Genome-Wide Identification and Functional Prediction of the GRAS Transcription Factor Family in Rice Under Abiotic Stress Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Characterization of Members of the GRAS Gene Family in Rice
2.2. Phylogenetic Tree of the GRAS Gene Family in Rice
2.3. Physicochemical Characterization of GRAS Proteins
2.4. Chromosomal Localization of the GRAS Gene Family in Rice
2.5. Analysis of Conserved Motifs and Conserved Domains of GRAS Proteins
2.6. Analysis of Cis-Acting Elements of Rice GRAS Gene Family Promoters
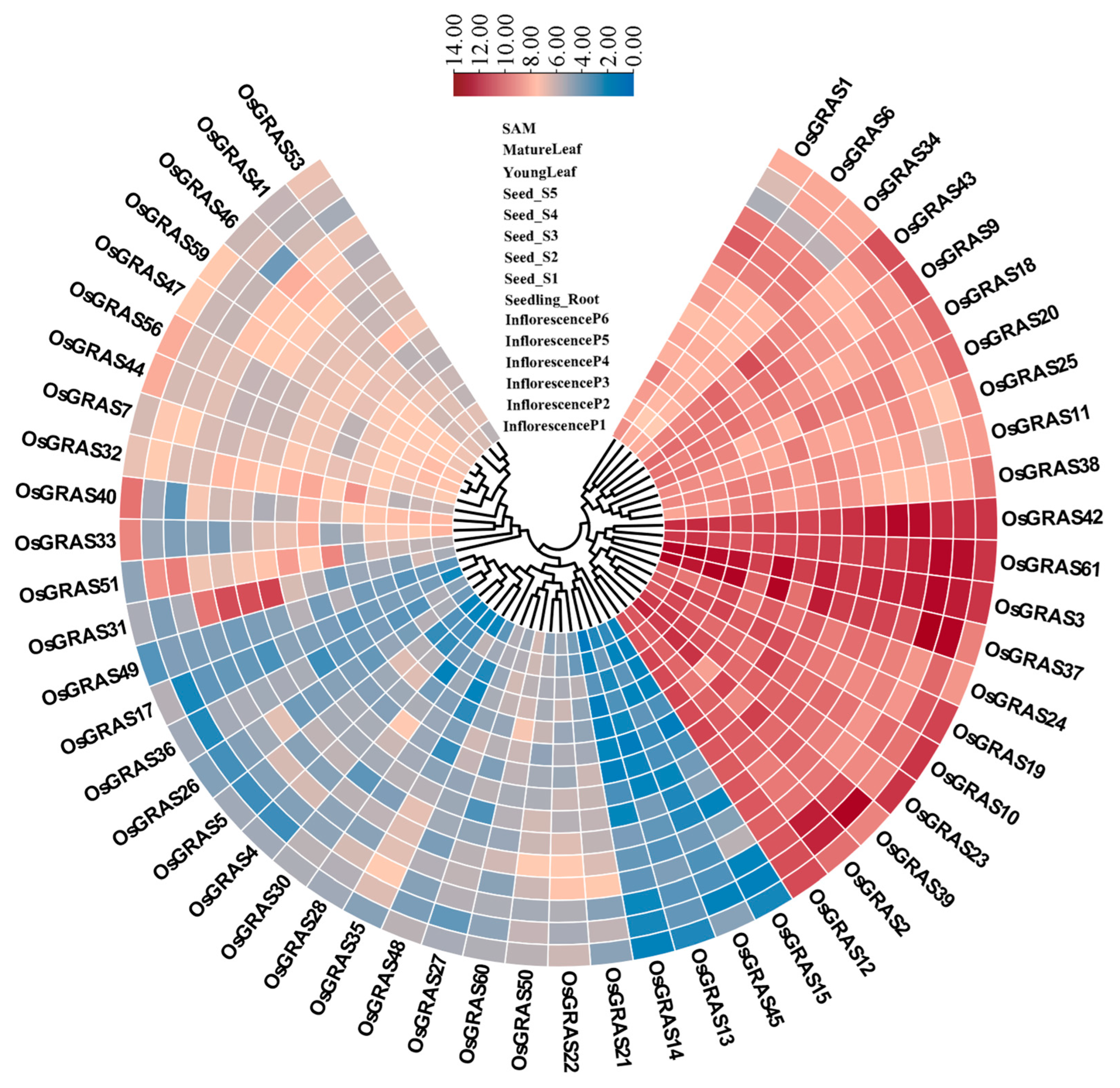
2.7. Tissue Expression Profiling
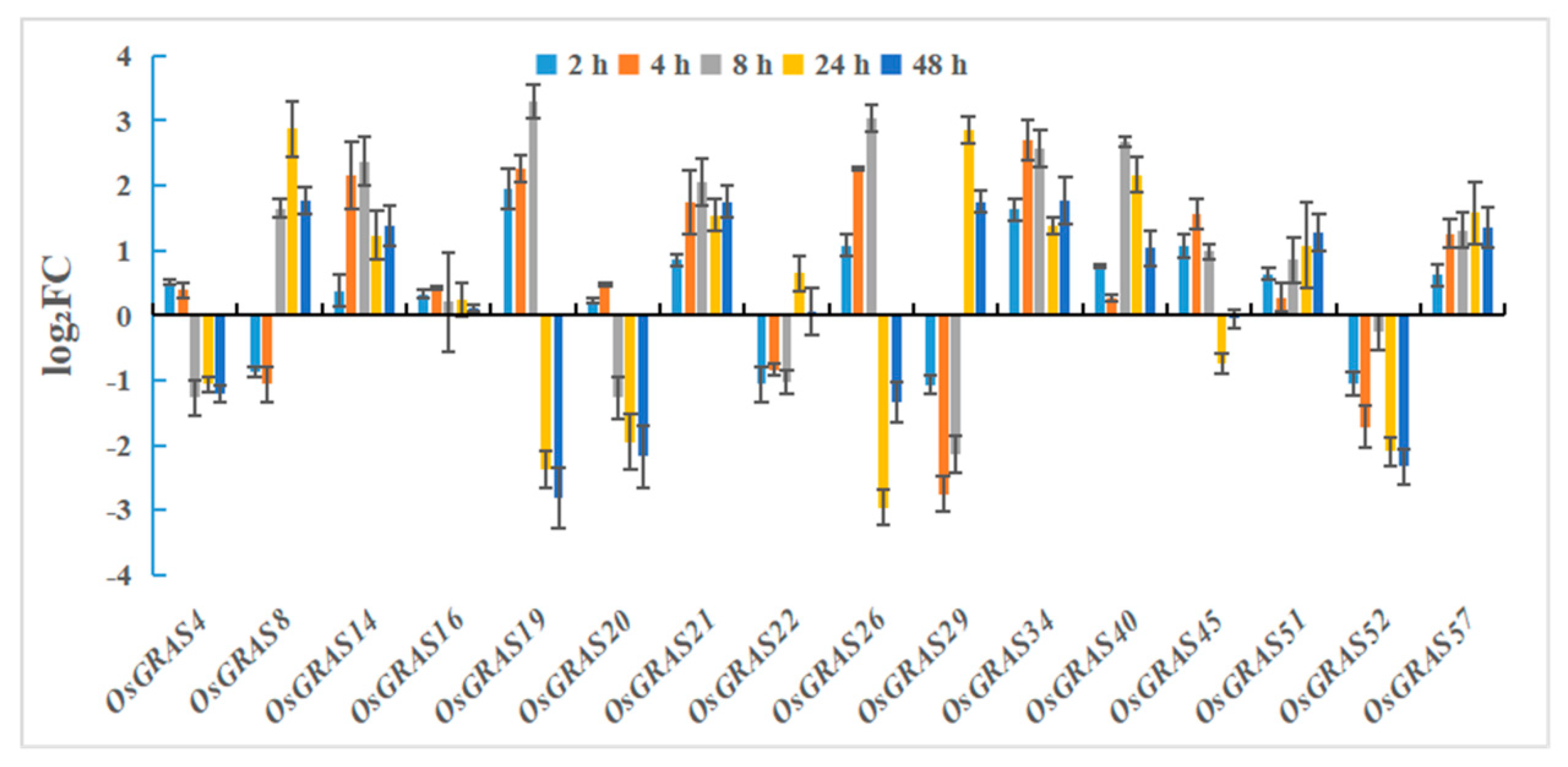
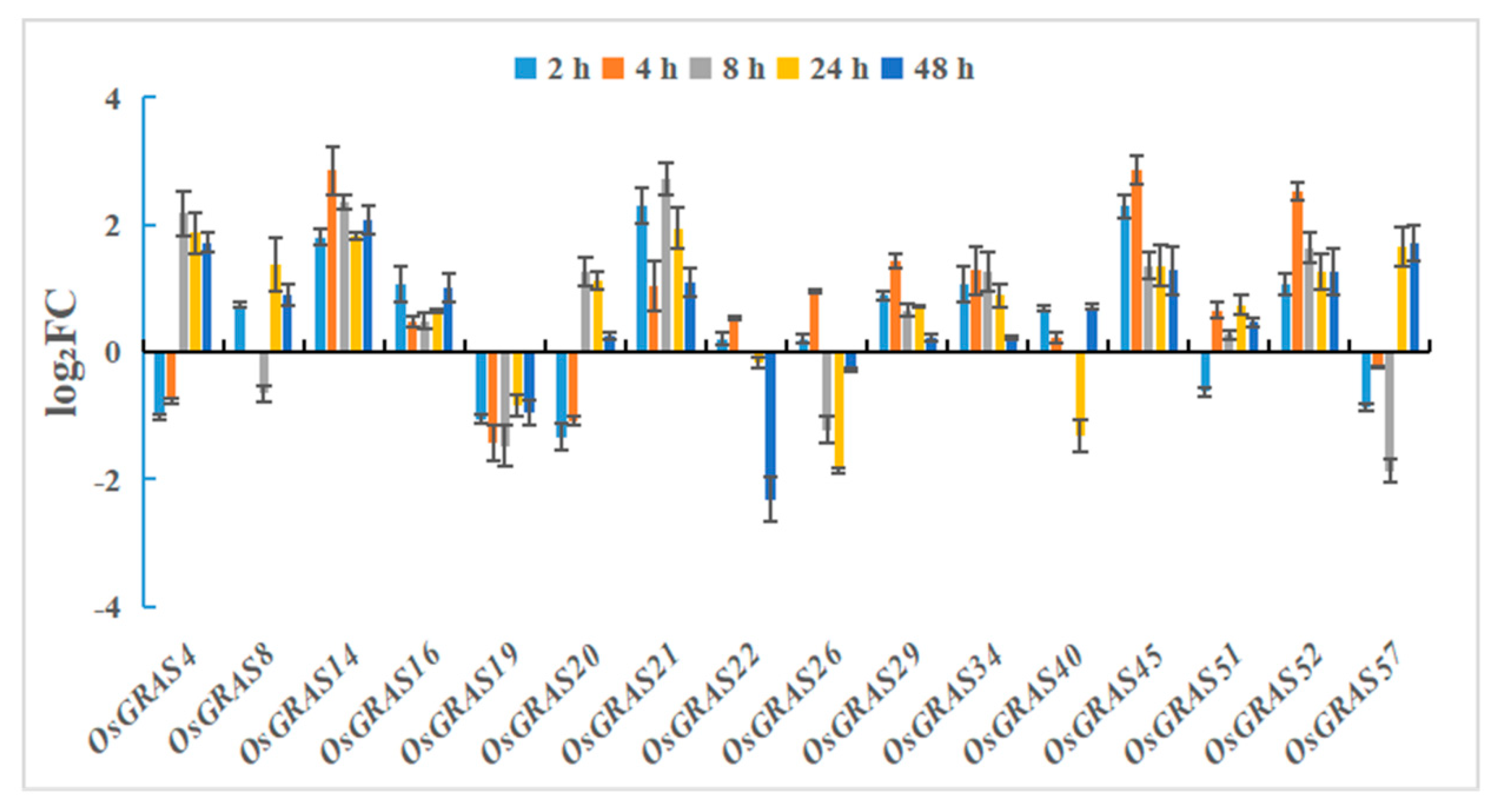
2.8. Expression Level Analysis After Drought and Salt Stress Treatments
3. Results and Analysis
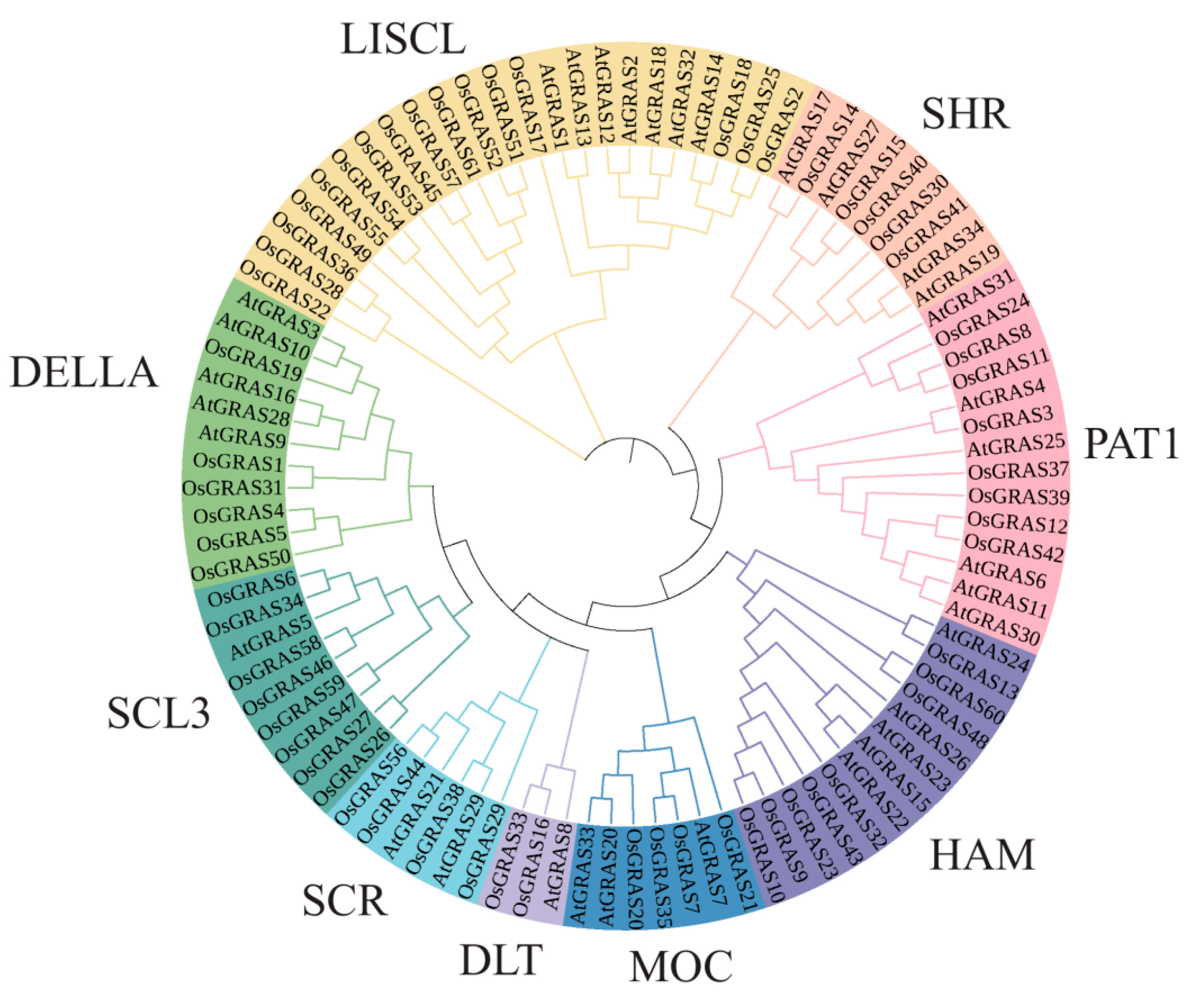
3.1. Genome-Wide Identification of Rice OsGRAS Gene Family Members and Construction of a Phylogenetic Tree
3.2. Analysis of Physicochemical Properties of Rice GRAS Proteins
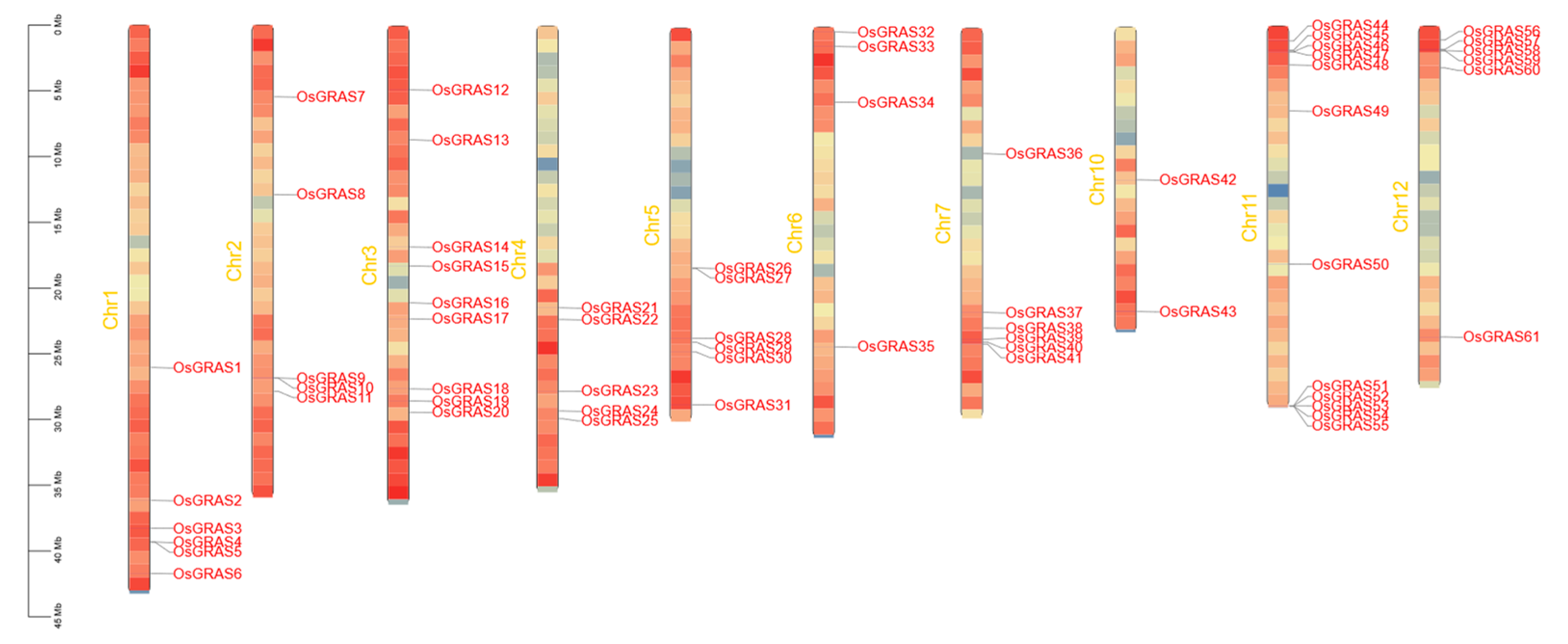
3.3. Chromosomal Localization of Members of the GRAS Gene Family in Rice
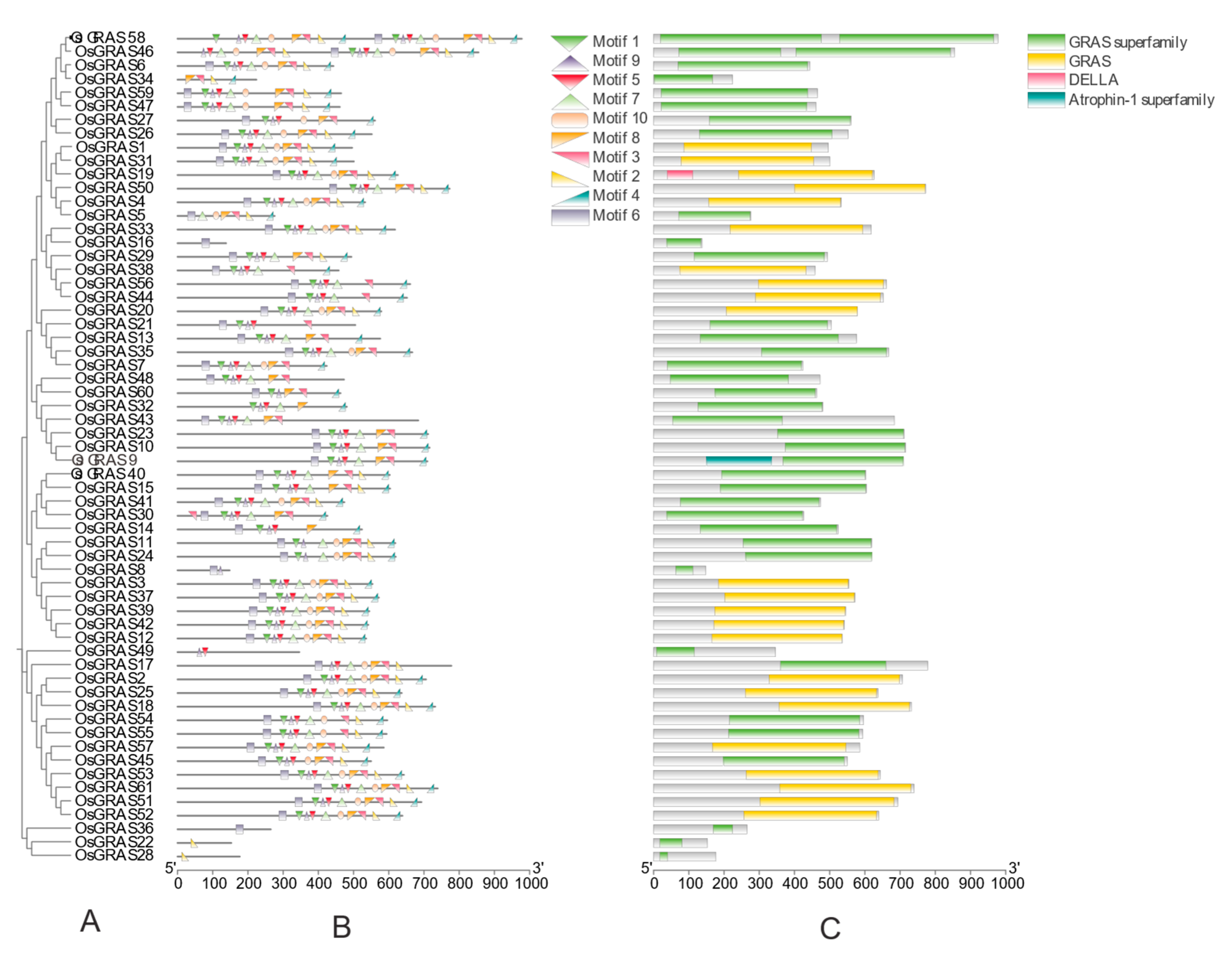
3.4. Analysis of Conserved Motifs and Conserved Structural Domains of the Rice OsGRAS Gene Family
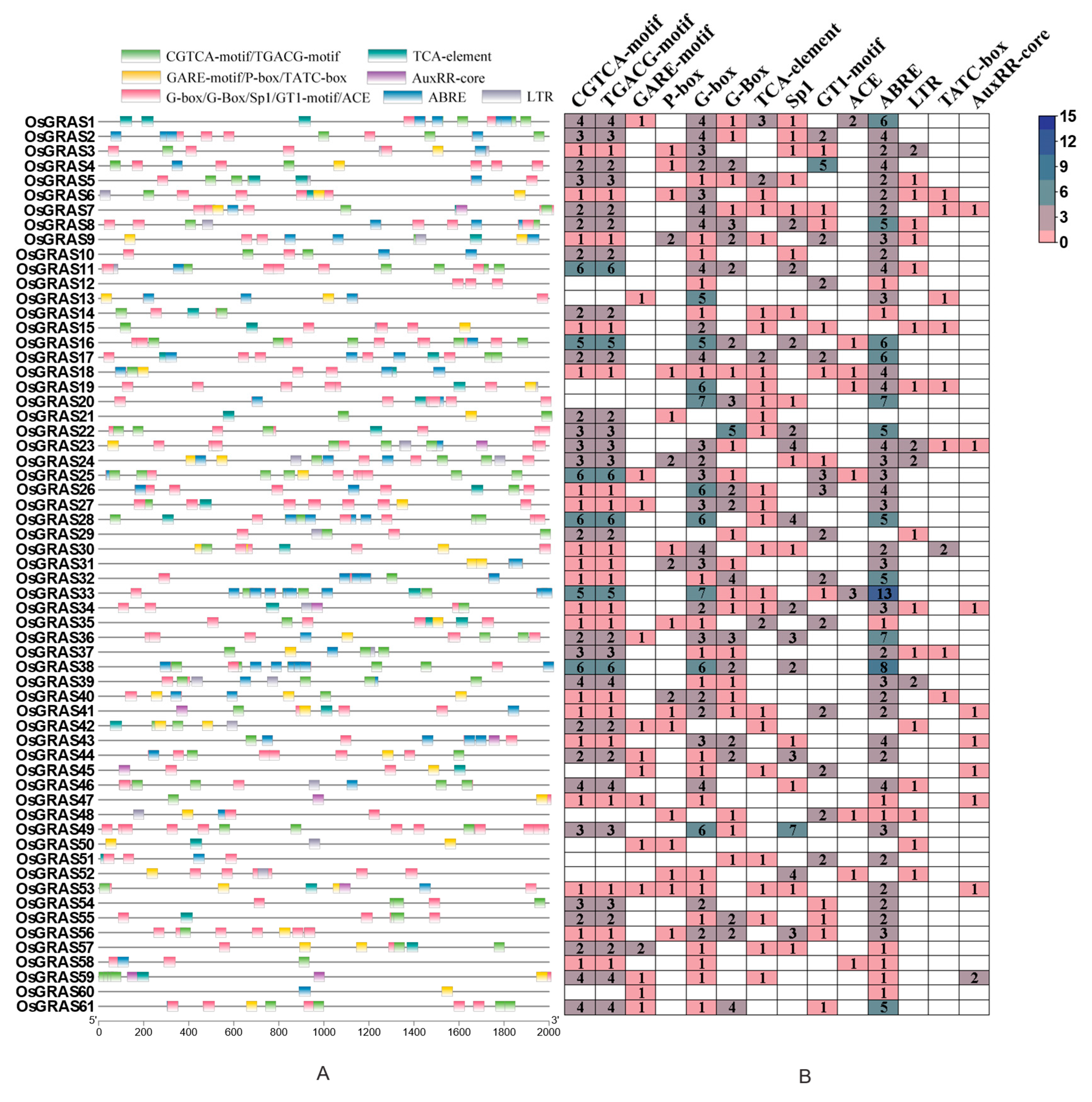
3.5. Functional Clustering of Cis-Acting Elements in the GRAS Gene Family of Rice
3.6. Tissue Expression Analysis of OsGRAS Gene
3.7. Analysis of the Expression Level of the GRAS Gene Family in Rice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kilian, J.; Peschke, F.; Berendzen, K.W.; Harter, K.; Wanke, D. Prerequisites, performance and profits of transcriptional profiling the abiotic stress response. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2012, 1819, 166–175. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T.; et al. The OsWRKY63–OsWRKY76–OsDREB1B module regulates chilling tolerance in rice. Plant J. 2022, 112, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Yao, A.; Li, X.; Li, W.; Gao, R.; Feng, Y.; Li, Z.; Guo, X.; Zhang, L.; Han, D. Overexpression of a Malus baccata (L.) Borkh WRKY transcription factor gene MbWRKY65 increased the tolerance to cold and drought in transgenic tomato. Vitr. Cell. Dev. Biol.—Plant 2024, 60, 620–633. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2013, 84, 19–36. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Liu, C.; Yin, H.; Liu, H.; Luo, H.; He, M.; Zhou, Y. CgbZIP1: A bZIP Transcription Factor from Chrysanthemum Grandiflora Confers Plant Tolerance to Salinity and Drought Stress. Agronomy 2022, 12, 556. [Google Scholar] [CrossRef]
- Jin, Y.; Pan, W.; Zheng, X.; Cheng, X.; Liu, M.; Ma, H.; Ge, X. OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues. Plant Mol. Biol. 2018, 98, 51–65. [Google Scholar] [CrossRef]
- She, M.; Zheng, D.; Zhang, S.; Ke, Z.; Wu, Z.; Zou, H.; Zhang, Z. Functional analysis of maize GRAS transcription factor gene ZmGRAS72 in response to drought and salt stresses. Agric. Commun. 2024, 2, 100054. [Google Scholar] [CrossRef]
- Cao, X.; Ding, L.; Liang, J.; Zhou, Y.; Chen, X.; Li, H.; Liu, T.; Yue, W.; Sui, J.; Jiang, L.; et al. LzSCL9, a Novel GRAS Transcription Factor in Lanzhou Lily (Lilium davidii var. unicolor), Participates in Regulation of Trichokonins-Primed Heat Stress Tolerance. Plants 2024, 13, 2330. [Google Scholar] [CrossRef]
- Ming, R.; Fang, T.; Ling, W.; Geng, J.; Qu, J.; Zhang, Y.; Chen, J.; Yao, S.; Li, L.; Huang, D.; et al. The GRAS transcription factor PtrPAT1 of Poncirus trifoliata functions in cold tolerance and modulates glycine betaine content by regulating the BADH-like gene. Hortic. Res. 2025, 12, uhae296. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, J.; Yang, Z.E.; Chen, E.Y.; Zhang, C.J.; Zhang, X.Y.; Li, F.G. Genome-wide analysis of GRAS transcription factor gene family in Gossypium hirsutum L. BMC Genom. 2018, 19, 348. [Google Scholar] [CrossRef] [PubMed]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef]
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef]
- Sun, X.; Jones, W.T.; Harvey, D.; Edwards, P.J.B.; Pascal, S.M.; Kirk, C.; Considine, T.; Sheerin, D.J.; Rakonjac, J.; Oldfield, C.J.; et al. N-terminal Domains of DELLA Proteins Are Intrinsically Unstructured in the Absence of Interaction with GID1/Gibberellic Acid Receptors. J. Biol. Chem. 2010, 285, 11557–11571. [Google Scholar] [CrossRef]
- Dutta, M.; Saha, A.; Moin, M.; Kirti, P.B. Genome-Wide Identification, Transcript Profiling and Bioinformatic Analyses of GRAS Transcription Factor Genes in Rice. Front. Plant Sci. 2021, 12, 777285. [Google Scholar] [CrossRef]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-Wide Analysis of the GRAS Gene Family in Rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, D1009–D1014. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; et al. CDD: Specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009, 37, D205–D210. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Cenci, A.; Rouard, M. Evolutionary Analyses of GRAS Transcription Factors in Angiosperms. Front. Plant Sci. 2017, 8, 273. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Widmer, A. Genome-wide Comparative Analysis of the GRAS Gene Family in Populus, Arabidopsis and Rice. Plant Mol. Biol. Report. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Waese, J.; Fan, J.; Pasha, A.; Yu, H.; Fucile, G.; Shi, R.; Cumming, M.; Kelley, L.A.; Sternberg, M.J.; Krishnakumar, V.; et al. ePlant: Visualizing and Exploring Multiple Levels of Data for Hypothesis Generation in Plant Biology. Plant Cell 2017, 29, 1806–1821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yao, T.; Wang, J.; Ji, G.; Cui, C.; Song, J.; Sun, N.; Qi, S.; Xu, N.; Zhang, H. Genome-wide identification of R2R3-MYB transcription factors in Betula platyphylla and functional analysis of BpMYB95 in salt tolerance. Int. J. Biol. Macromol. 2024, 279, 135193. [Google Scholar] [CrossRef]
- Jang, J.-C. Arginine-rich motif-tandem CCCH zinc finger proteins in plant stress responses and post-transcriptional regulation of gene expression. Plant Sci. 2016, 252, 118–124. [Google Scholar] [CrossRef]
- Shariatipour, N.; Heidari, B. Meta-Analysis of Expression of the Stress Tolerance Associated Genes and Uncover their Cis-Regulatory Elements in Rice (Oryza sativa L.). Open Bioinform. J. 2020, 13, 39–49. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Mu, Y.; Guan, L.; Wu, F.; Liu, K.; Li, M.; Wang, N.; Zhuang, Z.; Zhao, Y.; et al. PwuWRKY48 Confers Drought Tolerance in Populus wulianensis. Forests 2024, 15, 302. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, C.; Dong, H.; Liu, X.; Guo, H.; Tong, B.; Fang, F.; Zhao, Y.; Yu, Y.; Liu, Y.; et al. Activation and negative feedback regulation of SlHY5 transcription by the SlBBX20/21–SlHY5 transcription factor module in UV-B signaling. Plant Cell 2022, 34, 2038–2055. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, T.; Li, J.; Chen, S.; Grin, I.R.; Zharkov, D.O.; Yu, B.; Li, H. Genome-wide analysis of WD40 protein family and functional characterization of BvWD40-82 in sugar beet. Front. Plant Sci. 2023, 14, 1185440. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nakajima, S.; Morikami, A.; Nakamura, K. An Arabidopsis Protein with a Novel Calcium-binding Repeat Sequence Interacts with TONSOKU/MGOUN3/BRUSHY1 Involved in Meristem Maintenance. Plant Cell Physiol. 2005, 46, 1452–1461. [Google Scholar] [CrossRef]
- Qian, Z.; He, L.; Li, F. Understanding cold stress response mechanisms in plants: An overview. Front. Plant Sci. 2024, 15, 1443317. [Google Scholar] [CrossRef]
- Heidarzadeh, A. Role of amino acids in plant growth, development, and stress responses: A comprehensive review. Discov. Plants 2025, 2, 237. [Google Scholar] [CrossRef]
- Liu, X.; Wu, Y.; Gong, Q. Mechanism of NRP-mediated plant stress responses. Sci. Sin. Vitae 2019, 49, 1125–1132. [Google Scholar] [CrossRef]
- Ali, M.R.M.; Uemura, T.; Ramadan, A.; Adachi, K.; Nemoto, K.; Nozawa, A.; Hoshino, R.; Abe, H.; Sawasaki, T.; Arimura, G.-i. The Ring-Type E3 Ubiquitin Ligase JUL1 Targets the VQ-Motif Protein JAV1 to Coordinate Jasmonate Signaling. Plant Physiol. 2019, 179, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Chen, S.; Li, T.; Ma, X.; Liang, X.; Ding, X.; Liu, H.; Luo, L. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015, 15, 141. [Google Scholar] [CrossRef] [PubMed]
- Zinati, Z. Identification of novel genes potentially involved in rice (Oryza sativa L.) drought tolerance. BioTechnologia 2018, 98, 195–208. [Google Scholar] [CrossRef]
- Acharya, B.R.; Assmann, S.M. Hormone interactions in stomatal function. Plant Mol. Biol. 2009, 69, 451–462. [Google Scholar] [CrossRef]
- Cheong, J.-J.; Choi, Y.D. Methyl jasmonate as a vital substance in plants. Trends Genet. 2003, 19, 409–413. [Google Scholar] [CrossRef]
- Wasternack, C. Action of jasmonates in plant stress responses and development—Applied aspects. Biotechnol. Adv. 2014, 32, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, Z.Q.; Zhang, L.T.; Sun, M.M.; Lin, T.B. Photosynthetic responses of wheat (Triticum aestivum L.) to combined effects of drought and exogenous methyl jasmonate. Photosynthetica 2014, 52, 377–385. [Google Scholar] [CrossRef]
- Dreyer, I.; Uozumi, N. Potassium channels in plant cells. FEBS J. 2011, 278, 4293–4303. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Weiner, J.J.; Peterson, F.C.; Volkman, B.F.; Cutler, S.R. Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 2010, 13, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Yang, Z.; Xing, M.; Jing, Y.; Zhang, Y.; Zhang, K.; Zhou, Y.; Zhao, H.; Qiao, W.; Sun, J. TaBZR1 enhances wheat salt tolerance via promoting ABA biosynthesis and ROS scavenging. J. Genet. Genom. 2023, 50, 861–871. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional Regulatory Networks in Cellular Responses and Tolerance to Dehydration and Cold Stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Hirsch, S.; Oldroyd, G.E.D. GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav. 2009, 4, 698–700. [Google Scholar] [CrossRef]
- Bolle, C.; Koncz, C.; Chua, N.H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 2000, 14, 1269–1278. [Google Scholar] [CrossRef]
- Stuurman, J.; Jäggi, F.; Kuhlemeier, C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002, 16, 2213–2218. [Google Scholar] [CrossRef]
- Soma, F.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems. Plants 2021, 10, 756. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Li, M.; Ding, Z.; Qiao, F.; Jiang, X. Plant Hormone Signals Mediate Melatonin Synthesis to Enhance Osmotic Stress Tolerance in Watermelon Cells. Horticulturae 2023, 9, 927. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197. [Google Scholar] [CrossRef] [PubMed]
- Kaura, V.; Malhotra, P.K.; Mittal, A.; Sanghera, G.S.; Kaur, N.; Bhardwaj, R.D.; Cheema, R.S.; Kaur, G. Physiological, Biochemical, and Gene Expression Responses of Sugarcane Under Cold, Drought and Salt Stresses. J. Plant Growth Regul. 2022, 42, 6367–6376. [Google Scholar] [CrossRef]
- Atif, R.M.; Shahid, L.; Waqas, M.; Ali, B.; Rashid, M.A.R.; Azeem, F.; Nawaz, M.A.; Wani, S.H.; Chung, G. Insights on Calcium-Dependent Protein Kinases (CPKs) Signaling for Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2019, 20, 5298. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Xie, N.; Li, Y.; Zhao, Z.; Luo, P.; Cui, Y.; Rao, X.; Chen, W. Mediator Subunit RhMED15a Regulates Drought Tolerance in Rose. Horticulturae 2024, 10, 84. [Google Scholar] [CrossRef]
| Sequence ID | Protein Name | Number of Amino Acid | Molecular Weight | Theoretical pI |
|---|---|---|---|---|
| LOC_Os01g45860 | OsGRAS1 | 495 | 52,067.35 | 5.06 |
| LOC_Os01g62460 | OsGRAS2 | 705 | 78,998.08 | 6.1 |
| LOC_Os01g65900 | OsGRAS3 | 553 | 61,798.49 | 4.8 |
| LOC_Os01g67650 | OsGRAS4 | 532 | 57,449.17 | 6.09 |
| LOC_Os01g67670 | OsGRAS5 | 275 | 30,461.64 | 6.46 |
| LOC_Os01g71970 | OsGRAS6 | 442 | 48,023.77 | 6.15 |
| LOC_Os02g10360 | OsGRAS7 | 423 | 44,138.74 | 5.56 |
| LOC_Os02g21685 | OsGRAS8 | 147 | 15,305.11 | 9.18 |
| LOC_Os02g44360 | OsGRAS9 | 709 | 74,248.23 | 5.65 |
| LOC_Os02g44370 | OsGRAS10 | 715 | 74,131.93 | 5.82 |
| LOC_Os02g45760 | OsGRAS11 | 618 | 64,200.52 | 9.27 |
| LOC_Os03g09280 | OsGRAS12 | 535 | 59,647.91 | 5.86 |
| LOC_Os03g15680 | OsGRAS13 | 575 | 60,848.2 | 5.04 |
| LOC_Os03g29480 | OsGRAS14 | 523 | 54,077.65 | 5.99 |
| LOC_Os03g31880 | OsGRAS15 | 603 | 64,247.77 | 5.93 |
| LOC_Os03g37900 | OsGRAS16 | 137 | 15,192.7 | 7.87 |
| LOC_Os03g40080 | OsGRAS17 | 777 | 88,331.49 | 5.53 |
| LOC_Os03g48450 | OsGRAS18 | 731 | 82,218.82 | 6.44 |
| LOC_Os03g49990 | OsGRAS19 | 625 | 65406.24 | 5.14 |
| LOC_Os03g51330 | OsGRAS20 | 578 | 62,477.53 | 5.63 |
| LOC_Os04g35250 | OsGRAS21 | 504 | 52,579.87 | 8.67 |
| LOC_Os04g37440 | OsGRAS22 | 152 | 16,672.18 | 9.43 |
| LOC_Os04g46860 | OsGRAS23 | 711 | 74,007.88 | 5.57 |
| LOC_Os04g49110 | OsGRAS24 | 619 | 64,918.25 | 7.2 |
| LOC_Os04g50060 | OsGRAS25 | 636 | 71,655.98 | 5.51 |
| LOC_Os05g31380 | OsGRAS26 | 551 | 56,280.97 | 5.41 |
| LOC_Os05g31420 | OsGRAS27 | 560 | 58,183.56 | 7.85 |
| LOC_Os05g40130 | OsGRAS28 | 176 | 19,069.29 | 10.29 |
| LOC_Os05g40710 | OsGRAS29 | 493 | 51,644.41 | 5.75 |
| LOC_Os05g42130 | OsGRAS30 | 425 | 45,430.82 | 5.81 |
| LOC_Os05g49930 | OsGRAS31 | 500 | 52,994.99 | 5.91 |
| LOC_Os06g01620 | OsGRAS32 | 480 | 51,017.01 | 5.46 |
| LOC_Os06g03710 | OsGRAS33 | 617 | 65,846.84 | 6.02 |
| LOC_Os06g10900 | OsGRAS34 | 223 | 25,058.86 | 8.88 |
| LOC_Os06g40780 | OsGRAS35 | 666 | 69,779.11 | 5.76 |
| LOC_Os07g16330 | OsGRAS36 | 264 | 29,803.37 | 10.07 |
| LOC_Os07g36170 | OsGRAS37 | 571 | 64,602.19 | 5.87 |
| LOC_Os07g38030 | OsGRAS38 | 457 | 49,096.59 | 5.51 |
| LOC_Os07g39470 | OsGRAS39 | 544 | 60,108.16 | 6 |
| LOC_Os07g39820 | OsGRAS40 | 602 | 64,709.33 | 5.61 |
| LOC_Os07g40020 | OsGRAS41 | 473 | 50,596.45 | 5.31 |
| LOC_Os10g22430 | OsGRAS42 | 541 | 59,930.59 | 5.79 |
| LOC_Os10g40390 | OsGRAS43 | 683 | 74,280.4 | 8.85 |
| LOC_Os11g03110 | OsGRAS44 | 651 | 69,918.23 | 5.91 |
| LOC_Os11g04400 | OsGRAS45 | 549 | 59,950.36 | 4.98 |
| LOC_Os11g04570 | OsGRAS46 | 854 | 94,267.73 | 7.45 |
| LOC_Os11g04590 | OsGRAS47 | 460 | 50,420.42 | 6.92 |
| LOC_Os11g06180 | OsGRAS48 | 472 | 52,312.55 | 4.78 |
| LOC_Os11g11600 | OsGRAS49 | 345 | 37,286.47 | 10.13 |
| LOC_Os11g31100 | OsGRAS50 | 772 | 81,094.36 | 6.18 |
| LOC_Os11g47870 | OsGRAS51 | 692 | 77,442.34 | 5.01 |
| LOC_Os11g47890 | OsGRAS52 | 638 | 71,485.86 | 5.27 |
| LOC_Os11g47900 | OsGRAS53 | 642 | 72,083.57 | 6.26 |
| LOC_Os11g47910 | OsGRAS54 | 595 | 66,666.1 | 5.99 |
| LOC_Os11g47920 | OsGRAS55 | 593 | 66,636.95 | 5.98 |
| LOC_Os12g02870 | OsGRAS56 | 660 | 70,394.72 | 5.91 |
| LOC_Os12g04200 | OsGRAS57 | 585 | 63,953.19 | 5.23 |
| LOC_Os12g04370 | OsGRAS58 | 977 | 108,691.38 | 6.55 |
| LOC_Os12g04380 | OsGRAS59 | 464 | 50,674.5 | 5.99 |
| LOC_Os12g06540 | OsGRAS60 | 462 | 50,587.65 | 4.53 |
| LOC_Os12g38490 | OsGRAS61 | 738 | 81,365.04 | 5.17 |
| Conserved Motifs | Amino Acid Sequence | Length | E-Value | Nsites |
|---|---|---|---|---|
| Motif 1 | EACPFLKFAHFTANQAILEAVE | 22 | 2.8 × 10−368 | 48 |
| Motif 2 | NVVACEGAERVERHETYGQWR | 21 | 1.4 × 10−355 | 40 |
| Motif 3 | FLTRFREALHYYSALFDSLDATLP | 24 | 2.8 × 10−401 | 49 |
| Motif 4 | DGGWLLLGWKGRPLYAASAW | 20 | 3.7 × 10−302 | 42 |
| Motif 5 | LQWPSLLQALAARPGGPP | 18 | 1.0 × 10−266 | 48 |
| Motif 6 | PAGDAMQRLAAYFAEALAARL | 21 | 1.2 × 10−289 | 55 |
| Motif 7 | APADELEETGRRLSDFARSLGVPFEFRAV | 29 | 8.1 × 10−318 | 46 |
| Motif 8 | RDAVLRTVRSLSPKVVVLVEQEADHNAPF | 29 | 2.1 × 10−338 | 46 |
| Motif 9 | GERRVHIVDFGISYG | 15 | 1.5 × 10−240 | 56 |
| Motif 10 | PGEALVVNCVLQLHRLLDESV | 21 | 5.6 × 10−189 | 34 |
| Sequence ID | Gene Name | Forward Primer | Reverse Primer |
|---|---|---|---|
| LOC_Os01g67650 | OsGRAS4 | ACTACCGATTCTACGACGTG | CTCTCAAGAACGTCCCAGAA |
| LOC_Os02g21685 | OsGRAS8 | CCTCTTCATCCACGGCTTC | AAATGTTGAGACGGGCTTGTG |
| LOC_Os03g29480 | OsGRAS14 | TGGACTGGTTCGAGGAGTC | CGTAGAACAGGTGCTGCAC |
| LOC_Os03g37900 | OsGRAS16 | ATACGCACTACAGCACGTGT | TGGCTGATGTTGAACATGTGTG |
| LOC_Os03g49990 | OsGRAS19 | ATACGCACATACGCACTACAG | CGTTGAGCTCTGGAAAGCAT |
| LOC_Os03g51330 | OsGRAS20 | TTTCATGCTGCAGCTGTACC | AGAGCAAAAGATCAGCCTGG |
| LOC_Os04g35250 | OsGRAS21 | GCTTACATCAATACGCACACTC | TTAAAATCACCGACATCGCCG |
| LOC_Os04g37440 | OsGRAS22 | ATACGCACACTACCTACGGTA | CGTATGTGCTGTATGTGCGTATC |
| LOC_Os05g31380 | OsGRAS26 | CCTGACGGAGATACGCACA | TTAAAATCATCCGACCCACCAC |
| LOC_Os05g40710 | OsGRAS29 | ACCAGTATCTATACAGCAGCAG | TCCTCGGAGTACACCTCC |
| LOC_Os06g10900 | OsGRAS34 | TCCAGTTCAACCCGGTGG | GCAGTACGGATGGAGAGG |
| LOC_Os07g39820 | OsGRAS40 | TTTAGGTTGGTTAGCCTCCAAG | AGTCTTCATCCATGTAAAGCTGG |
| LOC_Os11g04400 | OsGRAS45 | ATGTTCTTCTGCCTGTACCC | GCATCATCTTCTCCAACATCTC |
| LOC_Os11g47870 | OsGRAS51 | GAAGAAGAGGTGCTCGTGG | AGTAGAAGAAGAGCGTCTCC |
| LOC_Os11g47890 | OsGRAS52 | ACAATACGACCAACTCCACC | GTGAAGGTGGGTTGAATATCC |
| LOC_Os12g04200 | OsGRAS57 | GCGCTCTTCTTCTTCTCGG | GTTGTTACTACTGTGAGCTCTC |
| Actin | TTCCAGCCTTCCTTCATA | AACGATGTTGCCATATAGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, M.; Liu, D.; Peng, Y.; Zhou, Y. Genome-Wide Identification and Functional Prediction of the GRAS Transcription Factor Family in Rice Under Abiotic Stress Conditions. Int. J. Plant Biol. 2025, 16, 95. https://doi.org/10.3390/ijpb16030095
Zhan M, Liu D, Peng Y, Zhou Y. Genome-Wide Identification and Functional Prediction of the GRAS Transcription Factor Family in Rice Under Abiotic Stress Conditions. International Journal of Plant Biology. 2025; 16(3):95. https://doi.org/10.3390/ijpb16030095
Chicago/Turabian StyleZhan, Meng, Daohe Liu, Yuxing Peng, and Yulu Zhou. 2025. "Genome-Wide Identification and Functional Prediction of the GRAS Transcription Factor Family in Rice Under Abiotic Stress Conditions" International Journal of Plant Biology 16, no. 3: 95. https://doi.org/10.3390/ijpb16030095
APA StyleZhan, M., Liu, D., Peng, Y., & Zhou, Y. (2025). Genome-Wide Identification and Functional Prediction of the GRAS Transcription Factor Family in Rice Under Abiotic Stress Conditions. International Journal of Plant Biology, 16(3), 95. https://doi.org/10.3390/ijpb16030095







