Genetic Variability, Broad-Sense Heritability, and Selection of Superior Genotypes for Fruit Improvement in Platonia insignis
Abstract
1. Introduction
2. Results
2.1. Genetic Parameters Based on Morpho-Agronomic Traits of Platonia insignis Fruits
2.2. Phenotypic Traits of Shape and Color in Platonia insignis Fruits
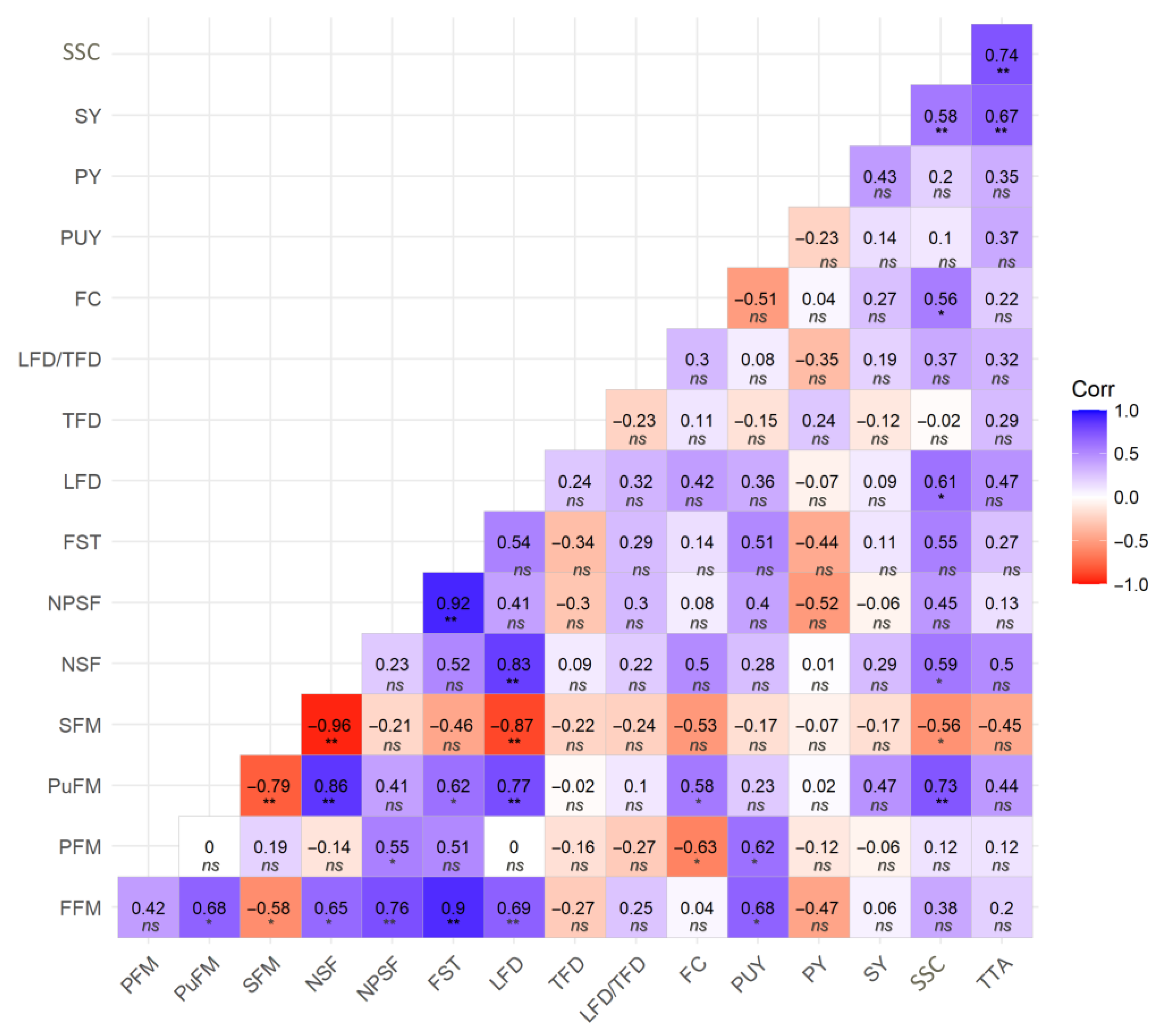
2.3. Genotypic Correlations Based on Morpho-Agronomic Traits of Platonia insignis Fruits
2.4. Genotypes Selection by Rank-Sum Index
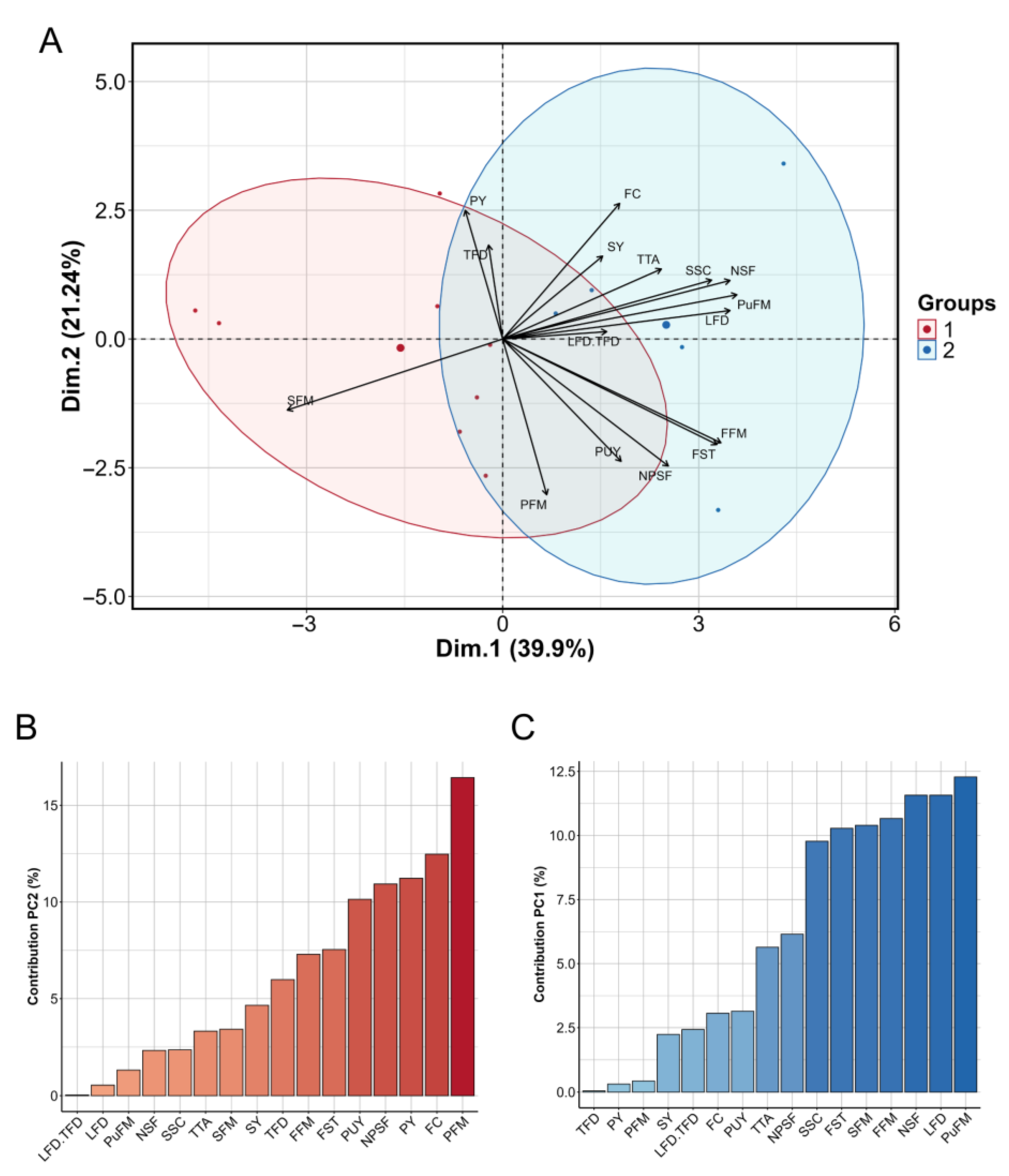
2.5. Principal Component Analysis of Morpho-Agronomic Traits
3. Discussion
4. Materials and Methods
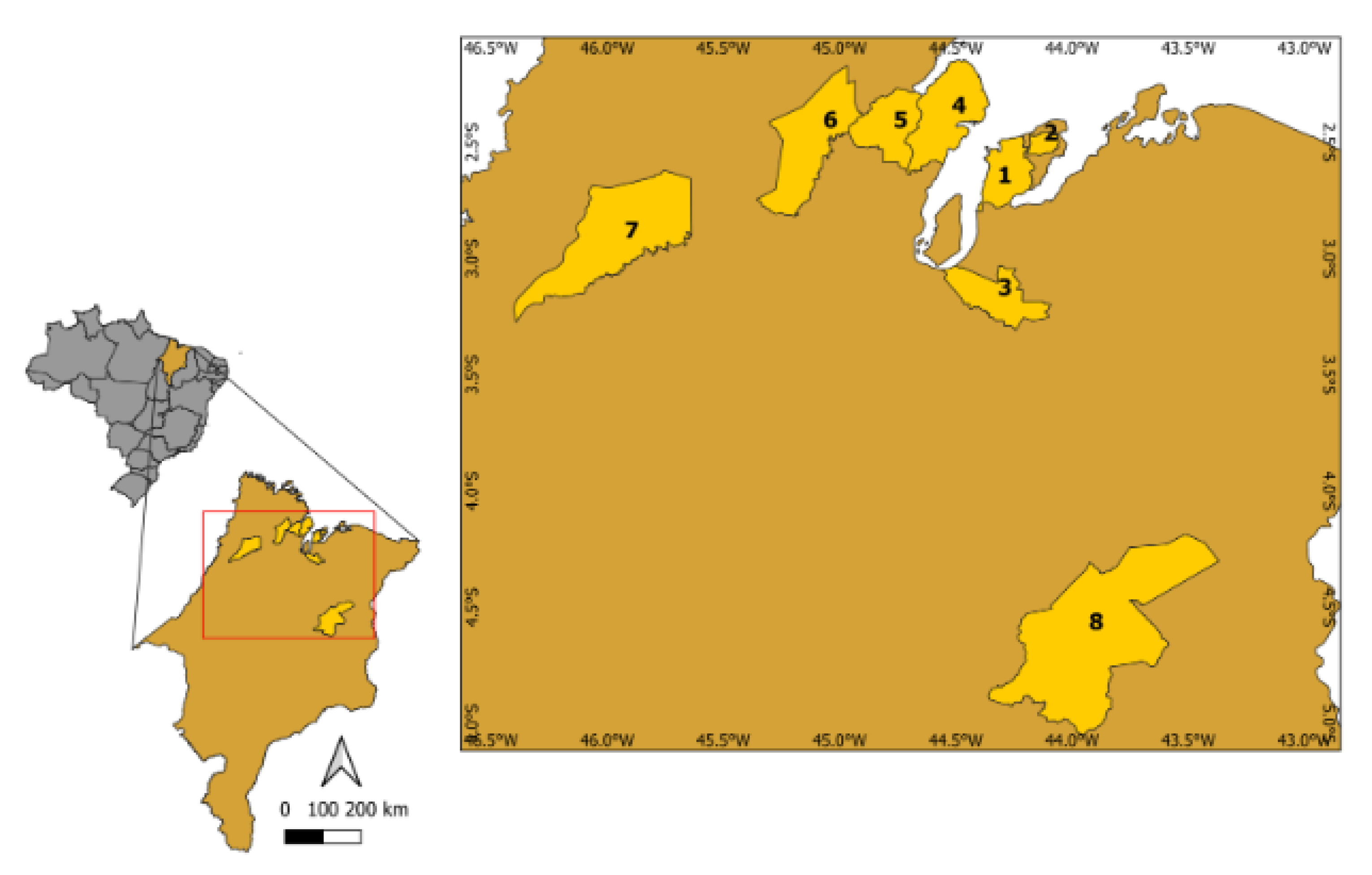
4.1. Study Area, Plant Material, and Fruit Harvesting
4.2. Determination of Morpho-Agronomic Traits
4.3. Determination of Soluble Solids and Titratable Acidity
4.4. Determination of Genetic Parameters
4.5. Statistical Analysis
4.6. Multivariate Analyses and Graphs
4.7. Selection Index
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sousa, H.M.S.; Leal, G.F.; Damiani, C.; Borges, S.V.; Freitas, B.C.; Martins, G.A.S. Some wild fruits from Amazon biodiversity: Composition, bioactive compounds, and characteristics. Food Res. 2021, 5, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Cuéllar-Álvarez, L.N.; García-Chacón, J.M.; Heredia, F.J.; González-Miret, M.L. Bioactive and agroindustrial potential of Amazonian fruit species: A review. Cogent Food Agric. 2025, 11, 2451062. [Google Scholar] [CrossRef]
- Rodrigues Lima, S.K.; Pereira, E.J.; Machado, G.D.; Silva, R.A.; Lucarini, M.; Durazzo, A.; Diele-Viegas, L.M.; Arcanjo, D.D. Systematic mapping of the production chain of “bacuri” (Platonia insignis Mart.) in Brazil. Sustainability 2022, 14, 15051. [Google Scholar] [CrossRef]
- Garcia, C.B.; Silva, A.V.; Carvalho, I.A.; Nascimento, W.F.; Ramos, S.L.; Rodrigues, D.P.; Zucchi, M.I.; Costa, F.M.; Alves-Pereira, A.; Batista, C.E.; et al. Low diversity and high genetic structure for Platonia insignis Mart., an endangered fruit tree species. Plants 2024, 13, 1033. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.E.U.; Homma, A.K.O.; Nascimento, W.M.O. Platonia insignis: Bacuri. In Espécies Nativas da Flora Brasileira de Valor Econômico Atual ou Potencial: Plantas Para o Futuro—Região Norte, 1st ed.; Coradin, L., Camillo, J., Vieira, L.C., Eds.; MMA: Brasília, Brazil, 2022; pp. 424–449. Available online: https://www.gov.br/mma/pt-br/livro-especies-nativas-da-flora-brasileira-de-valor-economico-atual-ou-potencial-2013-plantas-para-o-futuro-2013-regiao-norte.pdf (accessed on 12 June 2025).
- Alves, K.F.; Lima, A.D.; Rivas, P.M.; Albuquerque, I.C.; Pinheiro, J.F.; Catunda, P.H.; Felipe, S.H.; Reis, F.D.; Batista, D.S.; Henschel, J.M.; et al. Platonia insignis: A systematic synthesis of scientific studies on its biology, ecology, and potential applications. Plants 2025, 14, 884. [Google Scholar] [CrossRef] [PubMed]
- Menezes, A.J.E.A.; Schöffel, E.R.; Homma, A.K.O. Caracterização de sistemas de manejo de bacurizeiro (Platonia insignis Mart.) nas mesorregiões do Nordeste Paraense e do Marajó, Estado do Pará. Amaz. Ciênc. Desenv. 2010, 6, 49–62. Available online: http://www.alice.cnptia.embrapa.br/alice/handle/doc/905975 (accessed on 12 June 2025).
- Mendes, G.G.; de Gusmão, M.T.; Martins, T.G.; Rosado, R.D.; Sobrinho, R.S.; Nunes, A.C.; Ribeiro, W.S.; Zanuncio, J.C. Genetic divergence of native palms of Oenocarpus distichus considering biometric fruit variables. Sci. Rep. 2019, 9, 4943. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.P.; Nunes, A.C.P.; Garuzzo, M.S.P.B.; Corrêa, R.X.; Marques, F.G. Genetic variability and predicted gain in progeny tests of native Atlantic Forest timber species: Cariniana legalis, Cordia trichotoma, and Zeyheria tuberculosa. Ann. For. Res. 2022, 65, 85–96. [Google Scholar] [CrossRef]
- Tripathi, P.C.; Sane, A.; Kumar, P.; Chaturvedi, K.; Mishra, D.S.; Ravat, P. Phenotypic Diversity and Genetic Characterization of Cordia myxa L. using multivariate analysis. Flora 2025, 323, 152224. [Google Scholar] [CrossRef]
- Costa, R.J.C.; Rodrigues, J.I.M.; Gallo, R.; Corrêa, T.R.; Felipe, S.H.S.; Martins, W.B.R. Linking genetic variability to fruit and pyrene traits in Endopleura uchi genotypes from the Eastern Brazilian Amazon. Braz. J. Biol. 2025, 85, e290725. [Google Scholar] [CrossRef] [PubMed]
- Canal, G.B.; de Jesus Passos, A.B.; Péres, M.Z.; Ferreira, D.S.; de Arruda, V.C.; Moro, G.L.; Bernardes, C.D.; Nascimento, M.; Nascimento, A.C.; da Silva Ferreira, M.F.; et al. Selection of Euterpe edulis accessions based on fruit and pulp characteristics. Tree Genet. Genomes 2025, 21, 4. [Google Scholar] [CrossRef]
- Pinheiro, E.M.; Araujo, J.R.; Nobre, C.P.; Moraes, L.R.; Sousa, J.D.; Rivas, P.M.; Corrêa, T.R. Estimates of genetic parameters based on reproductive and fruit biometric traits of babassu (Attalea speciosa Mart.) populations in Maranhão State, Brazil. Ciênc. Agrotec. 2025, 49, e017124. [Google Scholar] [CrossRef]
- Leão, N.V.M.; Sousa Felipe, S.H.; Gallo, R.; Cordeiro Shimizu, E.S. Genetic variability of Tachigali vulgaris trees based on seed morphophysiological traits. South. For. 2023, 85, 185–193. [Google Scholar] [CrossRef]
- Silva, P.C.S.; Gallo, R.; Santos, M.M.; Nonato, E.R.L.; Santos, R.S.; Lira Júnior, J.S.; Batista, D.S. Analysis of genetic divergence in Psidium cattleyanum Sabine accessions based on morphological fruit descriptors. Genet. Resour. Crop Evol. 2024, 71, 5039–5054. [Google Scholar] [CrossRef]
- Ferrão, L.F.V.; Amadeu, R.R.; Benevenuto, J.; de Bem Oliveira, I.; Munoz, P.R. Genomic selection in an outcrossing autotetraploid fruit crop: Lessons from blueberry breeding. Front. Plant Sci. 2021, 12, 676326. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, J.S.; Peixoto, M.A.; Coelho, I.; Alves, R.; Resende, M.D.; Laviola, B.; Bhering, L.L. Genetic evaluation and selection in Jatropha curcas through Frequentist and Bayesian inferences. Bragantia 2022, 81, e2722. [Google Scholar] [CrossRef]
- Guimarães, A.D.G.; Mota, M.G.C.; Nazaré, R.F.R. Coleta de Germoplasma de Bacuri (Platonia insignis Mart.) na Amazônia. I: Microrregião Campos do Marajó (Soure/Salvaterra); EMBRAPA-CPATU: Belém, Brazil, 1992; p. 25. Available online: http://www.infoteca.cnptia.embrapa.br/infoteca/handle/doc/379659 (accessed on 12 June 2025).
- AOAC. Official Methods of Analysis of the AOAC International, 15th ed.; AOAC International: Arlington, VA, USA, 1992. [Google Scholar]
- Resende, M.A.V.D.; Freitas, J.A.D.; Lanza, M.A.; Resende, M.D.V.D.; Azevedo, C.F. Divergência genética e índice de seleção via BLUP em acessos de algodoeiro para características tecnológicas da fibra. Pesq. Agropecu. Trop. 2014, 44, 334–640. [Google Scholar] [CrossRef]
- de Resende, M.D.; Mora, A.L.; Higa, A.R.; Paludzyszyn Filho, E. Efeito do tamanho amostral na estimativa da herdabilidade em espécies perenes. Floresta 1998, 28, 51–63. Available online: http://www.alice.cnptia.embrapa.br/alice/handle/doc/288756 (accessed on 12 June 2025). [CrossRef][Green Version]
- Resende, M.D.V. Software Selegen-REML/BLUP: A useful tool for plant breeding. Crop Breed. Appl. Biotechnol. 2016, 16, 330–339. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, version 4; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Mulamba, N.N.; Mock, J.J. Improvement of yield potential of the ETO blanco maize (Zea mays L.) population by breeding for plant traits. Egypt. J. Genet. Cytol. 1978, 7, 40–51. [Google Scholar]
- Cruz, C.D.; Carneiro, P.C.S.; Regazzi, A.J. Modelos Biométricos Aplicados ao Melhoramento Genético; Editora UFV: Viçosa, Brazil, 2014; 668p. [Google Scholar]

| Trait * | Parameter ** | |||||||
|---|---|---|---|---|---|---|---|---|
| Vg | Ve | Vf | h2g | h2mc | Acc | CVr | M | |
| LFD (mm) | 291.93 | 69.76 | 362.00 | 0.81 | 0.99 | 0.99 | 27.47 | 96.11 |
| TFD (mm) | 0.09 | 0.25 | 0.35 | 0.27 | 0.99 | 0.99 | 11.83 | 1.80 |
| FST (mm) | 55.61 | 41.35 | 97.02 | 0.57 | 0.99 | 0.99 | 24.02 | 82.90 |
| FC | 17.41 | 56.22 | 73.74 | 0.23 | 0.93 | 0.96 | 1.65 | 15.31 |
| LFD/TFD | 3.62 | 3.01 | 6.68 | 0.54 | 0.99 | 0.99 | 8.99 | 4.03 |
| FFM (g) | 5933.70 | 3090.13 | 9027.80 | 0.66 | 0.99 | 0.99 | 27.70 | 240.24 |
| PFM (g) | 154.24 | 488.83 | 644.42 | 0.24 | 0.99 | 0.99 | 8.90 | 64.56 |
| PuFM (g) | 0.52 | 0.77 | 1.30 | 0.40 | 0.99 | 0.99 | 18.63 | 2.91 |
| SFM (g) | 559.47 | 181.42 | 741.94 | 0.75 | 0.99 | 0.99 | 21.02 | 46.05 |
| NPSF (unit) | 6.67 | 17.72 | 24.42 | 0.27 | 0.99 | 0.99 | 11.21 | 11.57 |
| NSF (unit) | 0.33 | 0.56 | 0.90 | 0.37 | 0.99 | 0.99 | 17.24 | 2.19 |
| PY (%) | 27.72 | 84.51 | 112.75 | 0.24 | 0.93 | 0.97 | 1.67 | 22.60 |
| PUY (%) | 14.63 | 297.67 | 312.51 | 0.05 | 0.68 | 0.83 | 0.66 | 63.15 |
| SY (%) | 0.57 | 89.37 | 126.26 | 0.00 | 0.06 | 0.24 | 0.11 | 19.89 |
| SSC (°Brix) | 1.00 | 3.33 | 10.17 | 0.10 | 0.44 | 0.67 | 0.40 | 3.16 |
| TTA | 1.07 | 107.93 | 310.73 | 0.00 | 0.02 | 0.16 | 0.07 | 19.40 |
| Trait | Deviance | LRT (Likelihood Ratio Test) |
|---|---|---|
| LFD (mm) | 1756.18 | 453.40 ** |
| TFD (mm) | −68.79 | 68.67 ** |
| FST (mm) | 1574.69 | 218.39 ** |
| FC | 1234.53 | 35.41 ** |
| LFD/TFD | 737.72 | 199.50 ** |
| FFM (g) | 2959.3 | 282.66 ** |
| PFM (g) | 2349.17 | 56.60 ** |
| PuFM (g) | 294.06 | 122.67 ** |
| SFM (g) | 2058.67 | 381.62 ** |
| NPSF (unit) | 1289.29 | 68.52 ** |
| NSF (unit) | 174.32 | 90.49 ** |
| PY (%) | 1359.89 | 42.11 ** |
| PUY (%) | 1654.63 | 2.98 ns |
| SY (%) | 1366.26 | 0.07 ns |
| SSC (°Brix) | 594.09 | 37.50 ** |
| TTA | 927.21 | 0.08 ns |
| Trait | Genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | G6 | G7 | G8 | G9 | G10 | G11 | G12 | G13 | |
| FFM (+) | 13 | 12 | 9 | 4 | 2 | 8 | 5 | 10 | 1 | 6 | 11 | 3 | 7 |
| PFM (−) | 4 | 6 | 2 | 8 | 5 | 9 | 13 | 7 | 12 | 10 | 1 | 3 | 11 |
| SFM (−) | 2 | 1 | 3 | 9 | 12 | 6 | 7 | 8 | 10 | 11 | 4 | 13 | 5 |
| PuFM (+) | 2 | 3 | 8 | 7 | 12 | 4 | 1 | 6 | 10 | 9 | 5 | 13 | 11 |
| NSF (−) | 3 | 2 | 9 | 6 | 11 | 5 | 1 | 7 | 12 | 10 | 4 | 13 | 8 |
| NPSF (+) | 13 | 12 | 10 | 3 | 4 | 5 | 1 | 9 | 2 | 8 | 11 | 7 | 6 |
| FST (−) | 1 | 2 | 4 | 11 | 9 | 8 | 1 | 5 | 13 | 7 | 3 | 10 | 6 |
| PUY (+) | 2 | 10 | 11 | 12 | 2 | 5 | 4 | 8 | 1 | 7 | 13 | 9 | 3 |
| PY (−) | 12 | 8 | 3 | 5 | 4 | 7 | 6 | 2 | 1 | 10 | 9 | 11 | 13 |
| SY (−) | 8 | 4 | 1 | 2 | 6 | 3 | 9 | 5 | 10 | 11 | 12 | 13 | 7 |
| SST (+) | 12 | 13 | 11 | 10 | 8 | 7 | 5 | 6 | 9 | 2 | 4 | 1 | 3 |
| ATT (−) | 2 | 3 | 4 | 1 | 5 | 7 | 6 | 8 | 10 | 11 | 9 | 12 | 13 |
| Average rank | 6.17 | 6.33 | 6.25 | 6.50 | 6.67 | 6.17 | 4.92 | 6.75 | 7.58 | 8.50 | 7.17 | 9.00 | 7.75 |
| Genotype | Municipality | Altitude | Latitude | Longitude |
|---|---|---|---|---|
| G1 | Alcântara | 515,510,403 | 2°32′2.34″ S | 44°38′23.61″ W |
| G2 | Bequimão | 233,110,114 | 2°30′2.06″ S | 44°44′39.56″ W |
| G3 | Bequimão | 223,636,639 | 2°30′2.65″ S | 44°52′48.15″ W |
| G4 | Bequimão | 221,215,877 | 2°30′1.78″ S | 44°52′47.23″ W |
| G5 | Codó | 634,813,685 | 4°27′39.93″ S | 43°47′7.22″ W |
| G6 | Codó | 73,011,913 | 4°27′42.23″ S | 43°47′15.48″ W |
| G7 | Paço do Lumiar | 246,098,218 | 2°31′55.82″ S | 44°10′32.97″ W |
| G8 | Nova Olinda | 581,240,947 | 2°41′21.30″ S | 45°42′32.90″ W |
| G9 | Pinheiros | 311,854,263 | 2°39′24.98″ S | 45°12′55.11″ W |
| G10 | Pinheiros | 204,556,364 | 2°25′21.78″ S | 45°8′46.12″ W |
| G11 | Pinheiros | 9,774,121 | 2°31′56.42″ S | 45°7′20.73″ W |
| G12 | São Luís | 489,158,026 | 2°36′2.48″ S | 44°12′ 44.43″ W |
| G13 | Santa Rita | 181,006,112 | 3°3′32.4″ S | 44°15′ 22.75″ W |
| Heritability Range (h2) | Heritability Classification | Accuracy Range (râa) | Accuracy Classification |
|---|---|---|---|
| 0.00–0.15 | Low | 0.10–0.40 | Low |
| 0.15–0.50 | Moderate | 0.40–0.70 | Moderate |
| ≥0.50 | High | ≥0.70 | High |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, S.S.M.; Felipe, S.H.S.; Alves, G.L.; Rivas, P.M.S.; Henschel, J.M.; de Moraes, L.R.R.; Reis, L.C.F.; Araújo, J.R.G.; Pinheiro, M.V.M.; Batista, D.S.; et al. Genetic Variability, Broad-Sense Heritability, and Selection of Superior Genotypes for Fruit Improvement in Platonia insignis. Int. J. Plant Biol. 2025, 16, 108. https://doi.org/10.3390/ijpb16030108
Ribeiro SSM, Felipe SHS, Alves GL, Rivas PMS, Henschel JM, de Moraes LRR, Reis LCF, Araújo JRG, Pinheiro MVM, Batista DS, et al. Genetic Variability, Broad-Sense Heritability, and Selection of Superior Genotypes for Fruit Improvement in Platonia insignis. International Journal of Plant Biology. 2025; 16(3):108. https://doi.org/10.3390/ijpb16030108
Chicago/Turabian StyleRibeiro, Suzane Sá Matos, Sérgio Heitor Sousa Felipe, Givago Lopes Alves, Priscila Marlys Sá Rivas, Juliane Maciel Henschel, Lúcio Rafael Rocha de Moraes, Luís Carlos Ferreira Reis, José Ribamar Gusmão Araújo, Marcos Vinícius Marques Pinheiro, Diego Silva Batista, and et al. 2025. "Genetic Variability, Broad-Sense Heritability, and Selection of Superior Genotypes for Fruit Improvement in Platonia insignis" International Journal of Plant Biology 16, no. 3: 108. https://doi.org/10.3390/ijpb16030108
APA StyleRibeiro, S. S. M., Felipe, S. H. S., Alves, G. L., Rivas, P. M. S., Henschel, J. M., de Moraes, L. R. R., Reis, L. C. F., Araújo, J. R. G., Pinheiro, M. V. M., Batista, D. S., & Corrêa, T. R. (2025). Genetic Variability, Broad-Sense Heritability, and Selection of Superior Genotypes for Fruit Improvement in Platonia insignis. International Journal of Plant Biology, 16(3), 108. https://doi.org/10.3390/ijpb16030108









