DFNA20/26 and Other ACTG1-Associated Phenotypes: A Case Report and Review of the Literature
Abstract
1. Introduction
2. Materials and Methods
3. Results
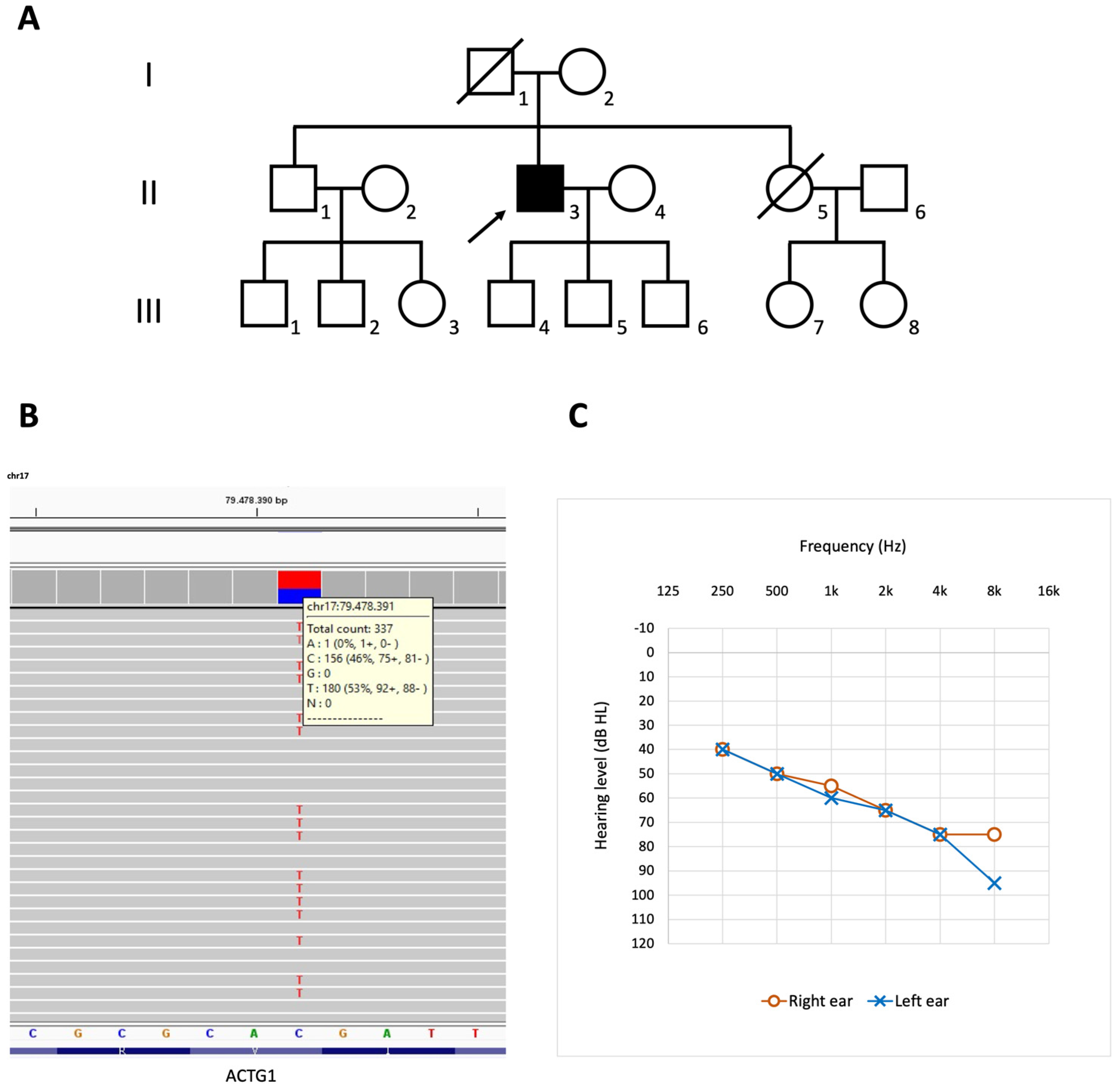
3.1. Case Description
3.2. Literature Review
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sonnemann, K.J.; Fitzsimons, D.P.; Patel, J.R.; Liu, Y.; Schneider, M.F.; Moss, R.L.; Ervasti, J.M. Cytoplasmic gamma-actin is not required for skeletal muscle development but its absence leads to a progressive myopathy. Dev. Cell 2006, 11, 387–397. [Google Scholar] [CrossRef]
- Khaitlina, S.Y. Functional specificity of actin isoforms. Int. Rev. Cytol. 2001, 202, 35–98. [Google Scholar] [CrossRef]
- Perrin, B.J.; Sonnemann, K.J.; Ervasti, J.M. β-actin and γ-actin are each dispensable for auditory hair cell development but required for Stereocilia maintenance. PLoS Genet. 2010, 6, e1001158. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.C.; Belyantseva, I.A.; Friderici, K.H.; Friedman, T.B. Actin in hair cells and hearing loss. Hear. Res. 2012, 288, 89–99. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, T.; Wei, S.; DeWan, A.T.; Morell, R.J.; Elfenbein, J.L.; Fisher, R.A.; Leal, S.M.; Smith, R.J.; Friderici, K.H. Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26). Am. J. Hum. Genet. 2003, 73, 1082–1091. [Google Scholar] [CrossRef] [PubMed]
- Rivière, J.B.; van Bon, B.W.; Hoischen, A.; Kholmanskikh, S.S.; O'Roak, B.J.; Gilissen, C.; Gijsen, S.; Sullivan, C.T.; Christian, S.L.; Abdul-Rahman, O.A.; et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 2012, 44, 440–444. [Google Scholar] [CrossRef]
- Verloes, A.; Di Donato, N.; Masliah-Planchon, J.; Jongmans, M.; Abdul-Raman, O.A.; Albrecht, B.; Allanson, J.; Brunner, H.; Bertola, D.; Chassaing, N.; et al. Baraitser-Winter cerebrofrontofacial syndrome: Delineation of the spectrum in 42 cases. Eur. J. Hum. Genet. 2015, 23, 292–301. [Google Scholar] [CrossRef] [PubMed]
- del Castillo, F.J.; Rodríguez-Ballesteros, M.; Alvarez, A.; Hutchin, T.; Leonardi, E.; de Oliveira, C.A.; Azaiez, H.; Brownstein, Z.; Avenarius, M.R.; Marlin, S.; et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 2005, 42, 588–594. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Oza, A.M.; DiStefano, M.T.; Hemphill, S.E.; Cushman, B.J.; Grant, A.R.; Siegert, R.K.; Shen, J.; Chapin, A.; Boczek, N.J.; Schimmenti, L.A.; et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 2018, 39, 1593–1613. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Jang, J.; Jin, H.S. A novel missense mutation in the ACTG1 gene in a family with congenital autosomal dominant deafness: A case report. Mol. Med. Rep. 2018, 17, 7611–7617. [Google Scholar] [CrossRef]
- Wang, H.; Guan, J.; Lan, L.; Yu, L.; Xie, L.; Liu, X.; Yang, J.; Zhao, C.; Wang, D.; Wang, Q. A novel de novo mutation of ACTG1 in two sporadic non-syndromic hearing loss cases. Sci. China Life Sci. 2018, 61, 729–732. [Google Scholar] [CrossRef] [PubMed]
- Miyajima, H.; Moteki, H.; Day, T.; Nishio, S.Y.; Murata, T.; Ikezono, T.; Takeda, H.; Abe, S.; Iwasaki, S.; Takahashi, M.; et al. Novel ACTG1 mutations in patients identified by massively parallel DNA sequencing cause progressive hearing loss. Sci. Rep. 2020, 10, 7056. [Google Scholar] [CrossRef]
- Miyagawa, M.; Nishio, S.Y.; Ichinose, A.; Iwasaki, S.; Murata, T.; Kitajiri, S.; Usami, S. Mutational spectrum and clinical features of patients with ACTG1 mutations identified by massively parallel DNA sequencing. Ann. Otol. Rhinol. Laryngol. 2015, 124, 84S–93S. [Google Scholar] [CrossRef] [PubMed]
- de Heer, A.M.; Huygen, P.L.; Collin, R.W.; Oostrik, J.; Kremer, H.; Cremers, C.W. Audiometric and vestibular features in a second Dutch DFNA20/26 family with a novel mutation in ACTG1. Ann. Otol. Rhinol. Laryngol. 2009, 118, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.; Lenarduzzi, S.; Cappellani, S.; Pecile, V.; Morgutti, M.; Orzan, E.; Ghiselli, S.; Ambrosetti, U.; Brumat, M.; Gajendrarao, P.; et al. Genomic Studies in a Large Cohort of Hearing Impaired Italian Patients Revealed Several New Alleles, a Rare Case of Uniparental Disomy (UPD) and the Importance to Search for Copy Number Variations. Front. Genet. 2018, 9, 681. [Google Scholar] [CrossRef]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef]
- Wang, L.; Yan, D.; Qin, L.; Li, T.; Liu, H.; Li, W.; Mittal, R.; Yong, F.; Chapagain, P.; Liao, S.; et al. Amino acid 118 in the Deafness Causing (DFNA20/26). Gene Rep. 2018, 11, 264–269. [Google Scholar] [CrossRef]
- Morín, M.; Bryan, K.E.; Mayo-Merino, F.; Goodyear, R.; Mencía, A.; Modamio-Høybjør, S.; del Castillo, I.; Cabalka, J.M.; Richardson, G.; Moreno, F.; et al. In vivo and in vitro effects of two novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum. Mol. Genet. 2009, 18, 3075–3089. [Google Scholar] [CrossRef]
- Liu, P.; Li, H.; Ren, X.; Mao, H.; Zhu, Q.; Zhu, Z.; Yang, R.; Yuan, W.; Liu, J.; Wang, Q.; et al. Novel ACTG1 mutation causing autosomal dominant non-syndromic hearing impairment in a Chinese family. J. Genet. Genom. 2008, 35, 553–558. [Google Scholar] [CrossRef]
- Cabanillas, R.; Diñeiro, M.; Cifuentes, G.A.; Castillo, D.; Pruneda, P.C.; Álvarez, R.; Sánchez-Durán, N.; Capín, R.; Plasencia, A.; Viejo-Díaz, M.; et al. Comprehensive genomic diagnosis of non-syndromic and syndromic hereditary hearing loss in Spanish patients. BMC Med. Genom. 2018, 11, 58. [Google Scholar] [CrossRef]
- Baux, D.; Vaché, C.; Blanchet, C.; Willems, M.; Baudoin, C.; Moclyn, M.; Faugère, V.; Touraine, R.; Isidor, B.; Dupin-Deguine, D.; et al. Combined genetic approaches yield a 48% diagnostic rate in a large cohort of French hearing-impaired patients. Sci. Rep. 2017, 7, 16783. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, M.; Naito, T.; Nishio, S.Y.; Kamatani, N.; Usami, S. Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of Japanese deafness patients. PLoS ONE 2013, 8, e71381. [Google Scholar] [CrossRef]
- Baek, J.I.; Oh, S.K.; Kim, D.B.; Choi, S.Y.; Kim, U.K.; Lee, K.Y.; Lee, S.H. Targeted massive parallel sequencing: The effective detection of novel causative mutations associated with hearing loss in small families. Orphanet J. Rare Dis. 2012, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Gao, X.; Huang, B.; Lu, J.; Wang, G.; Lin, X.; Qu, Y.; Dai, P. Phenotypic Heterogeneity in a DFNA20/26 family segregating a novel ACTG1 mutation. BMC Genet. 2016, 17, 33. [Google Scholar] [CrossRef]
- Mutai, H.; Suzuki, N.; Shimizu, A.; Torii, C.; Namba, K.; Morimoto, N.; Kudoh, J.; Kaga, K.; Kosaki, K.; Matsunaga, T. Diverse spectrum of rare deafness genes underlies early-childhood hearing loss in Japanese patients: A cross-sectional, multi-center next-generation sequencing study. Orphanet J. Rare Dis. 2013, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.H.; Chang, P.Y.; Chang, S.C.; Lu, J.J.; Wu, C.M. Mutation screening in non-syndromic hearing loss patients with cochlear implantation by massive parallel sequencing in Taiwan. PLoS ONE 2019, 14, e0211261. [Google Scholar] [CrossRef]
- van Wijk, E.; Krieger, E.; Kemperman, M.H.; De Leenheer, E.M.; Huygen, P.L.; Cremers, C.W.; Cremers, F.P.; Kremer, H. A mutation in the gamma actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26). J. Med. Genet. 2003, 40, 879–884. [Google Scholar] [CrossRef]
- Miyagawa, M.; Nishio, S.Y.; Ikeda, T.; Fukushima, K.; Usami, S. Massively parallel DNA sequencing successfully identifies new causative mutations in deafness genes in patients with cochlear implantation and EAS. PLoS ONE 2013, 8, e75793. [Google Scholar] [CrossRef]
- Park, G.; Gim, J.; Kim, A.R.; Han, K.H.; Kim, H.S.; Oh, S.H.; Park, T.; Park, W.Y.; Choi, B.Y. Multiphasic analysis of whole exome sequencing data identifies a novel mutation of ACTG1 in a nonsyndromic hearing loss family. BMC Genom. 2013, 14, 191. [Google Scholar] [CrossRef]
- Wei, Q.; Zhu, H.; Qian, X.; Chen, Z.; Yao, J.; Lu, Y.; Cao, X.; Xing, G. Targeted genomic capture and massively parallel sequencing to identify novel variants causing Chinese hereditary hearing loss. J. Transl. Med. 2014, 12, 311. [Google Scholar] [CrossRef] [PubMed]
- Vona, B.; Müller, T.; Nanda, I.; Neuner, C.; Hofrichter, M.A.; Schröder, J.; Bartsch, O.; Läßig, A.; Keilmann, A.; Schraven, S.; et al. Targeted next-generation sequencing of deafness genes in hearing-impaired individuals uncovers informative mutations. Genet. Med. 2014, 16, 945–953. [Google Scholar] [CrossRef]
- Rendtorff, N.D.; Zhu, M.; Fagerheim, T.; Antal, T.L.; Jones, M.; Teslovich, T.M.; Gillanders, E.M.; Barmada, M.; Teig, E.; Trent, J.M.; et al. A novel missense mutation in ACTG1 causes dominant deafness in a Norwegian DFNA20/26 family, but ACTG1 mutations are not frequent among families with hereditary hearing impairment. Eur. J. Hum. Genet. 2006, 14, 1097–1105. [Google Scholar] [CrossRef]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.F.; Holmes, K.C. Atomic structure of the actin:DNase I complex. Nature 1990, 347, 37–44. [Google Scholar] [CrossRef]
- Parker, F.; Baboolal, T.G.; Peckham, M. Actin Mutations and Their Role in Disease. Int. J. Mol. Sci. 2020, 21, 3371. [Google Scholar] [CrossRef]
- Morton, C.C.; Nance, W.E. Newborn hearing screening—A silent revolution. N. Engl. J. Med. 2006, 354, 2151–2164. [Google Scholar] [CrossRef] [PubMed]
- Morell, R.J.; Friderici, K.H.; Wei, S.; Elfenbein, J.L.; Friedman, T.B.; Fisher, R.A. A new locus for late-onset, progressive, hereditary hearing loss DFNA20 maps to 17q25. Genomics 2000, 63, 1–6. [Google Scholar] [CrossRef]
- Elfenbein, J.L.; Fisher, R.A.; Wei, S.; Morell, R.J.; Stewart, C.; Friedman, T.B.; Friderici, K. Audiologic aspects of the search for DFNA20: A gene causing late-onset, progressive, sensorineural hearing loss. Ear Hear. 2001, 22, 279–288. [Google Scholar] [CrossRef]
- Yang, T.; Smith, R. A novel locus DFNA26 maps to chromosome 17q25 in two unrelated families with progressive autosomal dominant hearing loss (Abstract). Am. J. Hum. Genet. 2000, 67, 300. [Google Scholar]
- DeWan, A.T.; Parrado, A.R.; Leal, S.M. A second kindred linked to DFNA20 (17q25.3) reduces the genetic interval. Clin. Genet. 2003, 63, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Belyantseva, I.A.; Perrin, B.J.; Sonnemann, K.J.; Zhu, M.; Stepanyan, R.; McGee, J.; Frolenkov, G.I.; Walsh, E.J.; Friderici, K.H.; Friedman, T.B.; et al. Gamma-actin is required for cytoskeletal maintenance but not development. Proc. Natl. Acad. Sci. USA 2009, 106, 9703–9708. [Google Scholar] [CrossRef]
- Nishio, S.Y.; Usami, S.I. Outcomes of cochlear implantation for the patients with specific genetic etiologies: A systematic literature review. Acta oto-Laryngol. 2017, 137, 730–742. [Google Scholar] [CrossRef]
- Bryan, K.E.; Wen, K.K.; Zhu, M.; Rendtorff, N.D.; Feldkamp, M.; Tranebjaerg, L.; Friderici, K.H.; Rubenstein, P.A. Effects of human deafness gamma-actin mutations (DFNA20/26) on actin function. J. Biol. Chem. 2006, 281, 20129–20139. [Google Scholar] [CrossRef] [PubMed]
- Bryan, K.E.; Rubenstein, P.A. Allele-specific effects of human deafness gamma-actin mutations (DFNA20/26) on the actin/cofilin interaction. J. Biol. Chem. 2009, 284, 18260–18269. [Google Scholar] [CrossRef] [PubMed]
- Kruth, K.A.; Rubenstein, P.A. Two deafness-causing (DFNA20/26) actin mutations affect Arp2/3-dependent actin regulation. J. Biol. Chem. 2012, 287, 27217–27226. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, L.; Kruth, K.A.; Rubenstein, P.A.; Sept, D. Two Deafness-Causing Actin Mutations (DFNA20/26) Have Allosteric Effects on the Actin Structure. Biophys. J. 2016, 111, 323–332. [Google Scholar] [CrossRef][Green Version]
- Di Donato, N.; Kuechler, A.; Vergano, S.; Heinritz, W.; Bodurtha, J.; Merchant, S.R.; Breningstall, G.; Ladda, R.; Sell, S.; Altmüller, J.; et al. Update on the ACTG1-associated Baraitser-Winter cerebrofrontofacial syndrome. Am. J. Med Genet. Part A 2016, 170, 2644–2651. [Google Scholar] [CrossRef]
- Di Donato, N.; Rump, A.; Koenig, R.; Der Kaloustian, V.M.; Halal, F.; Sonntag, K.; Krause, C.; Hackmann, K.; Hahn, G.; Schrock, E.; et al. Severe forms of Baraitser-Winter syndrome are caused by ACTB mutations rather than ACTG1 mutations. Eur. J. Hum. Genet. 2014, 22, 179–183. [Google Scholar] [CrossRef]
- Poirier, K.; Martinovic, J.; Laquerrière, A.; Cavallin, M.; Fallet-Bianco, C.; Desguerre, I.; Valence, S.; Grande-Goburghun, J.; Francannet, C.; Deleuze, J.F.; et al. Rare ACTG1 variants in fetal microlissencephaly. Eur. J. Med. Genet. 2015, 58, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Retterer, K.; Juusola, J.; Cho, M.T.; Vitazka, P.; Millan, F.; Gibellini, F.; Vertino-Bell, A.; Smaoui, N.; Neidich, J.; Monaghan, K.G.; et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016, 18, 696–704. [Google Scholar] [CrossRef]
- Allawh, T.C.; Brown, B.S. The Clinical Manifestations and Genetic Implications of Baraitser-Winter Syndrome Type 2. J. Pediatr. Genet. 2017, 6, 107–110. [Google Scholar] [CrossRef]
- Kemerley, A.; Sloan, C.; Pfeifer, W.; Smith, R.; Drack, A. A novel mutation in ACTG1 causing Baraitser-Winter syndrome with extremely variable expressivity in three generations. Ophthalmic Genet. 2017, 38, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Longoni, M.; High, F.A.; Qi, H.; Joy, M.P.; Hila, R.; Coletti, C.M.; Wynn, J.; Loscertales, M.; Shan, L.; Bult, C.J.; et al. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum. Genet. 2017, 136, 679–691. [Google Scholar] [CrossRef]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med. 2017, 376, 21–31. [Google Scholar] [CrossRef]
- Rainger, J.; Williamson, K.A.; Soares, D.C.; Truch, J.; Kurian, D.; Gillessen-Kaesbach, G.; Seawright, A.; Prendergast, J.; Halachev, M.; Wheeler, A.; et al. A recurrent de novo mutation in ACTG1 causes isolated ocular coloboma. Hum. Mutat. 2017, 38, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Zazo Seco, C.; Wesdorp, M.; Feenstra, I.; Pfundt, R.; Hehir-Kwa, J.Y.; Lelieveld, S.H.; Castelein, S.; Gilissen, C.; de Wijs, I.J.; Admiraal, R.J.; et al. The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands. Eur. J. Hum. Genet. 2017, 25, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, N.; Timms, A.E.; Aldinger, K.A.; Mirzaa, G.M.; Bennett, J.T.; Collins, S.; Olds, C.; Mei, D.; Chiari, S.; Carvill, G.; et al. Analysis of 17 genes detects mutations in 81% of 811 patients with lissencephaly. Genet. Med. 2018, 20, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Homma, T.K.; Freire, B.L.; Honjo Kawahira, R.S.; Dauber, A.; Funari, M.F.A.; Lerario, A.M.; Nishi, M.Y.; Albuquerque, E.V.; Vasques, G.A.; Collett-Solberg, P.F.; et al. Genetic Disorders in Prenatal Onset Syndromic Short Stature Identified by Exome Sequencing. J. Pediatr. 2019, 215, 192–198. [Google Scholar] [CrossRef]
- Thiffault, I.; Farrow, E.; Zellmer, L.; Berrios, C.; Miller, N.; Gibson, M.; Caylor, R.; Jenkins, J.; Faller, D.; Soden, S.; et al. Clinical genome sequencing in an unbiased pediatric cohort. Genet. Med. 2019, 21, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Vontell, R.; Supramaniam, V.G.; Davidson, A.; Thornton, C.; Marnerides, A.; Holder-Espinasse, M.; Lillis, S.; Yau, S.; Jansson, M.; Hagberg, H.E.; et al. Post-mortem Characterisation of a Case With an ACTG1 Variant, Agenesis of the Corpus Callosum and Neuronal Heterotopia. Front. Physiol. 2019, 10, 623. [Google Scholar] [CrossRef]
- Yamamoto, T.; Imaizumi, T.; Yamamoto-Shimojima, K.; Lu, Y.; Yanagishita, T.; Shimada, S.; Chong, P.F.; Kira, R.; Ueda, R.; Ishiyama, A.; et al. Genomic backgrounds of Japanese patients with undiagnosed neurodevelopmental disorders. Brain Dev. 2019, 41, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Accogli, A.; Severino, M.; Riva, A.; Madia, F.; Balagura, G.; Iacomino, M.; Carlini, B.; Baldassari, S.; Giacomini, T.; Croci, C.; et al. Targeted re-sequencing in malformations of cortical development: Genotype-phenotype correlations. Seizure 2020, 80, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Chacon-Camacho, O.F.; Barragán-Arévalo, T.; Villarroel, C.E.; Almanza-Monterrubio, M.; Zenteno, J.C. Previously undescribed phenotypic findings and novel ACTG1 gene pathogenic variants in Baraitser-Winter cerebrofrontofacial syndrome. Eur. J. Med. Genet. 2020, 63, 103877. [Google Scholar] [CrossRef] [PubMed]
- Gieldon, L.; Mackenroth, L.; Kahlert, A.K.; Lemke, J.R.; Porrmann, J.; Schallner, J.; von der Hagen, M.; Markus, S.; Weidensee, S.; Novotna, B.; et al. Diagnostic value of partial exome sequencing in developmental disorders. PLoS ONE 2018, 13, e0201041. [Google Scholar] [CrossRef]
- Stutterd, C.A.; Brock, S.; Stouffs, K.; Fanjul-Fernandez, M.; Lockhart, P.J.; McGillivray, G.; Mandelstam, S.; Pope, K.; Delatycki, M.B.; Jansen, A.; et al. Genetic heterogeneity of polymicrogyria: Study of 123 patients using deep sequencing. Brain Commun. 2021, 3, fcaa221. [Google Scholar] [CrossRef]
- Perea-Romero, I.; Blanco-Kelly, F.; Sanchez-Navarro, I.; Lorda-Sanchez, I.; Tahsin-Swafiri, S.; Avila-Fernandez, A.; Martin-Merida, I.; Trujillo-Tiebas, M.J.; Lopez-Rodriguez, R.; Rodriguez de Alba, M.; et al. NGS and phenotypic ontology-based approaches increase the diagnostic yield in syndromic retinal diseases. Hum. Genet. 2021, 1–14. [Google Scholar] [CrossRef]
| Exon 1 | Nucleotide Change 1 | Protein Change 1 | Protein Subdomain 2 | Number of Families | HL Onset | Age at HL Onset 3 | High Frequencies More Severily Affected | HL Progression | Tinnitus 4 | Vertigo and Other Vestibular Symptoms 4 | Other Associated Features | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | c.94C>T | p.Pro32Ser | 1 | 3 | Pre-lingual (congenital)/Post-lingual | Birth–3 years | + | NA | − | − | Complete bilateral CLP in a patient 5 | Lee et al. (2018) [11], Wang et al. (2018) [12] |
| 2 | c.102C>G | p.Ile34Met | 2 | 1 | Post-lingual | 15–18 years | + | + | + | + | − | Miyajima et al. (2020) [13] |
| 2 | c.110G>A | p.Arg37His | 2 | 1 | Post-lingual | 10 years | + | + | + | + | − | Miyajima et al. (2020) [13] |
| 3 | c.142G>C | p.Gly48Arg | 2 | 1 | Post-lingual | 6–11 years | + | + | + | − | − | Miyagawa et al. (2015) [14], Miyajima et al. (2020) [13] |
| 3 | c.151G>A | p.Asp51Asn | 2 | 1 | Post-lingual (?) | 1st decade | + | + | + | + | − | de Heer et al. (2009) [15] |
| 3 | c.197C>T | p.Thr66Ile | 2 | 1 | NA | NA (Early-onset) | + | NA | NA | NA | − | Morgan et al. (2018) [16] |
| 3 | c.244A>T | p.Met82Leu | 1 | 1 | Post-lingual | Adulthood | NA | NA | NA | NA | − | Sloan-Heggen et al. (2016) [17] |
| 3 | c.246G>A | p.Met82Ile | 1 | 1 | NA | NA | + | NA | − | − | − | Miyajima et al. (2020) [13] |
| 3 | c.266C>T | p.Thr89Ile | 1 | 3 | Post-lingual | Early teens/Adulthood | + | + | + | − | − | Zhu et al. (2003) [5], Sloan-Heggen et al. (2016) [17], Miyajima et al. (2020) [13] |
| 3 | c.353A>T | p.Lys118Met | 1 | 4 | Post-lingual | 15–26 years | + | + | + | + | − | Zhu et al. (2003) [5], Miyagawa et al. (2015) [14], Wang et al. (2018) [18], Miyajima et al. (2020) [13] |
| 3 | c.354G>C | p.Lys118Asn | 1 | 1 | Post-lingual | 3rd decade | + | + | NA | NA | − | Morín et al. (2009) [19] |
| 4 | c.364A>G | p.Ile122Val | 1 | 1 | Post-lingual | 6 years | + | + | NA | − | − | Liu et al. (2008) [20] |
| 4 | c.434C>G | Ser145Cys | 3 | 1 | Pre-lingual (congenital) | Birth | NA | NA | NA | NA | − | Cabanillas et al. (2018) [21] |
| 4 | c.434C>T | p.Ser145Phe | 3 | 1 | Pre-lingual (congenital) | Birth | NA | + | NA | NA | − | Baux et al. (2017) [22] |
| 4 | c.485C>T | p.Thr162Met | 3 | 1 | Post-lingual | NA (Late onset) | NA | NA | NA | NA | − | Miyagawa et al. (2013) [23] |
| 4 | c.493A>G | p.Ile165Val | 3 | 1 | Post-lingual | 11 years | + | + | − | − | − | Miyajima et al. (2020) [13] |
| 4 | c.542C>G | p.Ala181Gly | 4 | 1 | Post-lingual | Childhood | NA | NA | NA | NA | − | Sloan-Heggen et al. (2016) [17] |
| 4 | c.548G>A | p.Arg183Gln | 4 | 2 | Post-lingual (?) | Childhood (early-onset) | + | NA | NA | NA | − | Cabanillas et al. (2018) [21], Morgan et al. (2018) [16] |
| 4 | c.559G>C | p.Asp187His | 4 | 1 | Pre-lingual | 1st year | NA | NA | NA | NA | − | Baek et al. (2012) [24] |
| 4 | c.625G>A | p.Val209Met | 4 | 1 | Post-lingual | 6 years | + | + | + | − | − | This study |
| 4 | c.638A>G | p.Lys213Arg | 4 | 1 | Post-lingual | 2nd decade | + | + | − | NA | − | Yuan et al. (2016) [25] |
| 4 | c.721G>A | p.Glu241Lys | 4 | 2 | Post-lingual | 3–14 years | + | + | − | − | Developmental disorder; Moyamoya disease 6 | Morín et al. (2009) [19], Miyagawa et al. (2015) [14], Miyajima et al. (2020) [13] |
| 4 | c.791C>T | p.Pro264Leu | 4 | 2 | Post-lingual | 2nd decade | + | + | + | + | − | Zhu et al. (2003) [5], Miyajima et al. (2020) [13] |
| 4 | c.802G>A | p.Gly268Ser | 4 | 1 | Post-lingual | 10–45 years | + | + | NA | NA | − | Mutai et al. (2013) [26] |
| 5 | c.823C>T | p.His275Tyr | 3 | 1 | Post-lingual | 34 years | + | + | + | − | − | Miyajima et al. (2020) [13] |
| 5 | c.830C>T | p.Thr277Ile | 3 | 1 | Post-lingual | 1st–2nd decades | + | + | NA | NA | − | Liu et al. (2019) [27] |
| 5 | c.833C>T | p.Thr278Ile | 3 | 1 | Post-lingual | 5–25 years | + | + | NA | + | − | van Wijk et al. (2003) [28] |
| 5 | c.847A>G | p.Met283Val | 3 | 1 | Post-lingual | Adulthood | NA | NA | NA | NA | − | Morgan et al. (2018) [16] |
| 5 | c.848T>C | p.Met283Thr | 3 | 1 | Post-lingual (?) | Childhood (early-onset) | NA | NA | NA | NA | − | Cabanillas et al. (2018) [21] |
| 5 | c.895C>G | p.Leu299Val | 3 | 1 | Post-lingual | 1st–5th decades | + | + | + | − | − | Miyagawa et al. (2013) [29], Miyagawa et al. (2015) [14], Miyajima et al. (2020) [13] |
| 5 | c.914T>C | p.Met305Thr | 3 | 2 | Post-lingual | 6 years | + | + | + | − | − | Park et al. (2013) [30], Miyajima et al. (2020 [13] |
| 5 | c.946G>A | p.Glu316Lys | 3 | 1 | Post-lingual | NA | + | + | NA | − | − | Wei et al. (2014) [31] |
| 5 | c.974T>A | p.Met325Lys | 3 | 1 | Pre-lingual (congenital) 7 | Birth | + | NA | NA | NA | − | Vona et al. (2014) [32] |
| 6 | c.994C>G | p.Pro332Ala | 3 | 1 | Post-lingual | Before early teens/2nd decade | + | + | NA | − | − | Zhu et al. (2003) [5] |
| 6 | c.994C>T | p.Pro332Ser | 3 | 2 | Post-lingual | 35–59 years | + | + | + | − | − | Miyajima et al. (2020) [13] |
| 6 | c.1045C>A | p.Leu349Met | 1 | 1 | Pre-lingual (congenital) | Birth | NA | NA | NA | NA | NA | Sloan-Heggen et al. (2016) [17] |
| 6 | c.1109T>C | p.Val370Ala | 1 | 1 | Post-lingual | 1st–2nd decades | + | + | NA | + | − | Rendtorff et al. (2006) [33] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorrentino, U.; Piccolo, C.; Rigon, C.; Brasson, V.; Trevisson, E.; Boaretto, F.; Martini, A.; Cassina, M. DFNA20/26 and Other ACTG1-Associated Phenotypes: A Case Report and Review of the Literature. Audiol. Res. 2021, 11, 582-593. https://doi.org/10.3390/audiolres11040052
Sorrentino U, Piccolo C, Rigon C, Brasson V, Trevisson E, Boaretto F, Martini A, Cassina M. DFNA20/26 and Other ACTG1-Associated Phenotypes: A Case Report and Review of the Literature. Audiology Research. 2021; 11(4):582-593. https://doi.org/10.3390/audiolres11040052
Chicago/Turabian StyleSorrentino, Ugo, Chiara Piccolo, Chiara Rigon, Valeria Brasson, Eva Trevisson, Francesca Boaretto, Alessandro Martini, and Matteo Cassina. 2021. "DFNA20/26 and Other ACTG1-Associated Phenotypes: A Case Report and Review of the Literature" Audiology Research 11, no. 4: 582-593. https://doi.org/10.3390/audiolres11040052
APA StyleSorrentino, U., Piccolo, C., Rigon, C., Brasson, V., Trevisson, E., Boaretto, F., Martini, A., & Cassina, M. (2021). DFNA20/26 and Other ACTG1-Associated Phenotypes: A Case Report and Review of the Literature. Audiology Research, 11(4), 582-593. https://doi.org/10.3390/audiolres11040052









