Comparative Analysis of Chloroplast Genomes Across 20 Plant Species Reveals Evolutionary Patterns in Gene Content, Codon Usage, and Genome Structure
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Data Acquisition and Processing
2.2. Barcoding Gene Extraction and Annotation
2.3. Sequence Alignment and Supermatrix Construction
2.4. Phylogenetic Analysis
2.5. Genome Statistics Calculation
2.6. Genome Similarity Analysis
2.7. Gene Presence/Absence Profiling
2.8. Codon Usage Bias Analysis
2.9. Data Visualization and Figure Preparation
3. Results
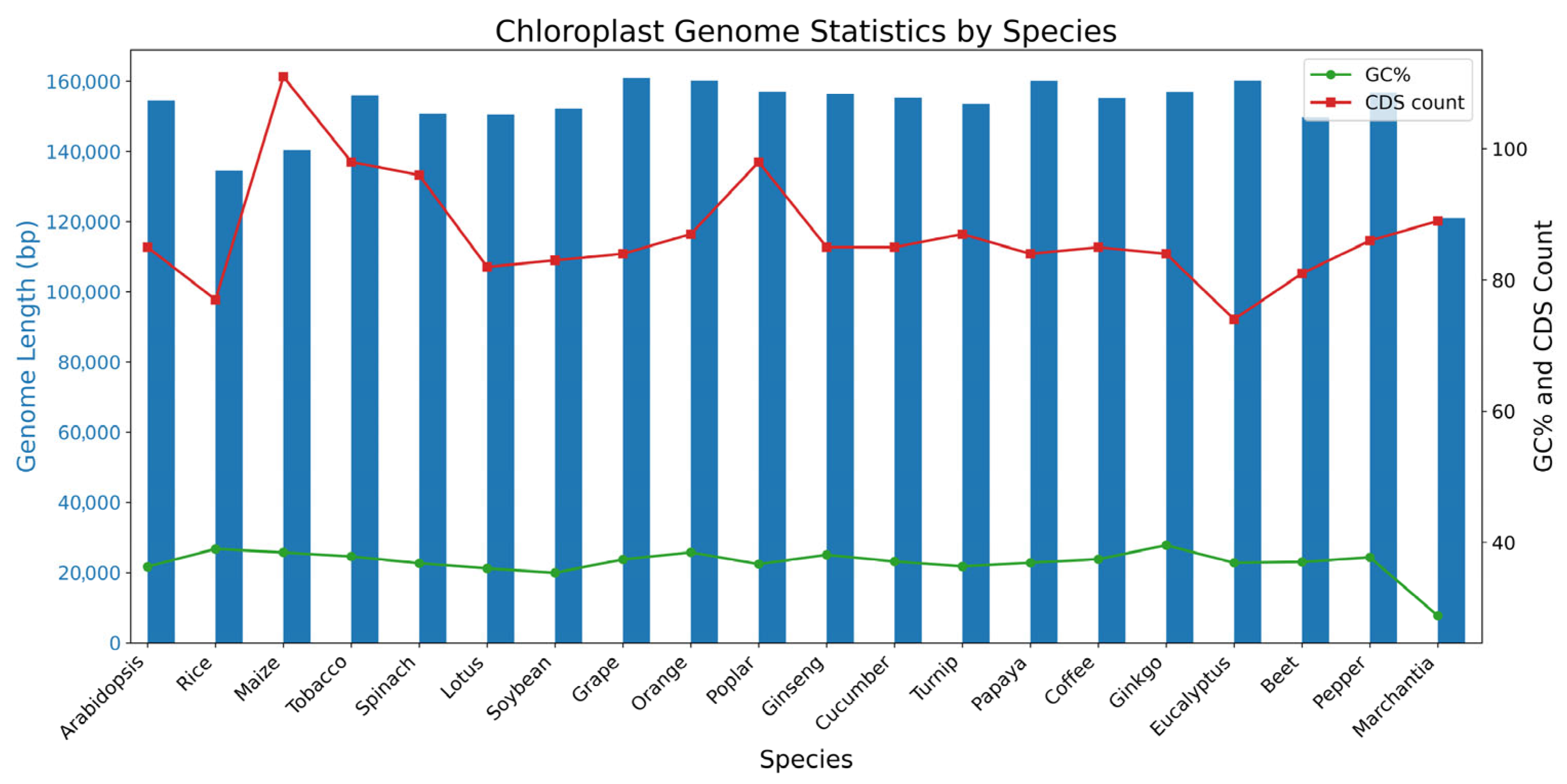
3.1. Chloroplast Genome Structure and General Features
3.2. Phylogenetic Relationships Based on Barcoding Genes
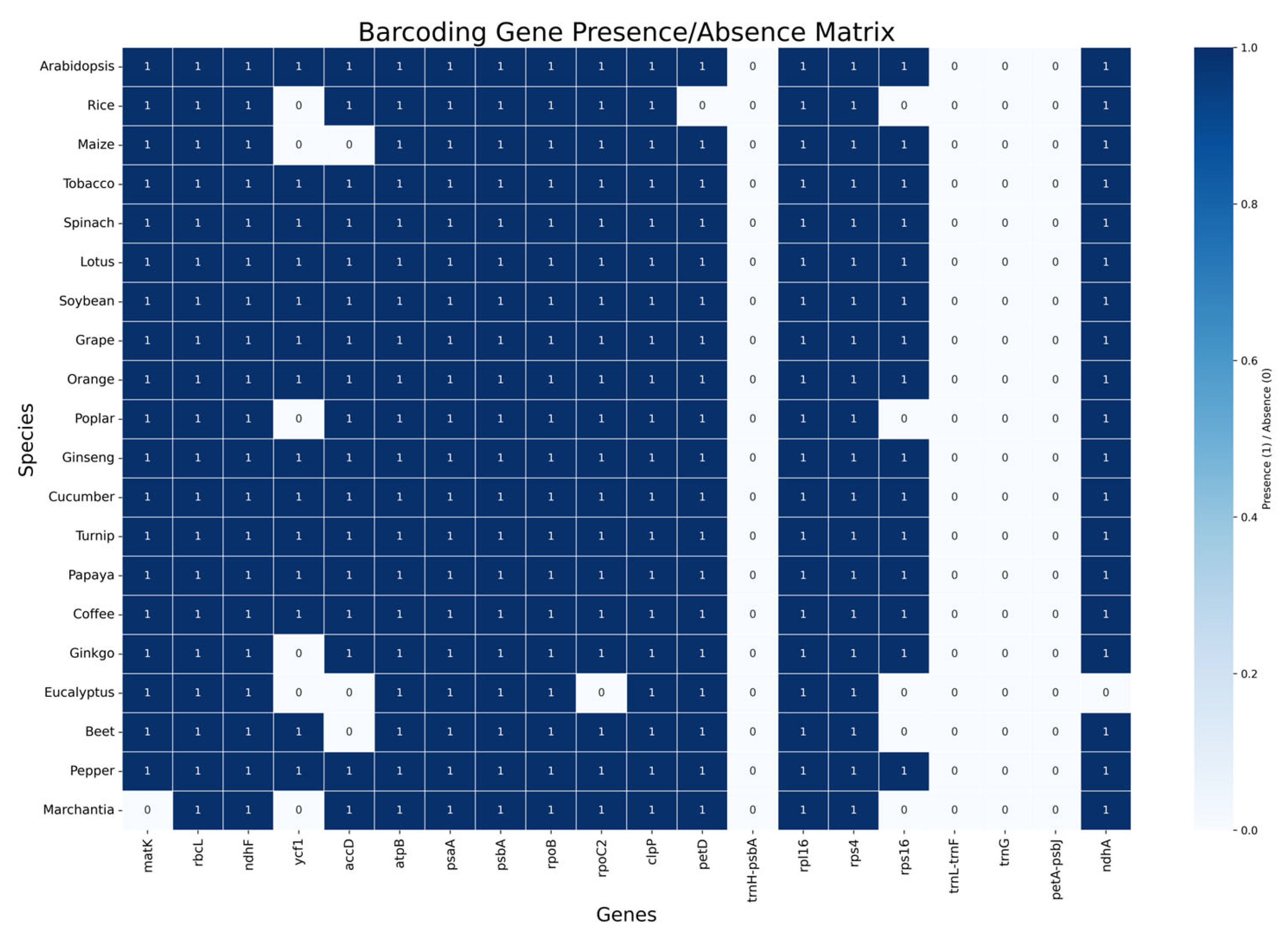
3.3. Gene Presence and Absence Across Species
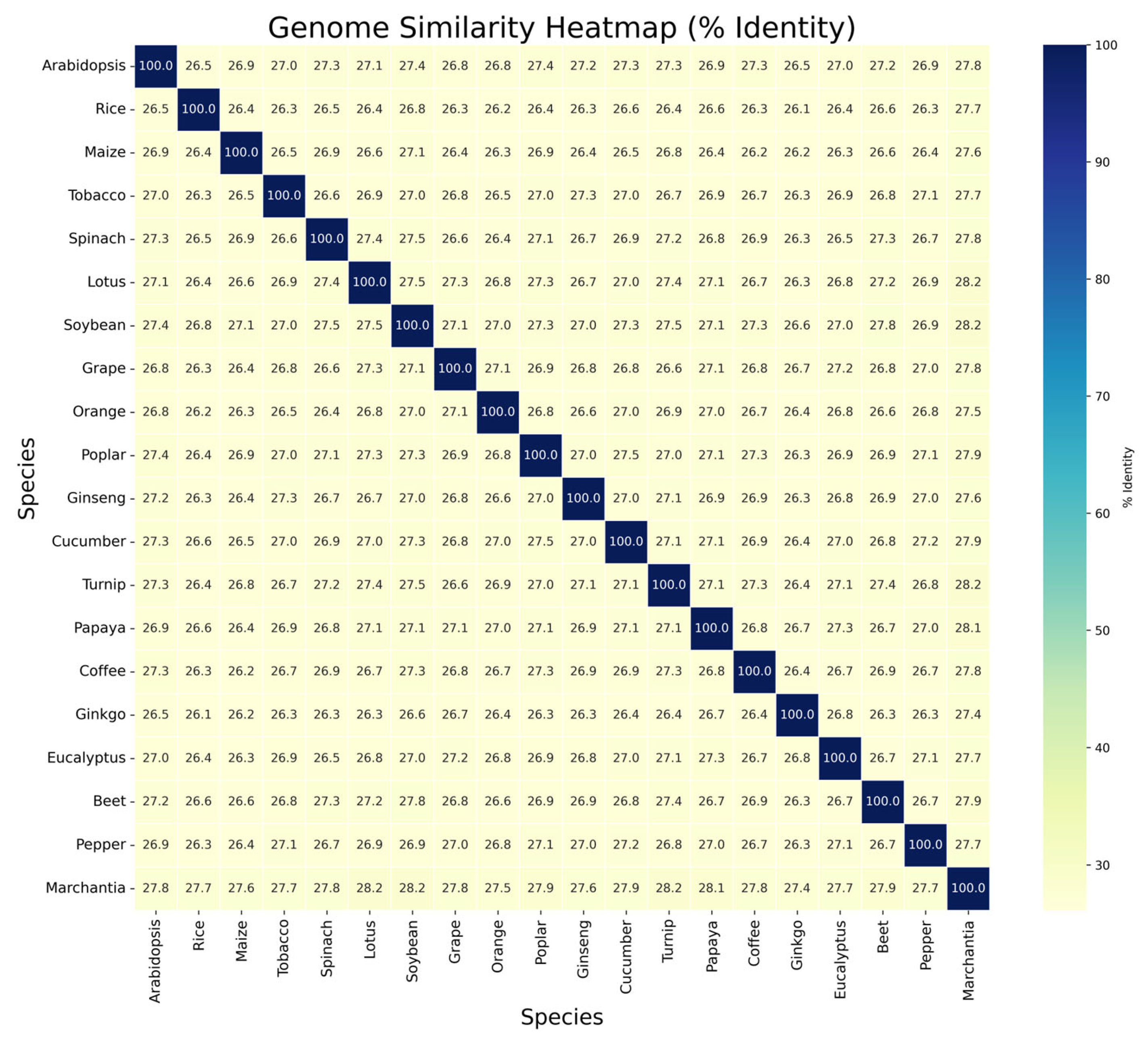
3.4. Genome-Wide Similarity Analysis
3.5. Core Versus Accessory Gene Content
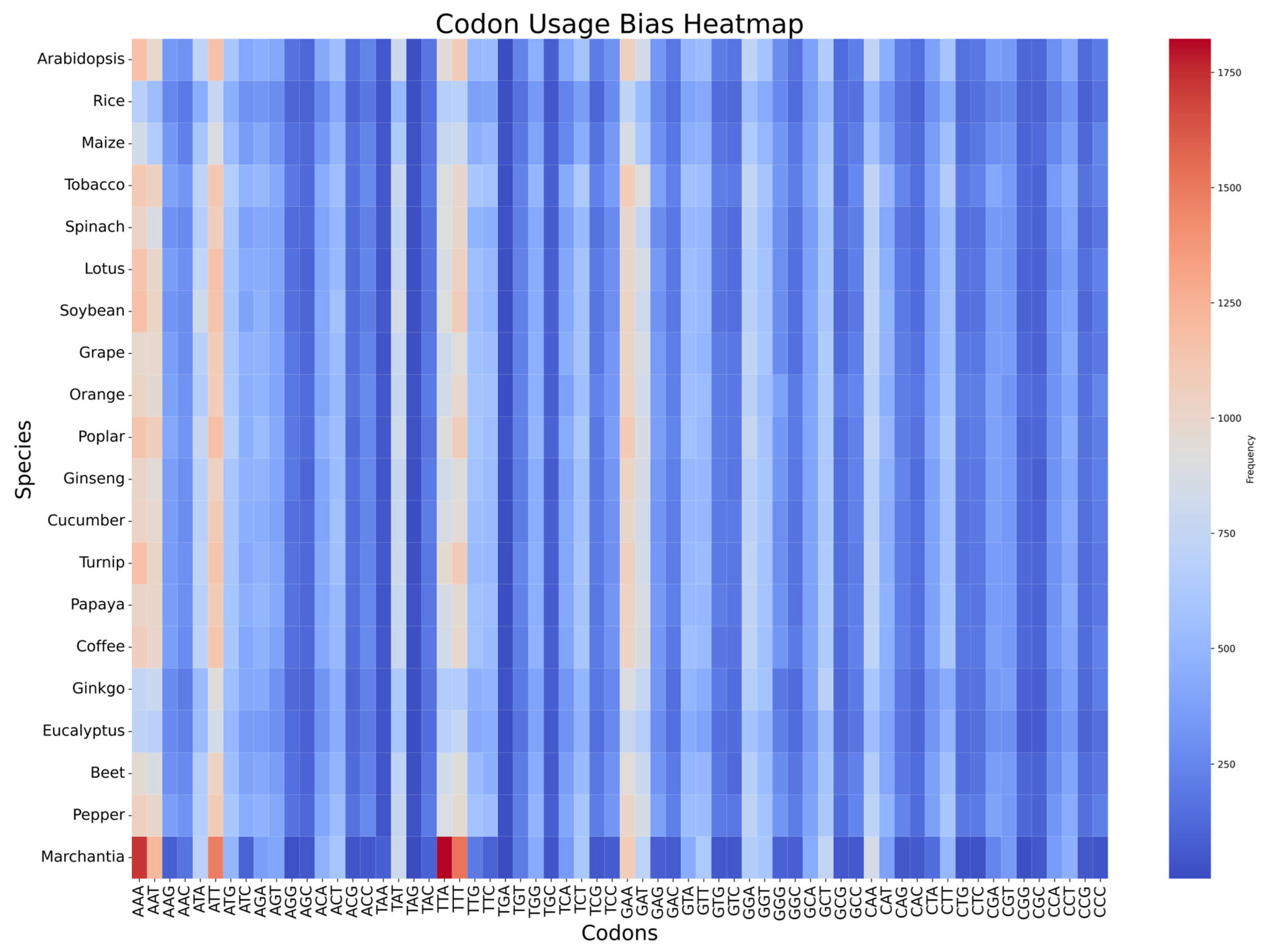
3.6. Codon Usage Bias Across Species
4. Discussion
4.1. Variation in Chloroplast Genome Size and Structure
4.2. Phylogenetic Relationships and Gene Evolution
4.3. Gene Presence/Absence Patterns
4.4. Genome-Wide Sequence Divergence
4.5. Codon Usage Bias and Evolution
4.6. Study Limitations and Future Directions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Description |
| matK | Maturase K |
| rbcL | Ribulose-bisphosphate carboxylase large chain |
| ndhF | NADH dehydrogenase subunit F |
| ycf1 | Hypothetical chloroplast reading frame 1 |
| accD | Acetyl-CoA carboxylase beta subunit |
| atpB | ATP synthase CF1 beta subunit |
| psaA | Photosystem I P700 chlorophyll a apoprotein A1 |
| psbA | Photosystem II protein D1 |
| rpoB | RNA polymerase beta subunit |
| rpoC2 | RNA polymerase beta’ subunit |
| clpP | ATP-dependent Clp protease proteolytic subunit |
| petD | Cytochrome b6/f complex subunit 4 |
| trnH-psbA | tRNA-His and photosystem II protein D1 intergenic spacer |
| rpl16 | Ribosomal protein L16 |
| rps4 | Ribosomal protein S4 |
| rps16 | Ribosomal protein S16 |
| trnL-trnF | tRNA-Leu and tRNA-Phe intergenic spacer |
| trnG | tRNA-Gly |
| petA-psbJ | Cytochrome f and photosystem II protein J intergenic spacer |
| ndhA | NADH dehydrogenase subunit A |
| LSC region | Large single-copy region |
| SSC region | Small single-copy region |
| IRs | Inverted repeats |
| CUB | Codon usage bias |
| NJ | Neighbor-Joining |
| CDS | Coding sequence(s) |
References
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Neuhauss, H.E.; Emes, M.J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Jansen, R.K. Chloroplast Genomes of Plants. In Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants; Henry, R.J., Ed.; CABI Publishing: Wallingford, UK, 2005; pp. 45–68. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M. Next-generation organellar genomics: Potentials and pitfalls of high-throughput technologies for molecular evolutionary studies and plant systematics. In Next-Generation Sequencing in Plant Systematics; Hörandl, E., Appelhans, M., Eds.; Koeltz Scientific Books: Königstein, Germany, 2015; pp. 1–18. [Google Scholar] [CrossRef]
- Wang, R.J.; Cheng, C.L.; Chang, C.C.; Wu, C.L.; Su, T.M.; Chaw, S.M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008, 8, 36. [Google Scholar] [CrossRef]
- Jansen, R.K.; Ruhlman, T.A. Plastid genomes of seed plants. In Genomics of Chloroplasts and Mitochondria, Advances in Photosynthesis and Respiration; Bock, R., Knoop, V., Eds.; Springer: Dordrecht, The Netherlands, 2012; Volume 35, pp. 103–126. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S.; Tate, J.A. Advances in the study of polyploidy since plant speciation. New Phytol. 2004, 161, 173–191. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 94, 275–288. [Google Scholar] [CrossRef]
- Ravi, V.; Khurana, J.P.; Tyagi, A.K.; Khurana, P. An update on chloroplast genomes. Plant Syst. Evol. 2008, 271, 101–122. [Google Scholar] [CrossRef]
- Zhang, S.D.; Jin, J.J.; Chen, S.Y.; Chase, M.W.; Soltis, D.E.; Li, H.T.; Yang, J.B.; Li, D.Z.; Yi, T.S. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 2017, 214, 1355–1367. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef]
- Barrett, C.F.; Davis, J.I.; Leebens-Mack, J.; Conran, J.G.; Stevenson, D.W. Plastid genomes and deep relationships among the commelinid monocot angiosperms. Cladistics 2013, 29, 65–87. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Cheng, T.; Zhou, S. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS ONE 2013, 8, e77965. [Google Scholar] [CrossRef] [PubMed]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef]
- Chumley, T.W.; Palmer, J.D.; Mower, J.P.; Fourcade, H.M.; Calie, P.J.; Boore, J.L.; Jansen, R.K. The complete chloroplast genome sequence of Pelargonium × hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006, 23, 2175–2190. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; dePamphilis, C.W.; Leebens-Mack, J.; Muller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef]
- Knox, E.B. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc. Natl. Acad. Sci. USA 2014, 111, 11097–11102. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Morden, C.W.; Palmer, J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 1992, 89, 10648–10652. [Google Scholar] [CrossRef]
- Morton, B.R. Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J. Mol. Evol. 1998, 46, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Morton, B.R. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J. Mol. Evol. 2003, 56, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2007, 4, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Grewe, F.; Guo, W.; Gubbels, E.A.; Hansen, A.K.; Mower, J.P. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol. Biol. 2013, 13, 8. [Google Scholar] [CrossRef]
- Hodkinson, T.R. Evolution and taxonomy of the grasses (Poaceae): A model family for the study of species-rich groups. Annu. Plant Rev. 2018, 1, 1–39. [Google Scholar] [CrossRef]
- Huo, Y.M.; Gao, L.M.; Liu, B.J.; Yang, Y.Y.; Kong, S.P.; Sun, Y.Q.; Yang, Y.H.; Wu, X. Complete chloroplast genome sequences of four Allium species: Comparative and phylogenetic analyses. Sci. Rep. 2019, 9, 12250. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Brister, J.R.; Chan, J.; Connor, R.; Feldgarden, M.; Fine, A.M.; Funk, K.; Hoffman, J.; et al. Database resources of the National Center for Biotechnology Information in 2025. Nucleic Acids Res. 2025, 53, D20–D29. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Evans, J.; Sheneman, L.; Foster, J. Relaxed neighbor joining: A fast distance-based phylogenetic tree construction method. J. Mol. Evol. 2006, 62, 785–792. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Downie, S.R.; Palmer, J.D. Use of chloroplast DNA rearrangements in reconstructing plant phylogeny. In Molecular Systematics of Plants; Soltis, P.S., Soltis, D.E., Doyle, J.J., Eds.; Springer: Boston, MA, USA, 1992; pp. 14–35. [Google Scholar] [CrossRef]
- Zhu, A.; Guo, W.; Gupta, S.; Fan, W.; Mower, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005, 15, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, J.; Hao, G.; Zhang, L.; Mao, K.; Wang, X.; Zhang, D.; Ma, T.; Hu, Q.; Al-Shehbaz, I.A.; et al. Plastome phylogeny and early diversification of Brassicaceae. BMC Genom. 2017, 18, 176. [Google Scholar] [CrossRef]
- Leaks, K.; El, A.; Alsaidi, Z.; Benton, K.; Chase, J.; Lewis, S.; Kassem, M.A. Comparative phylogenetic analysis of six angiosperm families using rbcL and matK chloroplast markers. J. Artif. Intell. Mach. Learn. Bioinform. 2025, 2025, 29–39. [Google Scholar] [CrossRef]
- Chaw, S.M.; Parkinson, C.L.; Cheng, Y.; Vincent, T.M.; Palmer, J.D. Seed plant phylogeny inferred from all three plant genomes: Monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc. Natl. Acad. Sci. USA 2000, 97, 4086–4091. [Google Scholar] [CrossRef] [PubMed]
- Grass Phylogeny Working Group; Barker, N.P.; Clark, L.G.; Davis, J.I.; Duvall, M.R.; Guala, G.F.; Hsiao, C.; Kellogg, E.A.; Linder, H.P.; Mason-Gamer, R.J.; et al. Phylogeny and subfamilial classification of the grasses (Poaceae). Ann. Mo. Bot. Gard. 2001, 88, 373–457. [Google Scholar] [CrossRef]
- Lanier, H.C.; Knowles, L.L. Applying species-tree analyses to deep phylogenetic histories: Challenges and potential suggested from a survey of empirical phylogenetic studies. Mol. Phylogenet. Evol. 2015, 83, 191–199. [Google Scholar] [CrossRef]
- Millen, R.S.; Olmstead, R.G.; Adams, K.L.; Palmer, J.D.; Lao, N.T.; Heggie, L.; Kavanagh, T.A.; Hibberd, J.M.; Gray, J.C.; Morden, C.W.; et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 2001, 13, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Qiu, Y.L.; Stoutemyer, M.; Palmer, J.D. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 9905–9912. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Zamir, D. Chloroplast DNA evolution and phylogenetic relationships in Lycopersicon. Proc. Natl. Acad. Sci. USA 1982, 79, 5006–5010. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.L.; Lee, J.; Bernasconi-Quadroni, F.; Soltis, D.E.; Soltis, P.S.; Zanis, M.; Zimmer, E.A.; Chen, Z.; Savolainen, V.; Chase, M.W. The earliest angiosperms: Evidence from mitochondrial, plastid and nuclear genomes. Nature 1999, 402, 404–407. [Google Scholar] [CrossRef]
- Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Carpenter, E.; Matasci, N.; Ayyampalayam, S.; Barker, M.S.; Burleigh, J.G.; Gitzendanner, M.A.; et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 2014, 111, E4859–E4868. [Google Scholar] [CrossRef]
- Parks, M.; Cronn, R.; Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009, 7, 84. [Google Scholar] [CrossRef]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef]
- de Oliveira, J.L.; Morales, A.C.; Hurst, L.D.; Urrutia, A.O.; Thompson, C.R.L.; Wolf, J.B. Inferring adaptive codon preference to understand sources of selection shaping codon usage bias. Mol. Biol. Evol. 2021, 38, 3247–3266. [Google Scholar] [CrossRef]
- Yengkhom, S.; Uddin, A.; Chakraborty, S. Deciphering codon usage patterns and evolutionary forces in chloroplast genes of Camellia sinensis var. assamica and Camellia sinensis var. sinensis in comparison to Camellia pubicosta. J. Integr. Agric. 2019, 18, 2771–2785. [Google Scholar] [CrossRef]
- Shen, L.; Chen, S.; Liang, M.; Qu, S.; Feng, S.; Wang, D.; Wang, G. Comparative analysis of codon usage bias in chloroplast genomes of ten medicinal species of Rutaceae. BMC Plant Biol. 2024, 24, 424. [Google Scholar] [CrossRef]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef]
- Smith, D.R. Mutation rates in plastid genomes: They are lower than you might think. Genome Biol. Evol. 2015, 7, 1227–1234. [Google Scholar] [CrossRef]
- Zhang, G.; Ma, H. Nuclear phylogenomics of angiosperms and insights into their relationships and evolution. J. Integr. Plant Biol. 2024, 66, 546–578. [Google Scholar] [CrossRef] [PubMed]
- Sebastin, R.; Kim, J.; Jo, I.H.; Yu, J.K.; Jang, W.; Han, S.; Park, H.S.; AlGarawi, A.M.; Hatamleh, A.A.; So, Y.S.; et al. Comparative chloroplast genome analyses of cultivated and wild Capsicum species shed light on evolution and phylogeny. BMC Plant Biol. 2024, 24, 797. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Han, S.; Zhou, J.; Zhao, M.; Zhang, S.; Kan, X. Codon Usage Analyses Reveal the Evolutionary Patterns among Plastid Genes of Saxifragales at a Larger-Sampling Scale. Genes 2023, 14, 694. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yu, D.; Kuo, W.; Huang, J.; Guo, J.; Sun, M.; Hu, Y.; Soltis, D.E.; Soltis, P.S.; Ma, H.; et al. Nuclear phylogenomics provide evidence to clarify key morphological evolution and whole-genome duplication across rosids. J. Integr. Plant Biol. 2025; Epub ahead of printing. [Google Scholar] [CrossRef]


| Species | Genus | Class | Accession Number | Genome Length (bp) | GC Content (%) | Number of CDSs |
|---|---|---|---|---|---|---|
| Arabidopsis thaliana | Arabidopsis | Magnoliopsida | NC_000932 | 154,478 | 36.3 | 85 |
| Oryza sativa (Rice) | Oryza | Liliopsida | KT289404 | 134,525 | 39.0 | 77 |
| Zea mays (Maize) | Zea | Liliopsida | NC_001666 | 140,384 | 38.5 | 111 |
| Nicotiana tabacum (Tobacco) | Nicotiana | Magnoliopsida | NC_001879 | 155,943 | 37.8 | 98 |
| Spinacia oleracea (Spinach) | Spinacia | Magnoliopsida | NC_002202 | 150,725 | 36.8 | 96 |
| Lotus japonicus (Lotus) | Lotus | Magnoliopsida | NC_002694 | 150,519 | 36.0 | 82 |
| Glycine max (Soybean) | Glycine | Magnoliopsida | NC_007942 | 152,218 | 35.4 | 83 |
| Vitis vinifera (Grape) | Vitis | Magnoliopsida | NC_007957 | 160,928 | 37.4 | 84 |
| Citrus sinensis (Orange) | Citrus | Magnoliopsida | NC_008334 | 160,129 | 38.5 | 87 |
| Populus trichocarpa (Poplar) | Populus | Magnoliopsida | NC_009143 | 157,033 | 36.7 | 98 |
| Panax ginseng (Ginseng) | Panax | Magnoliopsida | NC_006290 | 156,318 | 38.1 | 85 |
| Cucumis sativus (Cucumber) | Cucumis | Magnoliopsida | NC_007144 | 155,293 | 37.1 | 85 |
| Brassica rapa (Turnip) | Brassica | Magnoliopsida | NC_049891 | 153,621 | 36.3 | 87 |
| Carica papaya (Papaya) | Carica | Magnoliopsida | EU431223 | 160,100 | 36.9 | 84 |
| Coffea arabica (Coffee) | Coffea | Magnoliopsida | NC_008535 | 155,189 | 37.4 | 85 |
| Ginkgo biloba (Ginkgo) | Ginkgo | Ginkgoopsida | NC_016986 | 156,988 | 39.6 | 84 |
| Eucalyptus grandis (Eucalyptus) | Eucalyptus | Magnoliopsida | NC_014570 | 160,137 | 36.9 | 74 |
| Beta vulgaris (Beet) | Beta | Magnoliopsida | KR230391 | 149,722 | 37.0 | 81 |
| Capsicum annuum (Pepper) | Capsicum | Magnoliopsida | NC_018552 | 156,781 | 37.7 | 86 |
| Marchantia polymorpha (Mar.) | Marchantia | Marchantiopsida | NC_001319 | 121,024 | 28.8 | 89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kassem, M.A. Comparative Analysis of Chloroplast Genomes Across 20 Plant Species Reveals Evolutionary Patterns in Gene Content, Codon Usage, and Genome Structure. Int. J. Plant Biol. 2025, 16, 105. https://doi.org/10.3390/ijpb16030105
Kassem MA. Comparative Analysis of Chloroplast Genomes Across 20 Plant Species Reveals Evolutionary Patterns in Gene Content, Codon Usage, and Genome Structure. International Journal of Plant Biology. 2025; 16(3):105. https://doi.org/10.3390/ijpb16030105
Chicago/Turabian StyleKassem, My Abdelmajid. 2025. "Comparative Analysis of Chloroplast Genomes Across 20 Plant Species Reveals Evolutionary Patterns in Gene Content, Codon Usage, and Genome Structure" International Journal of Plant Biology 16, no. 3: 105. https://doi.org/10.3390/ijpb16030105
APA StyleKassem, M. A. (2025). Comparative Analysis of Chloroplast Genomes Across 20 Plant Species Reveals Evolutionary Patterns in Gene Content, Codon Usage, and Genome Structure. International Journal of Plant Biology, 16(3), 105. https://doi.org/10.3390/ijpb16030105








