table 2). The transconjugants showed multiplicative effect on the bioremediation process by reducing the biokinetic parameters in ten days while twenty days in immobilised cells.
There a lot of investigations on the effect of inoculation and subsequent transfer of catabolic genes on in situ biodegradation in soils and bioreactors as demonstrated in the present study, such as: Di Giovanni et al. [38] demonstrated transfer of the IncP-1L plasmid pJP4 from Ralstonia eutropha JMP134 to indigenous soil bacteria when high concentrations of 2,4-D were added to the soil.
Top and colleagues [6,39] studied the effect of conjugative transfer of a few different 2,4-D degradative plasmids on 2,4-D degradation in soil; while Mancini et al. [40] examined plasmid transfer in an activated sludge unit and found the highest numbers of transconjugants on the bottom of the primary clarifier and in the return-activated sludge from the secondary clarifier, where cell densities are the highest.
Diya’uddeen et al. [30] reported that the minimum amount of dissolved oxygen necessary for normal life in an aquatic environment is about 2 mg/L [41], and the discharge of high organic matter containing waste waters into water bodies results in the excess consumption of oxygen by the bacteria. This is oxidation of the effluent, thus depleting oxygen from the water faster than it dissolves back into the water from the air [41]. In addition, oxygen availability is important because the end products of chemical and biochemical reactions in anaerobic systems often produce aesthetically displeasing colours, tastes and odours in water [8].
Phenol is one of the major pollutants found in refinery effluents. It was found in large concentrations in both the raw wastewater. The major sources of phenol in the petroleum refinery wastewater are the thermal and catalytic cracking processes. The observed value for phenol in raw wastewater was 68.13 mg/L while both cells were able to reduce it significantly to (0.32 and 1.06 mg/L) in both cells. Its content in the treated sample was significantly higher than in transconjugant treated cells mg/L [19] or 0.1 mg/L [18] recommended for refinery effluents.
Phenols are very toxic to fish and other aquatic organisms and has the unique property of tainting fish if present in water at concentration between 0.1 to 1 mg/L [42,43]. The toxic concentration for fishes may range from <0.1 to >100 mg/L, depending on the chemical nature of the phenol, the fish species and the developmental stage, with embryo-larval stage being many times more susceptible than adults [44].
Oil and grease and total petroleum hydrocarbon were found to be (43.20±2.67) to (2.53±0.94) mg/L and (4.12±0.56) to (0.88±0.12) mg/L in both raw wastewater and treated wastewater respectively after 20 days. Petroleum hydrocarbons have been observed to be toxic to aquatic life. Pollino and Holdway [45] observed that a water accommodated fraction of crude oil or dispersed crude oil water accommodated fraction increased the activity of gill citrate synthase at a concentration of 14.5 mg/L. Lipophilic hydrocarbons have been observed to accumulate in the membrane lipid bilayers of microorganisms and interfere with their structural and functional properties [46]. Grant and Briggs [47] concluded that when oil containing hydrocarbons are discharged into water body can cause depletion of dissolved oxygen due to transformation of organic components into inorganic compounds, loss of that is important in food chain and eutrophication also has short term toxicity in fishes which includes: lymphocytosis, epidermal hyperplasia, hemorrhagic septicaemia [2,30,48]. At the end of 20 days treatment the isolates were able to reduce the physicochemical parameters of refinery waste water, therefore, mixed culture used in this research work confirmed microbial consortia as better degraders as reported by several authors [49,50,51,52,53] therefore, this microbial blend can be adopted in refinery for the treatment of their wastewater before being discharge into waterways.
Conclusions
The result of the present findings revealed that refinery sludge harbour hydrocarbon degraders and that such organism can be immobilised for reusable purposes and that they carry transmissible catabolic plasmid that can be acquired by other autochthonous bacteria with weak catabolic potentials. This can be used for bioremediation protocol in refinery wastewater treatment.
Acknowledgments
the authors thank the Board of Research and the Department of Microbiology, Ahmadu Bello University, Zaria for providing funds. Ajao thanks Engr. Alfred U. Amadi, Deputy Manager; process Engineering Dept., Kaduna Refining & Petrochemical Company (KRPC), Kaduna, Nigeria for his technical assistance.
References
- Suleimanov, A.Y. Conditions of waste fluid accumulation at petrochemical and processing enterprise prevention of their harm to water bodies. Meditsina Truda Promyswe Nnaia Ekologila 1995, 12, 31–36. [Google Scholar]
- Uzoekwe, S.A.; Oghosanine, F.A. The effect of refinery and petrochemical effluent on water quality of Ubeji Creek Warri, Southern Nigeria. Ethiopian J Environ Stud Manag 2011, 4, 107–116. [Google Scholar] [CrossRef]
- Coelho, A.; Castro, A.V.; Dezotti, M.; Sant’Anna, G.L., Jr. Treatment of petroleum refinery sourwater by advanced oxidation processes. J Hazard Mater 2006, 137, 178–184. [Google Scholar] [CrossRef]
- Doggett, T.; Rascoe, A. Global energy demand seen up 44 percent by 2030. Washington, DC: REUTERS; 2009. Available online: http://www.reuters.com/article/2009/05/27/us-eia-global-demand-idUSN2719528620090527 (accessed on 17 September 2009).
- Top, E.M.; Moenne-Loccoz, Y.; Pembroke, T.; Thomas, C.M. Phenotypic traits conferred by plasmids. In The horizontal gene pool bacterial plasmids and gene spread; Thomas, C.M., Ed.; Harwood Academic Publishers: Newark, NJ, USA, 2000; pp. 249–285. [Google Scholar]
- Top, E.M.; Maila, M.P.; Clerinx, M.; Goris, J.; De Vos, P.; Verstraete, W. Methane oxidation as a method to evaluate the removal of 2,4-dichlorophenoxyacetic acid (2,4-D) from soil by plasmid mediated bioaugmentation. FEMS Microbiol 1999, 28, 203–213. [Google Scholar] [CrossRef]
- Saien, J.; Nejati, H. Enhanced photocatalytic degradation of pollutants in petroleum refinery wastewater under mild conditions. J Hazard Mater 2007, 148, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Leahy, J.G.; Colwell, R.R. Microbial degradation of hydrocarbons in the environment. Microbiol Rev 1990, 45, 305–315. [Google Scholar] [CrossRef]
- Rosenberg, E.; Ron, E.Z. Bioremediation oi petroleum contamination. In Bioremediation: principles and applications; Crawford, R.L., Crawford, D.L., Eds.; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar]
- Balba, M.T.; Al-Awadhi, R.; Al-Daher, R. Bioremediation of oil contaminated soil: microbiological methods for feasibility and field evaluation. J Microbiol Methods 1998, 32, 155–164. [Google Scholar] [CrossRef]
- Bouchez–Naitali, M.; Rakatozafy, H.; Marchals, R.; Leveau, J.V.; Van Beilendecasteele, J.P. Diversity of bacterial strains degrading hexadecane in relation to the mode of substrate uptake. J Appl Microbiol 1999, 86, 421–428. [Google Scholar] [CrossRef]
- Margesin, R.; Labbé, D.; Schinner, F.; Greer, C.W.; Whyte, L.G. Characterization of hydrocarbon-degradative microbial populations in contaminated and pristine alpine soils. Appl Environ Microl 2003, 69, 3085–3092. [Google Scholar] [CrossRef]
- Nweke, C.O.; Okpokwasili, G.C. Effects of bioremediation treatments on the bacterial populations of soil at different depths. Nig J Microbiol 2004, 18, 363–372. [Google Scholar]
- Kaplan, C.W.; Kitts, C.L. Bacterial succession in a petroleum land treatment unit. Appl Environ Microbiol 2004, 70, 1777–1786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quatrini, P.; Scaglione, G.; De Pasquale, C.; Reila, S.; Puglia, A.M. Isolation of Gram-positive n-alkane degraders from a hydrocarbon contaminated Mediterranean shoreline. J Appl Microbiol 2008, 104, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Top, E.M.; Springael, D.; Boon, N. Erratum to catabolic mobile genetic elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol Ecol 2002, 42, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Sanni Gbemisola, O.; Ajisebutu, O. Biodegradation of Escravos light crude oil by some species of soil bacteria. Sci Focus 2003, 4, 85–87. [Google Scholar]
- Department of Petroleum Resources (DPR). Environmental Guidelines and standards for the petroleum industry in Nigeria, production and terminal operations; Department of Petroleum Resources, Ministry of Petroleum Resources: Abuja, Nigeria, 1991. [Google Scholar]
- FEPA. Guidelines and standards for environmental pollution control in Nigeria; Federal Enviromental Protection Agency (FEPA): Abuja, Nigeria, 1991. [Google Scholar]
- American Public Health Association (APHA). Standard methods for examination of water/wastewater; APHA-AWWA-WPCF: Washington, DC, USA, 1995. [Google Scholar]
- American Society for Testing and Materials (ASTM). Annual book of ASTM standards; ASTM: Philadelphia, PA, USA, 1979; p. 1527. [Google Scholar]
- Joel, O.F.; Amajuoyi, C.A. Physicochemical characteristics and microbial quality of an oil polluted site in Gokana, River State. J Appl Sci Environ Manage 2009, 13, 99–103. [Google Scholar] [CrossRef][Green Version]
- Ademoroti, C.M.A. Chapter 2: Environment microbiology and medical science on bioremediation. In Standard methods for water and effluent analysis, 1st ed.; Ademoroti, C.M.A., Ed.; Foludex press, Ltd.: Ibadan, Nigeria, 1996; pp. 20–50. [Google Scholar]
- Adinarayana, K.; Jyothi, B.; Ellaiah, P. Production of alkaline protease with immobilized cells of bacillus subtilis. AAPS Pharm Sci Tech 2005, 6, 391–397. [Google Scholar] [CrossRef]
- Margesin, R.; Schinner, F. (Eds.) Manual for soil analysis – monitoring andassessing soil bioremediation; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Bathe, S.; Hausner, M. Plasmid-mediated bioaugmentation of wastewater microbial communities to a laboatory-scale bioreaction. In Bioremediation, methods in molecular, biology; Cummings, S.P., Ed.; 599; Humma press: New York, NY, USA, 2010; pp. 185–200. [Google Scholar]
- Ojo, A.O.; Oso, B.A. Isolation of plasmid – DNA from synthetic detergent degraders in waste water from a tropical environment. Afr J Microbiol Res 2009, 3, 123–127. [Google Scholar]
- Ahrne, S.; Molun, G.; Staahl, S. plasmids in Lactobacillus strains isolated from meat and meat products system. Appl Microbial 1989, 11, 320–325. [Google Scholar]
- Bhalakia, N. Isolation and plasmid analysis of vancomycin resistant Staphylococcus aureus. J Young Invest 2006, 15, 15–24. [Google Scholar]
- Diya’uddeen Basheer, H.; Wan Mohd, A.; Wan, D.; Abdul Aziz, A.R. Treatment technologies for petroleum refinery effluents: a review. Proc Safe Environ Protect 2011, 89, 95–105. [Google Scholar] [CrossRef]
- Igbinosa, E.O.; Oko, A.I. Impact of discharge wastewater effluents on the physiscochemical qualities of a receiving watershed in a typical rural community. Int J Environ Sci Tech 2009, 6, 175–182. [Google Scholar] [CrossRef]
- Thavan, R.; Jayalakshmi, S.; Radha Krisnan, R.; Balasubramanian, T. Plasmid incidence infour species of hydrocarbonate bacteria isolated from oil polluted marine environment. Biotechnology 2007, 6, 349–352. [Google Scholar]
- Mirdamadian, S.H.; Entiazi, G.; Golabi, M.H.; Ghanavati, H. Biodegradation of petroleum and Aromatic hydrocarbons by bacteria isolated from petroleum contaminated soil. J Pet Environ Biotechnol 2010, 1, 102. [Google Scholar] [CrossRef]
- Toledo, F.L.; Calvo, C.; Rodelas, B.; González-López, J. Selection and identification of bacteria isolated from waste crude oil with polycyclic aromatic hydrocarbons removal capacities. System Appl Microbiol 2006, 29, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Whyte, L.G.; Bourbonnière, L.; Greer, C.W. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas Pseudomonas strains possessing both alkane (alkalk) and naphthalene (nah-nah) catabolic pathways. Appl Environ Microbiol 1997, 63, 3719–3723. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Singla, R.; Kumar, G. Plasmid associated anthracene degradation by Pseudomonas sp isolated from fitting stating site. Nature Sci 2010, 8, 89–94. [Google Scholar]
- Coral, G.; Karagoz, S. Isolation and characterization of phenan threne – degrading bacteria from a petroleum refinery soil. Annals Microbiol 2005, 55, 255–259. [Google Scholar]
- DiGiovanni, G.D.; Neilson, J.W.; Pepper, I.L.; Sinclair, N.A. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl Environ Microbiol 1996, 62, 2521–2526. [Google Scholar] [CrossRef]
- Top, E.M.; Van Daele, P.; De Saeyer, N.; Forney, L.J. Enhancement of 2,4-dichlorophenoxy-acetic acid (2,4-D) degradation in soil by dissemination of catabolic plasmids. Antonie Van Leeuwenhoek 1998, 73, 87–94. [Google Scholar] [CrossRef]
- Mancini, P.; Fertels, S.; Nave, D.; Gealt, M.A. Mobilization of plasmid pHSV106 from Escherichia coli HB101 in a laboratory-scale waste treatment facility. Appl Environ Microbiol 1987, 53, 665–671. [Google Scholar] [CrossRef]
- Attiogbe, F.K.; Glover-Amengor, M.; Nyadziehe, K.T. Correlating biochemical and chemical oxygen demand of effluents-a case study of selected industries in Kumasi, bacteria from coastal water. Biotechnol Bio Eng 2007, 14, 297–308. [Google Scholar]
- De-Bruin, D.B. A bacterial bioassay for assessment of waste-water toxicity. In Mechanism of toxicity for various compounds; Kenneth, J., Ed.; Wat-Res. G. Britain, 1976; pp. 383–390. [Google Scholar]
- Staples, C.A.; Dorn, P.B.; Klecka, G.M.; O’Block, S.T.; Harris, L.R. A review of the environmental fate, effects and exposures of bisphenol A. Chemosphere 1998, 36, 2149–2173. [Google Scholar] [CrossRef] [PubMed]
- DeGraeve, G.M.; Geiger, D.L.; Meyer, J.S.; Bergman, H.L. Acute and embryolarval toxicity of phenolic compounds to aquatic biota. Arch Environ Contam Toxicol 1980, 9, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Pollino, C.A.; Holdway, D.A. Hydrocarboninduced changes to metabolic and detoxification enzymes of the Australinan crimson-spotted rainbow fish (Melanotaeni fluviatilis). Environ Toxicol 2003, 18, 21–28. [Google Scholar] [CrossRef]
- Sikkema, J.A.; deBont, A.M.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microb Rev 1995, 59, 201–222. [Google Scholar] [CrossRef]
- Grant, A.; Briggs, A.D. Toxicity of sediments from around a North Sea oil platform: are metals or hydrocarbons responsible for ecological impacts? Marine Environ Res 2002, 53, 95–116. [Google Scholar] [CrossRef]
- Beeby, A. Measuring the effect of pollution. In Applying ecology; Beeby, A., Ed.; Chapman and Hall: London, UK; New York, NY, USA, 1993; pp. 220–420. [Google Scholar]
- Obire, O. Studies on the biodegradation potential of some microorganisms isolated from water systems of two petroleum-producing areas in Nigeria. Nig J Bot 1988, 1, 81–90. [Google Scholar]
- Amund, O.O.; Nwokaye, N. Hydrocarbon degradation potentials of yeast isolates from a polluted lagoon. J Sci Res Dev 1993, 1, 61–64. [Google Scholar]
- Facundo, J.M.R.; Vaness, H.R.; Teresa, M.L. Biodegradation of diesel oil in soil by a microbial consortium. Water Air Pollut 2001, 129, 313–320. [Google Scholar]
- Kulwadee, T.; Vithaya, M.; Prayad, P.; Attawut, I. Isolation and characterisation of crude oil, degrading bacteria in Thailand. In Proceedings of the International Conference on ‘New Horizons in Biotechnology’, Trivandruim, India, 18–21 April 2001. [Google Scholar]
- Ndip, R.N.; Akoachere, J.F.; Akeniji, T.N.; Yongabi, F.N.; Nkwekang, G. Lubrication oil degrading bacterial in soils from filling stations and auto mechanic workshops in Buea, Cameroon occurrence and characteristics of isolates. Afr J Biotechnol 2008, 11, 1700–1706. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
©Copyright A.T. Ajao et al., 2013 Licensee PAGEPress, Italy. This work is licensed under a Creative Commons Attribution NonCommercial 3.0 License (CC BYNC 3.0).