Screening of Ty1-copia Retrotransposons in Water Onion (Crinum thaianum), an Endangered Species in Thailand
Abstract
1. Introduction
2. Materials and Methods
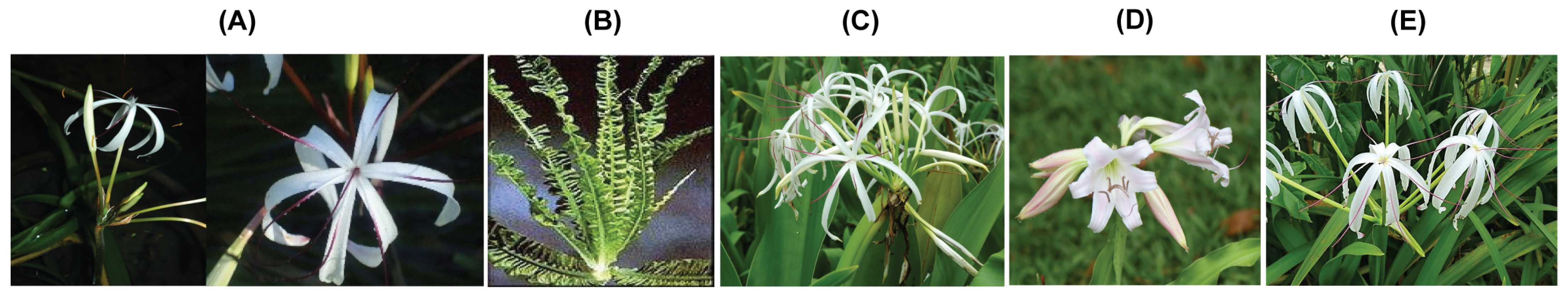
2.1. Plant Materials and DNA Extraction
2.2. Isolation of Ty1-copia rt Fragments
2.3. Ty1-copia rt Sequence Screening
2.4. Classification of Isolated Retrotransposons
2.5. Comparison with Ty1-copia Elements in Other Plants
3. Results
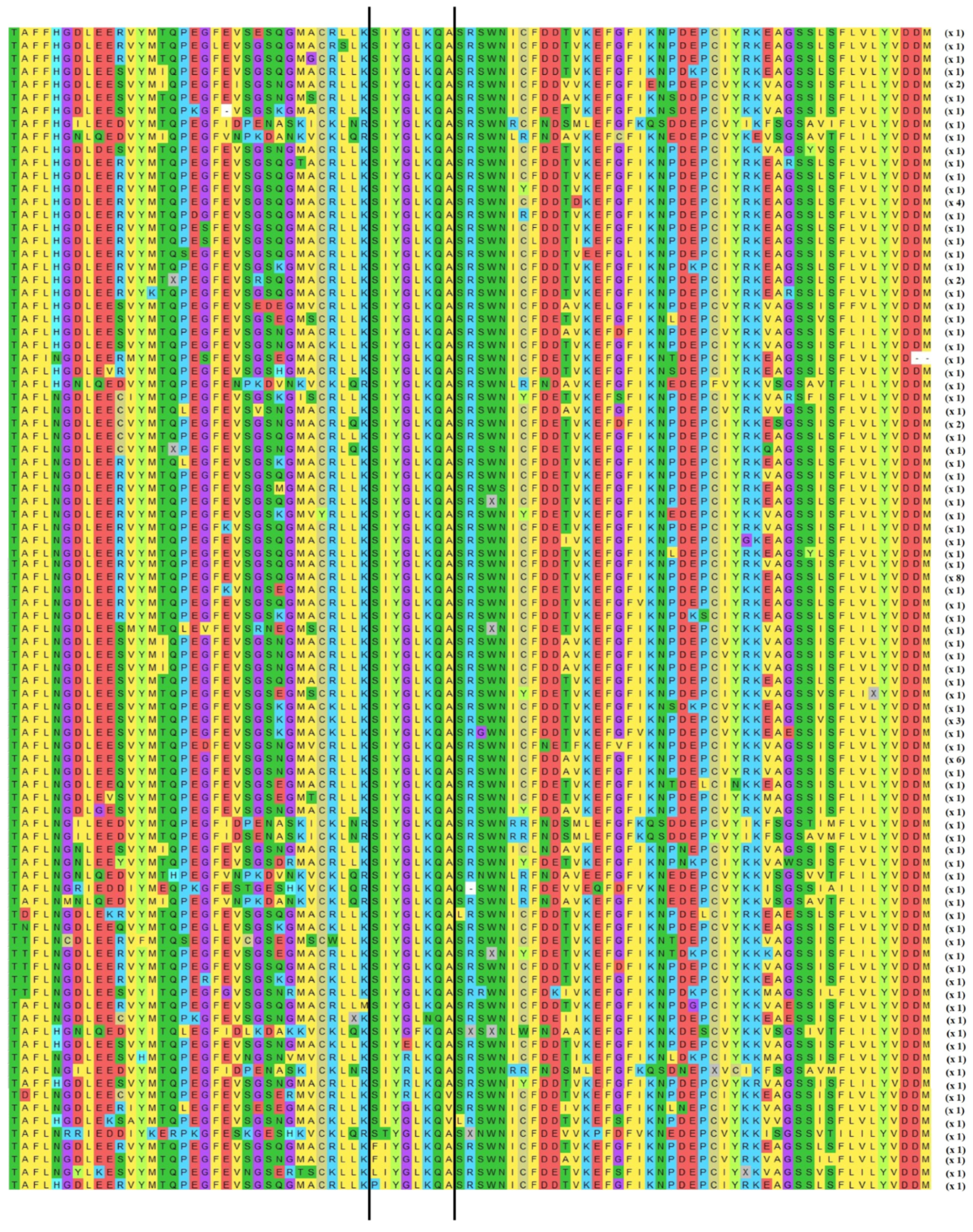
3.1. Ty1-copia rt Fragments of Crinum Species
3.2. Classification of Isolated rt Sequences
3.3. Comparative Analysis of Ty1-copia Factors
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Changcharoen, M.; Changtrakul, S.; Hongtrakul, V. Genetic variation of the endangered species, water-onion (Crinum thaianum) in Thailand and plants in family Amar. Turk. J. Gastroenterol. 2014, 7, 61–68. [Google Scholar]
- Athihirunwong, N.; Janekarnkij, P.; Sanglestsawai, S. Understanding youth motivation for water onion (Crinum thaianum J. Schulze) conservation in Thailand. Kasetsart J. Soc. Sci. 2018, 39, 42–50. [Google Scholar] [CrossRef]
- Hiscock, P. Encyclopedia of Aquarium Plants; Barron’s: New York, NY, USA, 2003. [Google Scholar]
- Flavell, A.J.; Smith, D.B.; Kumar, A. Extreme heterogeneity of Ty1-copia group retrotransposons in plants. Mol. Gen. Genet. MGG 1992, 231, 233–242. [Google Scholar] [CrossRef]
- Flavell, A.; Dunbar, E.; Anderson, R.; Pearce, S.; Hartley, R.; Kumar, A. Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 1992, 20, 3639–3644. [Google Scholar] [CrossRef][Green Version]
- Hirochika, H.; Hirochika, R. Ty1-copia group retrotransposons as ubiquitous components of plant genomes. Jpn. J. Genet. 1993, 68, 35–46. [Google Scholar] [CrossRef]
- Hansen, C.; Heslop-Harrison, J.S. Sequences and Phylogenies of Plant Pararetroviruses, Viruses, and Transposable Elements. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2004; Volume 41, pp. 165–193. [Google Scholar]
- Zhang, Q.-J.; Gao, L.-Z. Rapid and Recent Evolution of LTR Retrotransposons Drives Rice Genome Evolution During the Speciation of AA-Genome Oryza Species. G3 Genes Genomes Genet. 2017, 7, 1875–1885. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Voytas, D.F.; Ausubel, F.M. A copia-like transposable element family in Arabidopsis thaliana. Nature 1988, 336, 242–244. [Google Scholar] [CrossRef]
- Kamm, A.; Doudrick, R.; Heslop-Harrison, P.; Schmidt, T. The Genomic and Physical Organization of Ty1-copia-Like Sequences as a Component of Large Genomes in Pinus elliottii var. elliottii and Other Gymnosperms. Proc. Natl. Acad. Sci. USA 1996, 93, 2708–2713. [Google Scholar] [CrossRef]
- Friesen, N.; Brandes, A.; Heslop-Harrison, J.S. Diversity, Origin, and Distribution of Retrotransposons (gypsy and copia) in Conifers. Mol. Biol. Evol. 2001, 18, 1176–1188. [Google Scholar] [CrossRef]
- He, P.; Ma, Y.; Zhao, G.; Dai, H.; Li, H.; Chang, L.; Zhang, Z. FaRE1: A transcriptionally active Ty1-copia retrotransposon in strawberry. J. Plant Res. 2010, 123, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Wenke, T.; Frömmel, U.; Schmidt, T.; Heitkam, T. The Ty1-copia families SALIRE and Cotzilla populating the Beta vulgaris genome show remarkable differences in abundance, chromosomal distribution, and age. Chromosome Res. 2010, 18, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Flavell, A.J.; Ellis, T.H.N.; Sjakste, T.; Moisy, C.; Schulman, A.H. Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 2011, 106, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, M.G.; Alonso, S.B.; Cabrera, R.S.; Jiménez-Arias, D.; Pérez Pérez, J.A. Development of Retrotransposon-Based Molecular Markers for Characterization of Persea americana (Avocado) Cultivars and Horticultural Races. Agronomy 2022, 12, 1510. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Voytas, D.F.; Cummings, M.P.; Koniczny, A.; Ausubel, F.M.; Rodermel, S.R. copia-like retrotransposons are ubiquitous among plants. Proc. Natl. Acad. Sci. USA 1992, 89, 7124–7128. [Google Scholar] [CrossRef]
- Alipour, A.; Tsuchimoto, S.; Sakai, H.; Ohmido, N.; Fukui, K. Structural characterization of copia-type retrotransposons leads to insights into the marker development in a biofuel crop, Jatropha curcas L. Biotechnol. Biofuels 2013, 6, 129. [Google Scholar] [CrossRef]
- Kolano, B.; Bednara, E.; Weiss-Schneeweiss, H. Isolation and characterization of reverse transcriptase fragments of LTR retrotransposons from the genome of Chenopodium quinoa (Amaranthaceae). Plant Cell Rep. 2013, 32, 1575–1588. [Google Scholar] [CrossRef]
- Zedek, F.; Šmerda, J.; Šmarda, P.; Bureš, P. Correlated evolution of LTR retrotransposons and genome size in the genus Eleocharis. BMC Plant Biol. 2010, 10, 265. [Google Scholar] [CrossRef]
- Wicker, T.; Keller, B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007, 17, 1072–1081. [Google Scholar] [CrossRef]
- Lee, S.; Park, K.-C.; Son, J.-H.; Hwang, Y.-J.; Lim, K.-B.; Song, Y.; Kim, J.-H.; Kim, N.-S. Isolation and characterization of novel Ty1-copia-like retrotransposons from lily. Genome 2013, 56, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Llorens, C.; Muñoz-Pomer, A.; Bernad, L.; Botella, H.; Moya, A. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol. Direct 2009, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Kalendar, R.; Ford, R.; Macas, J.; Griga, M. Evolutionary conserved lineage of Angela-family retrotransposons as a genome-wide microsatellite repeat dispersal agent. Heredity 2009, 103, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.C.; Camayo, G.; De-La-Torre, G.; Galeano, N.; Salcedo, E.; Rivera, L.F.; Duran, A. Identification and chromosomal distribution of copia-like retrotransposon sequences in the coffee (Coffea L.) genome. Agron. Colomb. 2013, 31, 269–278. [Google Scholar]
- Du, J.; Tian, Z.; Hans, C.S.; Laten, H.M.; Cannon, S.B.; Jackson, S.A.; Shoemaker, R.C.; Ma, J. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: Insights from genome-wide analysis and multi-specific comparison. Plant J. 2010, 63, 584–598. [Google Scholar] [CrossRef]
- Tuntipaiboontana, R.; Kuleung, C.; Hongtrakul, V. Diverse Ty1-copia Retrotransposons Found in Waterlilies of the Genus Nymphaea. Hortic. J. 2018, 87, 524–531. [Google Scholar] [CrossRef]
- Stritt, C.; Thieme, M.; Roulin, A.C. Rare transposable elements challenge the prevailing view of transposition dynamics in plants. Am. J. Bot. 2021, 108, 1310–1314. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Arce, A.L.; Mencia, R.; Cambiagno, D.A.; Lang, P.L.; Liu, C.; Burbano, H.A.; Weigel, D.; Manavella, P.A. Polymorphic inverted repeats near coding genes impact chromatin topology and phenotypic traits in Arabidopsis thaliana. Cell Rep. 2023, 42, 112029. [Google Scholar] [CrossRef]
- Hassan, A.H.; Mokhtar, M.M.; Allali, A.E. Transposable elements: Multifunctional players in the plant genome. Front. Plant Sci. 2024, 14, 133012. [Google Scholar] [CrossRef]
- Badr, A.; Sherif, N.E.; Aly, S.; Ibrahim, S.D.; Ibrahim, M. Genetic Diversity among Selected Medicago sativa Cultivars Using Inter-Retrotransposon-Amplified Polymorphism, Chloroplast DNA Barcodes and Morpho-Agronomic Trait Analyses. Plants 2020, 9, 995. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Su, W.; Ma, Y.; Liu, L.; Gu, X.; Wu, D.; Shu, X.; Lai, O.; Tang, Y.; Wu, L.; et al. Assessment of genetic diversity and variety identification based on developed retrotransposon-based insertion polymorphism (RBIP) markers in sweet potato (Ipomoea batatas (L.) Lam.). Sci. Rep. 2021, 11, 17116. [Google Scholar] [CrossRef] [PubMed]
- Haliloğlu, K.; Türkoğlu, A.; Öztürk, H.I.; Özkan, G.; Elkoca, E.; Poczai, P. iPBS-Retrotransposon Markers in the Analysis of Genetic Diversity among Common Bean (Phaseolus vulgaris L.) Germplasm from Türkiye. Genes 2022, 13, 1147. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, A.; Dracatos, P.M.; Maqsood, L.; Yousafi, Q.; Hussain, A.; Jaskani, M.J.; Sajid, M.W.; Haider, M.S.; Hussain, M.M. Genetic Variability and Population Structure of Pakistani Potato Genotypes Using Retrotransposon-Based Markers. Agriculture 2023, 13, 185. [Google Scholar] [CrossRef]


| Sample | Total rt Sequences | Translated Sequences | Classification | |||
|---|---|---|---|---|---|---|
| Function | Non-Function | Ale-Like | TAR-Like | Angela-Like | ||
| C. thaianum_1 | 13 | 11 | 2 | 1 | 1 | 11 |
| C. thaianum_2 | 11 | 7 | 4 | 0 | 0 | 11 |
| C. thaianum_3 | 13 | 12 | 1 | 0 | 0 | 13 |
| C. thaianum_4 | 12 | 12 | 0 | 0 | 0 | 12 |
| C. thaianum_5 | 16 | 13 | 3 | 1 | 1 | 14 |
| C. thaianum_6 | 11 | 9 | 2 | 2 | 0 | 9 |
| C. natans | 14 | 10 | 4 | 1 | 2 | 11 |
| C. asiaticum L. | 12 | 9 | 3 | 3 | 0 | 9 |
| C. latifolium L. | 9 | 6 | 3 | 1 | 0 | 8 |
| C. erubescens L.f. ex Aiton | 12 | 11 | 1 | 0 | 0 | 12 |
| All samples | 123 | 100 | 23 | 9 | 4 | 110 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Putanyawiwat, P.; Kuleung, C.; Veerana, M.; Hongtrakul, V. Screening of Ty1-copia Retrotransposons in Water Onion (Crinum thaianum), an Endangered Species in Thailand. Int. J. Plant Biol. 2025, 16, 71. https://doi.org/10.3390/ijpb16030071
Putanyawiwat P, Kuleung C, Veerana M, Hongtrakul V. Screening of Ty1-copia Retrotransposons in Water Onion (Crinum thaianum), an Endangered Species in Thailand. International Journal of Plant Biology. 2025; 16(3):71. https://doi.org/10.3390/ijpb16030071
Chicago/Turabian StylePutanyawiwat, Piriya, Chatuporn Kuleung, Mayura Veerana, and Vipa Hongtrakul. 2025. "Screening of Ty1-copia Retrotransposons in Water Onion (Crinum thaianum), an Endangered Species in Thailand" International Journal of Plant Biology 16, no. 3: 71. https://doi.org/10.3390/ijpb16030071
APA StylePutanyawiwat, P., Kuleung, C., Veerana, M., & Hongtrakul, V. (2025). Screening of Ty1-copia Retrotransposons in Water Onion (Crinum thaianum), an Endangered Species in Thailand. International Journal of Plant Biology, 16(3), 71. https://doi.org/10.3390/ijpb16030071





