The Oral Bacteriome
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iyer, P. Oral Cavity Is the Gateway to the Body: Role of Oral Health Professionals: A Narrative Review. J. Calif. Dent. Assoc. 2023, 51, 2193372. [Google Scholar] [CrossRef]
- Benn, A.M.L.; Heng, N.C.K.; Broadbent, J.M.; Thomson, W.M. Studying the Human Oral Microbiome: Challenges and the Evolution of Solutions. Aust. Dent. J. 2018, 63, 14–24. [Google Scholar] [CrossRef]
- Byrne, S.J.; Butler, C.A.; Reynolds, E.C.; Dashper, S.G. Taxonomy of Oral Bacteria. Methods Microbiol. 2018, 45, 171–201. [Google Scholar]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Braga, P.A.C.; Tata, A.; Gonçalves Dos Santos, V.; Barreiro, J.R.; Schwab, N.V.; Veiga Dos Santos, M.; Eberlin, M.N.; Ferreira, C.R. Bacterial Identification: From the Agar Plate to the Mass Spectrometer. RSC Adv. 2013, 3, 994–1008. [Google Scholar] [CrossRef]
- Escapa, I.F.; Chen, T.; Huang, Y.; Gajare, P.; Dewhirst, F.E.; Lemon, K.P. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (EHOMD): A Resource for the Microbiome of the Human Aerodigestive Tract. mSystems 2018, 3, e00187-18. [Google Scholar] [CrossRef]
- Zaura, E.; Keijser, B.J.; Huse, S.M.; Crielaard, W. Defining the Healthy “Core Microbiome” of Oral Microbial Communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J.A. Biology of Oral Streptococci. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Kolenbrander, P.E.; Palmer, R.J.; Rickard, A.H.; Jakubovics, N.S.; Chalmers, N.I.; Diaz, P.I. Bacterial Interactions and Successions during Plaque Development. Periodontol 2000 2006, 42, 47–79. [Google Scholar] [CrossRef] [PubMed]
- Simon-Soro, A.; Ren, Z.; Krom, B.P.; Hoogenkamp, M.A.; Cabello-Yeves, P.J.; Daniel, S.G.; Bittinger, K.; Tomas, I.; Koo, H.; Mira, A. Polymicrobial Aggregates in Human Saliva Build the Oral Biofilm. mBio 2022, 13, e00131-22. [Google Scholar] [CrossRef] [PubMed]
- Zuber, P.; Kreth, J. Aspects of Oral Streptococcal Metabolic Diversity: Imagining the Landscape Beneath the Fog. Mol. Microbiol. 2023, 120, 508–524. [Google Scholar] [CrossRef]
- Erttmann, S.F.; Gekara, N.O. Hydrogen Peroxide Release by Bacteria Suppresses Inflammasome-Dependent Innate Immunity. Nat. Commun. 2019, 10, 3493. [Google Scholar] [CrossRef]
- Medapati, M.R.; Singh, N.; Bhagirath, A.Y.; Duan, K.; Triggs-Raine, B.; Batista, E.L.; Chelikani, P. Bitter Taste Receptor T2R14 Detects Quorum Sensing Molecules from Cariogenic Streptococcus mutans and Mediates Innate Immune Responses in Gingival Epithelial Cells. FASEB J. 2021, 35, e21375. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.S.; Gunda, V.; Reinartz, D.M.; Pond, K.W.; Thorne, C.A.; Santiago Raj, P.V.; Johnson, M.D.L.; Wilson, J.E. Oral Streptococci S. anginosus and S. mitis Induce Distinct Morphological, Inflammatory, and Metabolic Signatures in Macrophages. Infect. Immun. 2024, 92, e00536-23. [Google Scholar] [CrossRef]
- Tang, Y.L.; Sim, T.S.; Tan, K.S. Oral Streptococci Subvert the Host Innate Immune Response through Hydrogen Peroxide. Sci. Rep. 2022, 12, 656. [Google Scholar] [CrossRef]
- Matsushima, H.; Kumagai, Y.; Vandenbon, A.; Kataoka, H.; Kadena, M.; Fukamachi, H.; Arimoto, T.; Morisaki, H.; Fujiwara, N.; Okahashi, N.; et al. Microarray Analysis of Macrophage Response to Infection with Streptococcus Oralis Reveals the Immunosuppressive Effect of Hydrogen Peroxide. Biochem. Biophys. Res. Commun. 2017, 485, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Myers, S.; Do, T.; Meade, J.L.; Tugnait, A.; Vernon, J.J.; Pistolic, J.; Hancock, R.E.W.; Marsh, P.D.; Trivedi, H.M.; Chen, D.; et al. Immunomodulatory Streptococci That Inhibit CXCL8 Secretion and NFκB Activation Are Common Members of the Oral Microbiota. J. Med. Microbiol. 2021, 70, 001329. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.R.; Ahn, K.B.; Kim, H.Y.; Seo, H.S.; Kum, K.Y.; Yun, C.H.; Han, S.H. Streptococcus Gordonii Lipoproteins Induce IL-8 in Human Periodontal Ligament Cells. Mol. Immunol. 2017, 91, 218–224. [Google Scholar] [CrossRef]
- Peyyala, R.; Kirakodu, S.; Novak, K.F.; Ebersole, J.L. Epithelial Interleukin-8 Responses to Oral Bacterial Biofilms. Clin. Vaccine Immunol. 2011, 18, 1770–1772. [Google Scholar] [CrossRef]
- de Toledo, A.; Nagata, E.; Yoshida, Y.; Oho, T. Streptococcus Oralis Coaggregation Receptor Polysaccharides Induce Inflammatory Responses in Human Aortic Endothelial Cells. Mol. Oral Microbiol. 2012, 27, 295–307. [Google Scholar] [CrossRef]
- Wicaksono, D.P.; Washio, J.; Abiko, Y.; Domon, H.; Takahashi, N. Nitrite Production from Nitrate and Its Link with Lactate Metabolism in Oral Veillonella Spp. Appl. Environ. Microbiol. 2020, 86, e01255-20. [Google Scholar] [CrossRef]
- Rajasekaran, J.J.; Krishnamurthy, H.K.; Bosco, J.; Jayaraman, V.; Krishna, K.; Wang, T.; Bei, K. Oral Microbiome: A Review of Its Impact on Oral and Systemic Health. Microorganisms 2024, 12, 1797. [Google Scholar] [CrossRef]
- Giacomini, J.J.; Torres-Morales, J.; Dewhirst, F.E.; Borisy, G.G.; Mark Welch, J.L. Site Specialization of Human Oral Veillonella Species. Microbiol. Spectr. 2023, 11, e04042-22. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Washio, J.; Shimizu, K.; Takahashi, N. Effects of Nitrate and Nitrite on Plaque PH Decrease and Nitrite-Producing and -Degrading Activities of Plaque In Vitro. Caries Res. 2024, 58, 552–561. [Google Scholar] [CrossRef]
- Sato-Suzuki, Y.; Washio, J.; Wicaksono, D.P.; Sato, T.; Fukumoto, S.; Takahashi, N. Nitrite-Producing Oral Microbiome in Adults and Children. Sci. Rep. 2020, 10, 16652. [Google Scholar] [CrossRef]
- Radcliffe, C.E.; Akram, N.C.; Hurrell, F.; Drucker, D.B. Effects of Nitrite and Nitrate on the Growth and Acidogenicity of Streptococcus mutans. J. Dent. 2002, 30, 325–331. [Google Scholar] [CrossRef]
- Liu, S.; Chen, M.; Wang, Y.; Zhou, X.; Peng, X.; Ren, B.; Li, M.; Cheng, L. Effect of Veillonella parvula on the Physiological Activity of Streptococcus mutans. Arch. Oral Biol. 2020, 109, 104578. [Google Scholar] [CrossRef]
- Li, G.; Liu, Y.; Zhang, M.; Ning, J.; Wu, L.; Jian, L.; Wu, H.; Cheng, X. Veillonella parvula Promotes Root Caries Development through Interactions with Streptococcus mutans and Candida albicans. Microb. Biotechnol. 2024, 17, e14547. [Google Scholar] [CrossRef]
- Luppens, S.B.I.; Kara, D.; Bandounas, L.; Jonker, M.J.; Wittink, F.R.A.; Bruning, O.; Breit, T.M.; Ten Cate, J.M.; Crielaard, W. Effect of Veillonella parvula on the Antimicrobial Resistance and Gene Expression of Streptococcus mutans Grown in a Dual-Species Biofilm. Oral Microbiol. Immunol. 2008, 23, 183–189. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, Y.; Zhuang, Y.; Tang, Y.; Nie, H.; Haung, Y.; Liu, T.; Yang, W.; Yan, F.; Zhu, Y. Veillonella parvula Acts as a Pathobiont Promoting the Biofilm Virulence and Cariogenicity of Streptococcus mutans in Adult Severe Caries. Microbiol. Spectr. 2024, 12, e04318-23. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, J.; McDaniel, S.; Samiano, B.J.; Marrujo, M.; Kingsley, K.; Howard, K.M. Microbial Screening Reveals Oral Site-Specific Locations of the Periodontal Pathogen Selenomonas Noxia. Curr. Issues Mol. Biol. 2021, 43, 353–364. [Google Scholar] [CrossRef]
- Drescher, J.; Schlafer, S.; Schaudinn, C.; Riep, B.; Neumann, K.; Friedmann, A.; Petrich, A.; Göbel, U.B.; Moter, A. Molecular Epidemiology and Spatial Distribution of Selenomonas Spp. in Subgingival Biofilms. Eur. J. Oral Sci. 2010, 118, 466–474. [Google Scholar] [CrossRef]
- Oliveira, R.R.D.S.; Fermiano, D.; Feres, M.; Figueiredo, L.C.; Teles, F.R.F.; Soares, G.M.S.; Faveri, M. Levels of Candidate Periodontal Pathogens in Subgingival Biofilm. J. Dent. Res. 2016, 95, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, C.G.; Hinson, A.N.; Vashishta, A.; Read, C.B.; Carlyon, J.A.; Lamont, R.J.; Uriarte, S.M.; Miller, D.P. Selenomonas sputigena Interactions with Gingival Epithelial Cells That Promote Inflammation. Infect. Immun. 2023, 91, e00319-22. [Google Scholar] [CrossRef]
- Yang, Z.; Cai, T.; Li, Y.; Jiang, D.; Luo, J.; Zhou, Z. Oral Microbial Communities in 5-Year-Old Children with versus without Dental Caries. BMC Oral Health 2023, 23, 400. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, N.; Wang, S.; Hu, X.; Jiao, K.; He, X.; Li, Z.; Wang, J. Exploration of Human Salivary Microbiomes—Insights into the Novel Characteristics of Microbial Community Structure in Caries and Caries-Free Subjects. PLoS ONE 2016, 11, e0147039. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.R.; Paster, B.J.; Leys, E.J.; Moeschberger, M.L.; Kenyon, S.G.; Galvin, J.L.; Boches, S.K.; Dewhirst, F.E.; Griffen, A.L. Molecular Analysis of Bacterial Species Associated with Childhood Caries. J. Clin. Microbiol. 2002, 40, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Shouche, Y.S.; Dighe, A.S.; Dhotre, D.P.; Patole, M.S.; Ranade, D.R. Selenomonas. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–12. [Google Scholar] [CrossRef]
- Cho, H.; Ren, Z.; Divaris, K.; Roach, J.; Lin, B.M.; Liu, C.; Azcarate-Peril, M.A.; Simancas-Pallares, M.A.; Shrestha, P.; Orlenko, A.; et al. Selenomonas sputigena Acts as a Pathobiont Mediating Spatial Structure and Biofilm Virulence in Early Childhood Caries. Nat. Commun. 2023, 14, 2919. [Google Scholar] [CrossRef]
- Tett, A.; Pasolli, E.; Masetti, G.; Ercolini, D.; Segata, N. Prevotella Diversity, Niches and Interactions with the Human Host. Nat. Rev. Microbiol. 2021, 19, 585–599. [Google Scholar] [CrossRef]
- Sharma, G.; Garg, N.; Hasan, S.; Shirodkar, S. Prevotella: An Insight into Its Characteristics and Associated Virulence Factors. Microb. Pathog. 2022, 169, 105673. [Google Scholar] [CrossRef]
- Accetto, T.; Avguštin, G. Polysaccharide Utilization Locus and CAZYme Genome Repertoires Reveal Diverse Ecological Adaptation of Prevotella Species. Syst. Appl. Microbiol. 2015, 38, 453–461. [Google Scholar] [CrossRef]
- Ibrahim, M.; Subramanian, A.; Anishetty, S. Comparative Pan Genome Analysis of Oral Prevotella Species Implicated in Periodontitis. Funct. Integr. Genom. 2017, 17, 513–536. [Google Scholar] [CrossRef]
- Kressirer, C.A.; Chen, T.; Lake Harriman, K.; Frias-Lopez, J.; Dewhirst, F.E.; Tavares, M.A.; Tanner, A.C.R. Functional Profiles of Coronal and Dentin Caries in Children. J. Oral Microbiol. 2018, 10, 1495976. [Google Scholar] [CrossRef]
- Könönen, E.; Fteita, D.; Gursoy, U.K.; Gursoy, M. Prevotella Species as Oral Residents and Infectious Agents with Potential Impact on Systemic Conditions. J. Oral Microbiol. 2022, 14, 2079814. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, C.; Sun, Y.; Dong, L.; Si, Y.; Yang, J.; Zhu, P.; Yang, F. Symbiotic Relationship between Prevotella Denticola and Streptococcus mutans Enhances Virulence of Plaque Biofilms. Arch. Oral Biol. 2023, 151, 105714. [Google Scholar] [CrossRef] [PubMed]
- Kianoush, N.; Adler, C.J.; Nguyen, K.A.T.; Browne, G.V.; Simonian, M.; Hunter, N. Bacterial Profile of Dentine Caries and the Impact of PH on Bacterial Population Diversity. PLoS ONE 2014, 9, e92940. [Google Scholar] [CrossRef]
- Chen, L.; Qin, B.; Du, M.; Zhong, H.; Xu, Q.; Li, Y.; Zhang, P.; Fan, M. Extensive Description and Comparison of Human Supra-Gingival Microbiome in Root Caries and Health. PLoS ONE 2015, 10, e0117064. [Google Scholar] [CrossRef]
- Chen, W.A.; Dou, Y.; Fletcher, H.M.; Boskovic, D.S. Local and Systemic Effects of Porphyromonas gingivalis Infection. Microorganisms 2023, 11, 470. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Diaz, P.I. Porphyromonas gingivalis: Immune Subversion Activities and Role in Periodontal Dysbiosis. Curr. Oral Health Rep. 2020, 7, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Tribble, G.D.; Kerr, J.E.; Wang, B.Y. Genetic Diversity in the Oral Pathogen Porphyromonas gingivalis: Molecular Mechanisms and Biological Consequences. Futur. Microbiol. 2013, 8, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Shibasaki, M.; Maruyama, F.; Sekizaki, T.; Nakagawa, I. Investigation of Potential Targets of Porphyromonas CRISPRs among the Genomes of Porphyromonas Species. PLoS ONE 2017, 12, e0183752. [Google Scholar] [CrossRef]
- Tribble, G.D.; Rigney, T.W.; Dao, D.H.V.; Wong, C.T.; Kerr, J.E.; Taylor, B.E.; Pacha, S.; Kaplan, H.B. Natural Competence Is a Major Mechanism for Horizontal DNA Transfer in the Oral Pathogen Porphyromonas gingivalis. mBio 2012, 3, e00231-11. [Google Scholar] [CrossRef]
- Ingalagi, P.; Bhat, K.; Kulkarni, R.; Kotrashetti, V.; Kumbar, V.; Kugaji, M. Detection and Comparison of Prevalence of Porphyromonas gingivalis through Culture and Real Time-Polymerase Chain Reaction in Subgingival Plaque Samples of Chronic Periodontitis and Healthy Individuals. J. Oral Maxillofac. Pathol. 2022, 26, 288. [Google Scholar] [CrossRef]
- Zhao, L.; Wu, Y.F.; Meng, S.; Yang, H.; OuYang, Y.L.; Zhou, X.D. Prevalence of FimA Genotypes of Porphyromonas gingivalis and Periodontal Health Status in Chinese Adults. J. Periodontal. Res. 2007, 42, 511–517. [Google Scholar] [CrossRef]
- Kugaji, M.; Muddapur, U.; Bhat, K.; Joshi, V.; Manubolu, M.; Pathakoti, K.; Peram, M.R.; Kumbar, V. Variation in the Occurrence of Fima Genotypes of Porphyromonas gingivalis in Periodontal Health and Disease. Int. J. Environ. Res. Public Health 2020, 17, 1826. [Google Scholar] [CrossRef]
- Bartnicka, D.; Karkowska-Kuleta, J.; Zawrotniak, M.; Satała, D.; Michalik, K.; Zielinska, G.; Bochenska, O.; Kozik, A.; Ciaston, I.; Koziel, J.; et al. Adhesive Protein-Mediated Cross-Talk between Candida albicans and Porphyromonas gingivalis in Dual Species Biofilm Protects the Anaerobic Bacterium in Unfavorable Oxic Environment. Sci. Rep. 2019, 9, 4376. [Google Scholar] [CrossRef]
- Soto, C.; Rojas, V.; Yáñez, L.; Hidalgo, A.; Olivera, M.; Pacheco, M.; Venegas, D.; Salinas, D.; Bravo, D.; Quest, A.F.G. Porphyromonas gingivalis-Helicobacter Pylori Co-Incubation Enhances Porphyromonas gingivalis Virulence and Increases Migration of Infected Human Oral Keratinocytes. J. Oral Microbiol. 2022, 14, 2107691. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Ikegami, A.; Kuramitsu, H.K. Synergistic Biofilm Formation by Treponema Denticola and Porphyromonas gingivalis. FEMS Microbiol. Lett. 2005, 250, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, S.; Kolenbrander, P.E. Mutualistic Biofilm Communities Develop with Porphyromonas gingivalis and Initial, Early, and Late Colonizers of Enamel. J. Bacteriol. 2009, 191, 6804–6811. [Google Scholar] [CrossRef]
- Hajishengallis, G. Porphyromonas gingivalis-Host Interactions: Open War or Intelligent Guerilla Tactics? Microbes Infect. 2009, 11, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Van Hoogmoed, C.G.; Geertsema-Doornbusch, G.I.; Teughels, W.; Quirynen, M.; Busscher, H.J.; Van Der Mei, H.C. Reduction of Periodontal Pathogens Adhesion by Antagonistic Strains. Oral Microbiol. Immunol. 2008, 23, 43–48. [Google Scholar] [CrossRef]
- Tenorio, E.L.; Klein, B.A.; Cheung, W.S.; Hu, L.T. Identification of Interspecies Interactions Affecting Porphyromonas gingivalis Virulence Phenotypes. J. Oral Microbiol. 2011, 3, 8396. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, B.-Y.; Pratap, S.; Xie, H. Oral Microbiome Associated with Differential Ratios of Porphyromonas gingivalis and Streptococcus cristatus. Microbiol. Spectr. 2024, 12, e03482-23. [Google Scholar] [CrossRef]
- Aleksijević, L.H.; Aleksijević, M.; Škrlec, I.; Šram, M.; Šram, M.; Talapko, J. Porphyromonas gingivalis Virulence Factors and Clinical Significance in Periodontal Disease and Coronary Artery Diseases. Pathogens 2022, 11, 1173. [Google Scholar] [CrossRef] [PubMed]
- de Jongh, C.A.; de Vries, T.J.; Bikker, F.J.; Gibbs, S.; Krom, B.P. Mechanisms of Porphyromonas gingivalis to Translocate over the Oral Mucosa and Other Tissue Barriers. J. Oral Microbiol. 2023, 15, 2205291. [Google Scholar] [CrossRef]
- Mei, F.; Xie, M.; Huang, X.; Long, Y.; Lu, X.; Wang, X.; Chen, L. Porphyromonas gingivalis and Its Systemic Impact: Current Status. Pathogens 2020, 9, 944. [Google Scholar] [CrossRef]
- Huang, Z.; Hao, M.; Shi, N.; Wang, X.; Yuan, L.; Yuan, H.; Wang, X. Porphyromonas gingivalis: A Potential Trigger of Neurodegenerative Disease. Front. Immunol. 2025, 16, 1482033. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, F.; Okada, M.; Zhong, X.; Miura, K. PCR Detection of Capnocytophaga Species in Dental Plaque Samples from Children Aged 2 to 12 Years. Microbiol. Immunol. 2001, 45, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Hosohama-Saito, K.; Kokubu, E.; Okamoto-Shibayama, K.; Kita, D.; Katakura, A.; Ishihara, K. Involvement of LuxS in Biofilm Formation by Capnocytophaga ochracea. PLoS ONE 2016, 11, e0147114. [Google Scholar] [CrossRef]
- Kita, D.; Shibata, S.; Kikuchi, Y.; Kokubu, E.; Nakayama, K.; Saito, A.; Ishihara, K. Involvement of the Type IX Secretion System in Capnocytophaga ochracea Gliding Motility and Biofilm Formation. Appl. Environ. Microbiol. 2016, 82, 1756–1766. [Google Scholar] [CrossRef]
- Ratheesh, N.K.; Zdimal, A.M.; Calderon, C.A.; Shrivastava, A. Bacterial Swarm-Mediated Phage Transportation Disrupts a Biofilm Inherently Protected from Phage Penetration. Microbiol. Spectr. 2023, 11, e00937-23. [Google Scholar] [CrossRef]
- Shrivastava, A.; Patel, V.K.; Tang, Y.; Yost, S.C.; Dewhirst, F.E.; Berg, H.C. Cargo Transport Shapes the Spatial Organization of a Microbial Community. Proc. Natl. Acad. Sci. USA 2018, 115, 8633–8638. [Google Scholar] [CrossRef]
- Idate, U.; Bhat, K.; Kotrashetti, V.; Kugaji, M.; Kumbar, V. Molecular Identification of Capnocytophaga Species from the Oral Cavity of Patients with Chronic Periodontitis and Healthy Individuals. J. Oral Maxillofac. Pathol. 2020, 24, 397. [Google Scholar] [CrossRef]
- Pudakalkatti, P.; Baheti, A.; Hattarki, S.; Kambali, S.; Naik, R. Detection and Prevalence of Capnocytophaga in Periodontal Health and Disease. J. Orofac. Sci. 2016, 8, 92. [Google Scholar] [CrossRef]
- Shigeishi, H.; Hamada, N.; Kaneyasu, Y.; Niitani, Y.; Takemoto, T.; Ohta, K. Prevalence of Oral Capnocytophaga Species and Their Association with Dental Plaque Accumulation and Periodontal Inflammation in Middle-Aged and Older People. Biomed. Rep. 2024, 20, 99. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Shen, W.; Wang, J.; Xu, Y.; Zhai, R.; Zhang, J.; Wang, M.; Wang, M.; Liu, L. Capnocytophaga Gingivalis Is a Potential Tumor Promotor in Oral Cancer. Oral Dis. 2022, 30, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Helmerhorst, E.J.; Leone, C.W.; Troxler, R.F.; Yaskell, T.; Haffajee, A.D.; Socransky, S.S.; Oppenheim, F.G. Identification of Early Microbial Colonizers in Human Dental Biofilm. J. Appl. Microbiol. 2004, 97, 1311–1318. [Google Scholar] [CrossRef]
- Soria, S.; Angulo-Bejarano, P.I.; Sharma, A. Biofilms: Development and Molecular Interaction of Microbiome in the Human Oral Cavity. In New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms Current Research and Future Trends in Microbial Biofilms; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Zijnge, V.; Van Leeuwen, M.B.M.; Degener, J.E.; Abbas, F.; Thurnheer, T.; Gmür, R.; Harmsen, H.J.M. Oral Biofilm Architecture on Natural Teeth. PLoS ONE 2010, 5, e9321. [Google Scholar] [CrossRef]
- Yang, R.; Zou, J.; Li, J.Y. Study of the Relationship between Oral Actinomyces and Childhood Caries. West China J. Stomatol. 2007, 25, 568–570. [Google Scholar]
- Struzycka, I. The Oral Microbiome in Dental Caries. Pol. J. Microbiol. 2014, 63, 127–135. [Google Scholar] [CrossRef]
- Dame-Teixeira, N.; Parolo, C.C.F.; Maltz, M.; Tugnait, A.; Devine, D.; Do, T. Actinomyces Spp. Gene Expression in Root Caries Lesions. J. Oral Microbiol. 2016, 8, 32383. [Google Scholar] [CrossRef] [PubMed]
- Brailsford, S.R.; Lynch, E.; Beighton, D. The Isolation of Actinomyces Naeslundii from Sound Root Surfaces and Root Carious Lesions. Caries Res. 1998, 32, 100–106. [Google Scholar] [CrossRef]
- Tang, G.; Samaranayake, L.P.; Yip, H.K.; Chu, F.C.S.; Tsang, P.C.S.; Cheung, B.P.K. Direct Detection of Actinomyces Spp. from Infected Root Canals in a Chinese Population: A Study Using PCR-Based, Oligonucleotide-DNA Hybridization Technique. J. Dent. 2003, 31, 559–568. [Google Scholar] [CrossRef]
- Isaac, R.D.; Sanjeev, K.; Subbulakshmi, C.L.; Amirtharaj, L.V.; Sekar, M. Identification of a Novel Bacterium Scardovia Wiggsiae in High Caries Risk Adolescence: A Metagenomic and Melt Curve Analysis. J. Conserv. Dent. 2022, 25, 297–305. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, S.; McDaniel, J.; Howard, K.M.; Kingsley, K. Molecular Screening and Analysis Reveal Novel Oral Site-Specific Locations for the Cariogenic Pathogen Scardovia Wiggsiae. Dent. J. 2021, 9, 73. [Google Scholar] [CrossRef] [PubMed]
- Kressirer, C.A.; Smith, D.J.; King, W.F.; Dobeck, J.M.; Starr, J.R.; Tanner, A.C.R. Scardovia Wiggsiae and Its Potential Role as a Caries Pathogen. J. Oral Biosci. 2017, 59, 135–141. [Google Scholar] [CrossRef]
- Kameda, M.; Abiko, Y.; Washio, J.; Tanner, A.C.R.; Kressirer, C.A.; Mizoguchi, I.; Takahashi, N. Sugar Metabolism of Scardovia Wiggsiae, a Novel Caries-Associated Bacterium. Front. Microbiol. 2020, 11, 479. [Google Scholar] [CrossRef]
- Shao, Q.; Feng, D.; Yu, Z.; Chen, D.; Ji, Y.; Ye, Q.; Cheng, D. The Role of Microbial Interactions in Dental Caries: Dental Plaque Microbiota Analysis. Microb. Pathog. 2023, 185, 106390. [Google Scholar] [CrossRef]
- Welch, J.L.M.; Rossetti, B.J.; Rieken, C.W.; Dewhirst, F.E.; Borisy, G.G. Biogeography of a Human Oral Microbiome at the Micron Scale. Proc. Natl. Acad. Sci. USA 2016, 113, E791–E800. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, F.; Su, Z.; Li, Y.; Li, J. Corynebacterium matruchotii: A Confirmed Calcifying Bacterium with a Potentially Important Role in the Supragingival Plaque. Front. Microbiol. 2022, 13, 940643. [Google Scholar] [CrossRef]
- Morillo-Lopez, V.; Sjaarda, A.; Islam, I.; Borisy, G.G.; Mark Welch, J.L. Corncob Structures in Dental Plaque Reveal Microhabitat Taxon Specificity. Microbiome 2022, 10, 145. [Google Scholar] [CrossRef]
- Kreth, J.; Helliwell, E.; Treerat, P.; Merritt, J. Molecular Commensalism: How Oral Corynebacteria and Their Extracellular Membrane Vesicles Shape Microbiome Interactions. Front. Oral Health 2024, 5, 1410786. [Google Scholar] [CrossRef]
- Almeida, E.; Puri, S.; Labossiere, A.; Elangovan, S.; Kim, J.; Ramsey, M. Bacterial Multispecies Interaction Mechanisms Dictate Biogeographic Arrangement between the Oral Commensals Corynebacterium matruchotii and Streptococcus mitis. mSystems 2023, 8, e00115-23. [Google Scholar] [CrossRef]
- Briaud, P.; Carroll, R.K. Extracellular Vesicle Biogenesis and Functions in Gram-Positive Bacteria. Infect. Immun. 2020, 88, e00433-20. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, J.J.; Torres-Morales, J.; Dewhirst, F.E.; Borisy, G.G.; Mark Welch, J.L. Spatial Ecology of the Neisseriaceae Family in the Human Oral Cavity. Microbiol. Spectr. 2025, 13, e03275-24. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Liu, J.; Jiang, M.; Chen, Q.; Zhang, Y.; Yang, M.; Zhai, Y. The Role of Oral Nitrate-Reducing Bacteria in the Prevention of Caries: A Review Related to Caries and Nitrate Metabolism. Caries Res. 2023, 57, 119–132. [Google Scholar] [CrossRef]
- Moran, S.P.; Rosier, B.T.; Henriquez, F.L.; Burleigh, M.C. The Effects of Nitrate on the Oral Microbiome: A Systematic Review Investigating Prebiotic Potential. J. Oral Microbiol. 2024, 16, 2322228. [Google Scholar] [CrossRef] [PubMed]
- Richards, V.P.; Alvarez, A.J.; Luce, A.R.; Bedenbaugh, M.; Mitchell, M.L.; Burne, R.A.; Nascimento, M.M. Microbiomes of Sitespecific Dental Plaques from Children with Different Caries Status. Infect. Immun. 2017, 85, e00106-17. [Google Scholar] [CrossRef]
- Gross, E.L.; Beall, C.J.; Kutsch, S.R.; Firestone, N.D.; Leys, E.J.; Griffen, A.L. Beyond Streptococcus mutans: Dental Caries Onset Linked to Multiple Species by 16S RRNA Community Analysis. PLoS ONE 2012, 7, e47722. [Google Scholar] [CrossRef]
- Peterson, S.N.; Snesrud, E.; Liu, J.; Ong, A.C.; Kilian, M.; Schork, N.J.; Bretz, W. The Dental Plaque Microbiome in Health and Disease. PLoS ONE 2013, 8, e58487. [Google Scholar] [CrossRef]
- Tanner, A.C.R.; Kressirer, C.A.; Faller, L.L. Understanding Caries From the Oral Microbiome Perspective. J. Calif. Dent. Assoc. 2016, 44, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Akatsu, T.; Fujii, A.; Kawano, S.; Minegishi, Y.; Ota, N. Commensal Neisseria Inhibit Porphyromonas gingivalis Invasion of Gingival Epithelial Cells. Oral Health Prev. Dent. 2024, 22, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.L.; Leys, E.J.; Gasparovich, S.R.; Firestone, N.D.; Schwartzbaum, J.A.; Janies, D.A.; Asnani, K.; Griffen, A.L. Bacterial 16S Sequence Analysis of Severe Caries in Young Permanent Teeth. J. Clin. Microbiol. 2010, 48, 4121–4128. [Google Scholar] [CrossRef] [PubMed]
- Gerner-Smidt, P.; Keiser-Nielsen, H.; Dorsch, M.; Stackebrandt, E.; Ursing, J.; Blom, J.; Christensen, A.C.; Christensen, J.J.; Frederiksen, W.; Hoffmann, S.; et al. Lautropia mirabilis Gen. Nov., Sp. Nov., a Gram-Negative Motile Coccus with Unusual Morphology Isolated from the Human Mouth. Microbiology 1994, 140, 1787–1797. [Google Scholar] [CrossRef]
- Lim, Y.K.; Park, S.N.; Lee, W.P.; Shin, J.H.; Jo, E.; Shin, Y.; Paek, J.; Chang, Y.H.; Kim, H.; Kook, J.K. Lautropia Dentalis Sp. Nov., Isolated from Human Dental Plaque of a Gingivitis Lesion. Curr. Microbiol. 2019, 76, 1369–1373. [Google Scholar] [CrossRef]
- Calheiros Cruz, G.; Sousa, M.; Vilela, S.; Teixeira E Costa, F.; Silva, F.J. Lautropia mirabilis: An Exceedingly Rare Cause of Peritoneal Dialysis-Associated Peritonitis. Case Rep. Nephrol. Dial. 2022, 12, 81–84. [Google Scholar] [CrossRef]
- Tan, R.Y.P.; Tan, B.Q.; Rao, N. The First Case of Peritoneal Dialysis-Associated Peritonitis Due to Lautropia mirabilis. Nephrology 2021, 26, 634–635. [Google Scholar] [CrossRef]
- Perera, D.; McLean, A.; Morillo-López, V.; Cloutier-Leblanc, K.; Almeida, E.; Cabana, K.; Mark Welch, J.; Ramsey, M. Mechanisms Underlying Interactions between Two Abundant Oral Commensal Bacteria. ISME J. 2021, 16, 948–957. [Google Scholar] [CrossRef]
- de Palma, T.H.; Powers, C.; McPartland, M.J.; Mark Welch, J.; Ramsey, M. Essential Genes for Haemophilus parainfluenzae Survival and Biofilm Growth. mSystems 2024, 9, e00674-24. [Google Scholar] [CrossRef]
- Giacomini, J.J.; Torres-Morales, J.; Tang, J.; Dewhirst, F.E.; Borisy, G.G.; Mark Welch, J.L. Spatial Ecology of Haemophilus and Aggregatibacter in the Human Oral Cavity. Microbiol. Spectr. 2024, 12, e04017-23. [Google Scholar] [CrossRef] [PubMed]
- Utter, D.R.; Borisy, G.G.; Eren, A.M.; Cavanaugh, C.M.; Mark Welch, J.L. Metapangenomics of the Oral Microbiome Provides Insights into Habitat Adaptation and Cultivar Diversity. Genome Biol. 2020, 21, 293. [Google Scholar] [CrossRef]
- Tseng, Y.C.; Yang, H.Y.; Lin, W.T.; Chang, C.B.; Chien, H.C.; Wang, H.P.; Chen, C.M.; Wang, J.T.; Li, C.; Wu, S.F.; et al. Salivary Dysbiosis in Sjögren’s Syndrome and a Commensal-Mediated Immunomodulatory Effect of Salivary Gland Epithelial Cells. npj Biofilms Microbiomes 2021, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.C.; Liao, K.S.; Lin, W.T.; Li, C.; Chang, C.B.; Hsu, J.W.; Chan, C.P.; Chen, C.M.; Wang, H.P.; Chien, H.C.; et al. A Human Oral Commensal-Mediated Protection against Sjögren’s Syndrome with Maintenance of T Cell Immune Homeostasis and Improved Oral Microbiota. Npj Biofilms Microbiomes 2025, 11, 18. [Google Scholar] [CrossRef]
- Diao, J.; Yuan, C.; Tong, P.; Ma, Z.; Sun, X.; Zheng, S. Potential Roles of the Free Salivary Microbiome Dysbiosis in Periodontal Diseases. Front. Cell. Infect. Microbiol. 2021, 11, 711282. [Google Scholar] [CrossRef]
- Wajid, S.; Farrukh, L.; Rosenberg, L.; Faiz, M.; Singh, G. Systemic Haemophilus parainfluenzae Infection Manifesting with Endocarditis and Membranoproliferative Glomerulonephritis. Cureus 2023, 15, e41086. [Google Scholar] [CrossRef]
- Jenkinson, H.F.; Lamont, R.J. Oral Microbial Communities in Sickness and in Health. Trends Microbiol. 2005, 13, 589–595. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, T.; Li, Y.; Huang, L.; Yin, D. Fusobacterium Nucleatum: The Opportunistic Pathogen of Periodontal and Peri-Implant Diseases. Front. Microbiol. 2022, 13, 860149. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, C.W.; Lux, R.; Haake, S.K.; Shi, W. The Fusobacterium Nucleatum Outer Membrane Protein RadD Is an Arginine-Inhibitable Adhesin Required for Inter-Species Adherence and the Structured Architecture of Multispecies Biofilm. Mol. Microbiol. 2009, 71, 35–47. [Google Scholar] [CrossRef]
- Coppenhagen-Glazer, S.; Sol, A.; Abed, J.; Naor, R.; Zhang, X.; Han, Y.W.; Bachrach, G. Fap2 of Fusobacterium Nucleatum Is a Galactose-Inhibitable Adhesin Involved in Coaggregation, Cell Adhesion, and Preterm Birth. Infect. Immun. 2015, 83, 1104–1113. [Google Scholar] [CrossRef]
- Thurnheer, T.; Karygianni, L.; Flury, M.; Belibasakis, G.N. Fusobacterium Species and Subspecies Differentially Affect the Composition and Architecture of Supra- and Subgingival Biofilms Models. Front. Microbiol. 2019, 10, 1716. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Nambu, T.; Liang, Y.; Kato, J.; Shinkawa, A.; Uchida, H.; Zhang, R.; Zheng, J.; Okinaga, T.; Takahashi, K. Phylogenetic Analysis and Subspecies Distribution of Fusobacterium Nucleatum Isolated from Interdental Plaque. Nano Biomed. 2024, 16, 109–115. [Google Scholar] [CrossRef]
- Muchova, M.; Balacco, D.L.; Grant, M.M.; Chapple, I.L.C.; Kuehne, S.A.; Hirschfeld, J. Fusobacterium Nucleatum Subspecies Differ in Biofilm Forming Ability In Vitro. Front. Oral Health 2022, 3, 853618. [Google Scholar] [CrossRef] [PubMed]
- Krieger, M.; AbdelRahman, Y.M.; Choi, D.; Palmer, E.A.; Yoo, A.; McGuire, S.; Kreth, J.; Merritt, J. Stratification of Fusobacterium Nucleatum by Local Health Status in the Oral Cavity Defines Its Subspecies Disease Association. Cell Host Microbe 2024, 32, 479–488.e4. [Google Scholar] [CrossRef]
- Muchova, M.; Kuehne, S.A.; Grant, M.M.; Smith, P.P.; Nagi, M.; Chapple, I.L.C.; Hirschfeld, J. Fusobacterium Nucleatum Elicits Subspecies-Specific Responses in Human Neutrophils. Front. Cell. Infect. Microbiol. 2024, 14, 1449539. [Google Scholar] [CrossRef]
- Kook, J.K.; Park, S.N.; Lim, Y.K.; Cho, E.; Jo, E.; Roh, H.; Shin, Y.; Paek, J.; Kim, H.S.; Kim, H.; et al. Genome-Based Reclassification of Fusobacterium Nucleatum Subspecies at the Species Level. Curr. Microbiol. 2017, 74, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Sun, T.; Zhou, J.; Zhi, M.; Shen, S.; Wang, Y.; Gu, X.; Li, Z.; Gao, H.; Wang, P.; et al. Pangenomic Study of Fusobacterium Nucleatum Reveals the Distribution of Pathogenic Genes and Functional Clusters at the Subspecies and Strain Levels. Microbiol. Spectr. 2023, 11, e05184-22. [Google Scholar] [CrossRef]
- Krieger, M.; Guo, M.; Merritt, J. Reexamining the Role of Fusobacterium Nucleatum Subspecies in Clinical and Experimental Studies. Gut Microbes 2024, 16, 2415490. [Google Scholar] [CrossRef]
- Eribe, E.R.K.; Olsen, I. Leptotrichia Species in Human Infections II. J. Oral Microbiol. 2017, 9, 1368848. [Google Scholar] [CrossRef]
- Thompson, J.; Pikis, A. Metabolism of Sugars by Genetically Diverse Species of Oral Leptotrichia. Mol. Oral Microbiol. 2012, 27, 34–44. [Google Scholar] [CrossRef]
- Khelaifia, S.; Virginie, P.; Belkacemi, S.; Tassery, H.; Terrer, E.; Aboudharam, G. Culturing the Human Oral Microbiota, Updating Methodologies and Cultivation Techniques. Microorganisms 2023, 11, 836. [Google Scholar] [CrossRef]
- Bhandary, R.; Venugopalan, G.; Ramesh, A.; Tartaglia, G.M.; Singhal, I.; Khijmatgar, S. Microbial Symphony: Navigating the Intricacies of the Human Oral Microbiome and Its Impact on Health. Microorganisms 2024, 12, 571. [Google Scholar] [CrossRef] [PubMed]
- He, X.; McLean, J.S.; Edlund, A.; Yooseph, S.; Hall, A.P.; Liu, S.Y.; Dorrestein, P.C.; Esquenazi, E.; Hunter, R.C.; Cheng, G.; et al. Cultivation of a Human-Associated TM7 Phylotype Reveals a Reduced Genome and Epibiotic Parasitic Lifestyle. Proc. Natl. Acad. Sci. USA 2015, 112, 244–249. [Google Scholar] [CrossRef]
- Bor, B.; Poweleit, N.; Bois, J.S.; Cen, L.; Bedree, J.K.; Zhou, Z.H.; Gunsalus, R.P.; Lux, R.; McLean, J.S.; He, X.; et al. Phenotypic and Physiological Characterization of the Epibiotic Interaction Between TM7x and Its Basibiont Actinomyces. Microb. Ecol. 2016, 71, 243–255. [Google Scholar] [CrossRef]
- Bedree, J.K.; Bor, B.; Cen, L.; Edlund, A.; Lux, R.; Mclean, J.S.; Shi, W.; He, X. Quorum Sensing Modulates the Epibiotic-Parasitic Relationship Between Actinomyces Odontolyticusand Its Saccharibacteria Epibiont, a Nanosynbacter LyticusStrain, TM7x. Front. Microbiol. 2018, 9, 2049. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, E.L.; Bor, B.; Kerns, K.A.; Cen, L.; Shi, W.; He, X.; McLean, J.S. Ultrasmall Epibiont Nanosynbacter Lyticus Strain TM7x and Host Bacteria Transcriptional Activity after Initial Host Parasitism. J. Oral Microbiol. 2024, 16, 2287349. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, E.L.; Bor, B.; Kerns, K.A.; Lamont, E.I.; Chang, Y.; Liu, J.; Cen, L.; Schulte, F.; Hardt, M.; Shi, W.; et al. Transcriptome of Epibiont Saccharibacteria Nanosynbacter Lyticus Strain TM7x During the Establishment of Symbiosis. J. Bacteriol. 2022, 204, e00112-22. [Google Scholar] [CrossRef]
- Bor, B.; McLean, J.S.; Foster, K.R.; Cen, L.; To, T.T.; Serrato-Guillen, A.; Dewhirst, F.E.; Shi, W.; He, X. Rapid Evolution of Decreased Host Susceptibility Drives a Stable Relationship between Ultrasmall Parasite TM7x and Its Bacterial Host. Proc. Natl. Acad. Sci. USA 2018, 115, 12277–12282. [Google Scholar] [CrossRef]
- Dong, P.T.; Tian, J.; Kobayashi-Kirschvink, K.J.; Cen, L.; McLean, J.S.; Bor, B.; Shi, W.; He, X. Episymbiotic Saccharibacteria Induce Intracellular Lipid Droplet Production in Their Host Bacteria. ISME J. 2024, 18, wrad034. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Park, H.; Cheng, B.; Kokaras, A.; Paster, B.; Burkett, S.; Watson, C.W.M.; Annavajhala, M.K.; Uhlemann, A.C.; Noble, J.M. Subgingival Microbiome and Clinical Periodontal Status in an Elderly Cohort: The WHICAP Ancillary Study of Oral Health. J. Periodontol. 2020, 91, S56–S67. [Google Scholar] [CrossRef]
- Kharitonova, M.; Vankov, P.; Abdrakhmanov, A.; Mamaeva, E.; Yakovleva, G.; Ilinskaya, O. The Composition of Microbial Communities in Inflammatory Periodontal Diseases in Young Adults Tatars. AIMS Microbiol. 2021, 7, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Brinig, M.M.; Lepp, P.W.; Ouverney, C.C.; Armitage, G.C.; Relman, D.A. Prevalence of Bacteria of Division TM7 in Human Subgingival Plaque and Their Association with Disease. Appl. Environ. Microbiol. 2003, 69, 1687–1694. [Google Scholar] [CrossRef] [PubMed]
- Chipashvili, O.; Utter, D.R.; Bedree, J.K.; Ma, Y.; Schulte, F.; Mascarin, G.; Alayyoubi, Y.; Chouhan, D.; Hardt, M.; Bidlack, F.; et al. Episymbiotic Saccharibacteria Suppresses Gingival Inflammation and Bone Loss in Mice through Host Bacterial Modulation. Cell Host Microbe 2021, 29, 1649–1662.e7. [Google Scholar] [CrossRef] [PubMed]
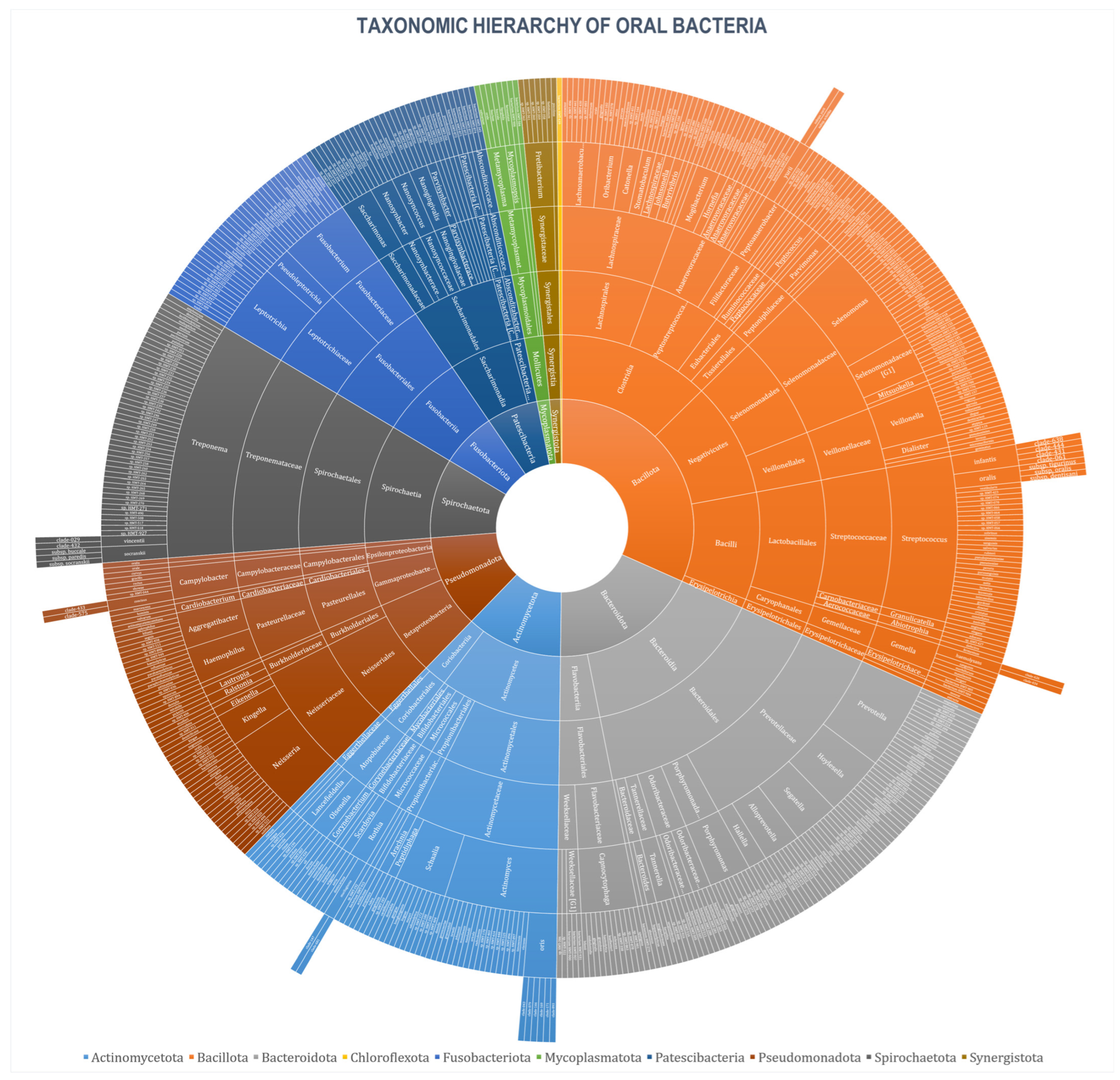

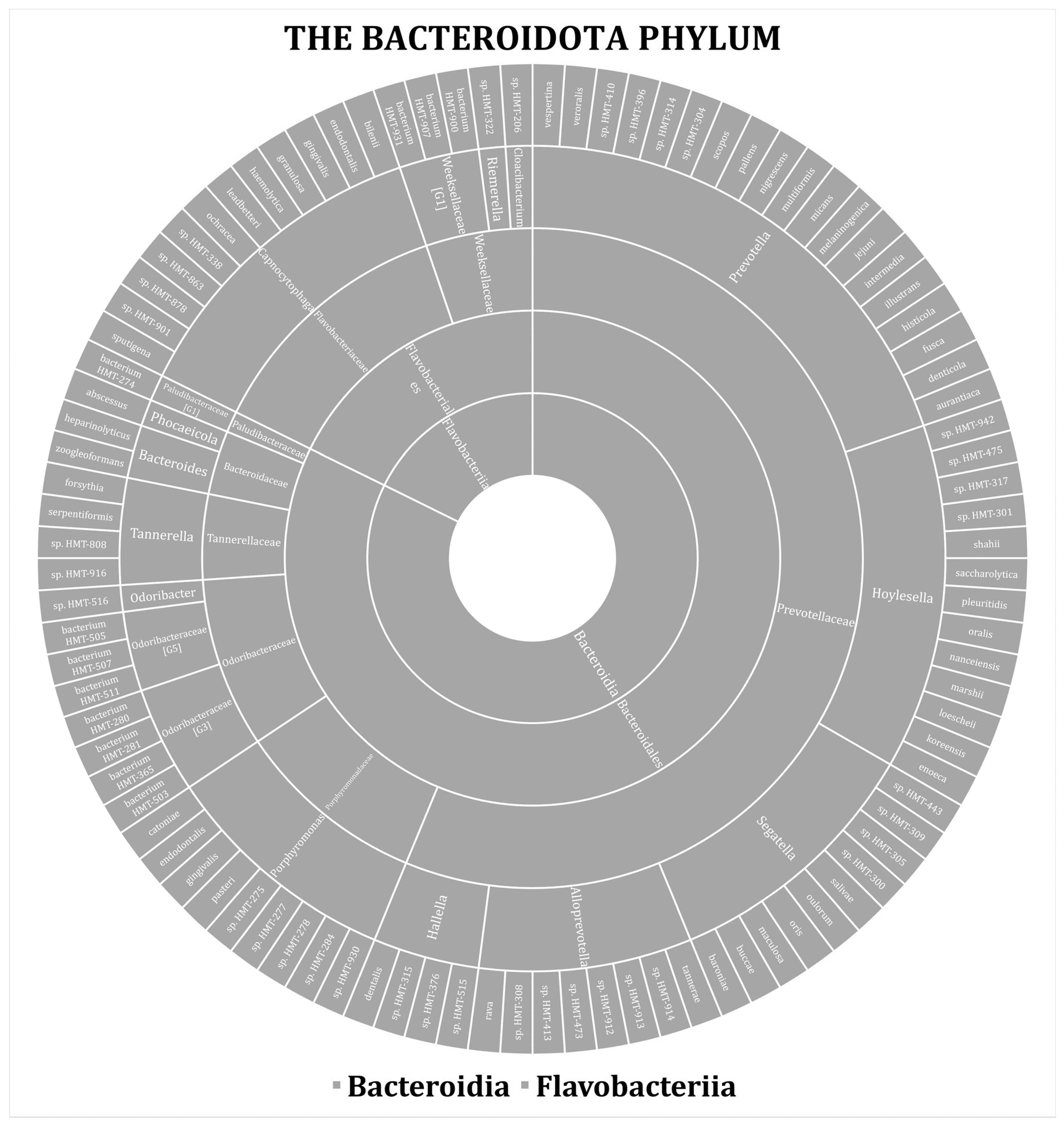
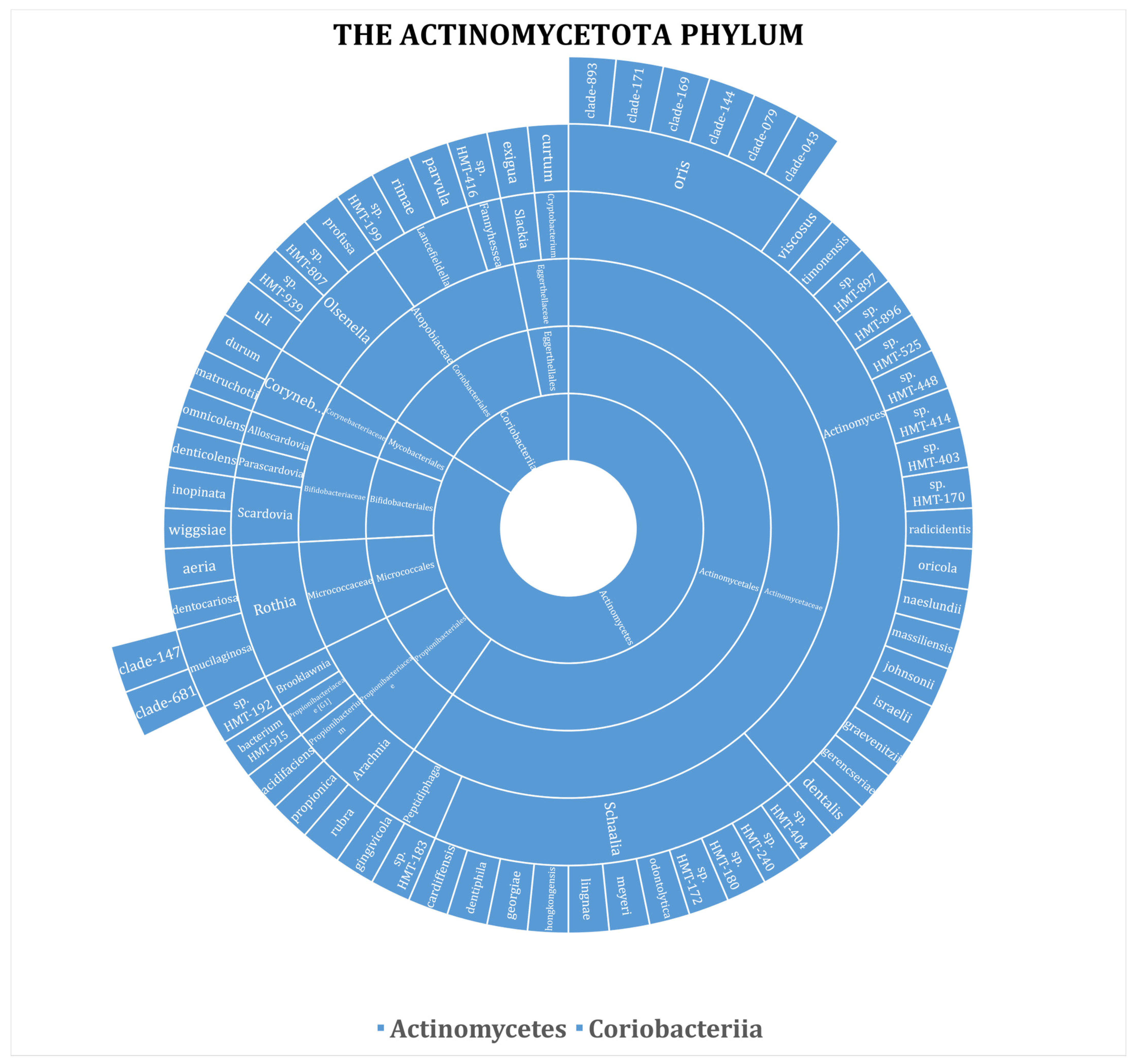

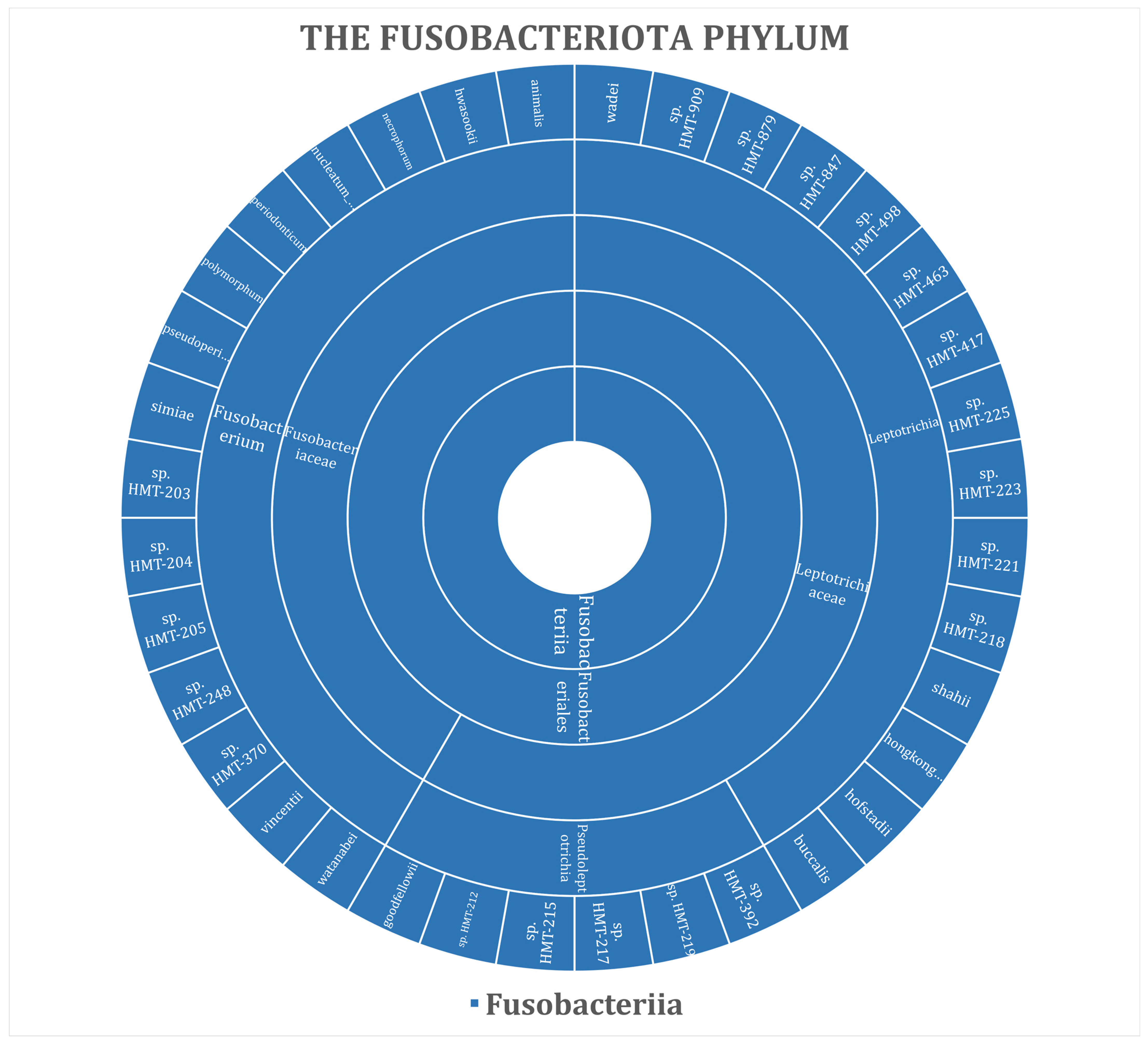
| Phylum | No. (%) | ||||||
|---|---|---|---|---|---|---|---|
| Taxa 1 | Status (Name and Culture) | Abundance 3 | |||||
| Named 2 | Unnamed Cultivated | Uncultivated Phylotypes | High | Medium | Low | ||
| Bacillota | 166 (31.80%) | 85 (51.20%) | 36 (21.69%) | 40 (24.10%) | 24 (14.46%) | 55 (33.13%) | 37 (22.29%) |
| Bacteroidota | 96 (18.39%) | 49 (51.04%) | 14 (14.58%) | 32 (33.33%) | 19 (19.79%) | 41 (42.71%) | 15 (15.63%) |
| Actinomycetota | 62 (11.88%) | 41 (66.13%) | 14 (22.58%) | 4 (6.45%) | 18 (29.03%) | 17 (27.42%) | 9 (14.52%) |
| Pseudomonadota | 61 (11.69%) | 45 (73.77%) | 6 (9.84%) | 10 (16.39%) | 19 (31.15%) | 23 (37.70%) | 5 (8.20%) |
| Spirochaetota | 52 (9.96%) | 13 (25%) | 4 (7.69%) | 35 (67.31%) | - | 14 (26.92%) | 18 (34.62%) |
| Fusobacteriota | 36 (6.90%) | 15 (41.67%) | 13 (36.11%) | 7 (19.44%) | 7 (19.44%) | 17 (47.22%) | 6 (16.67%) |
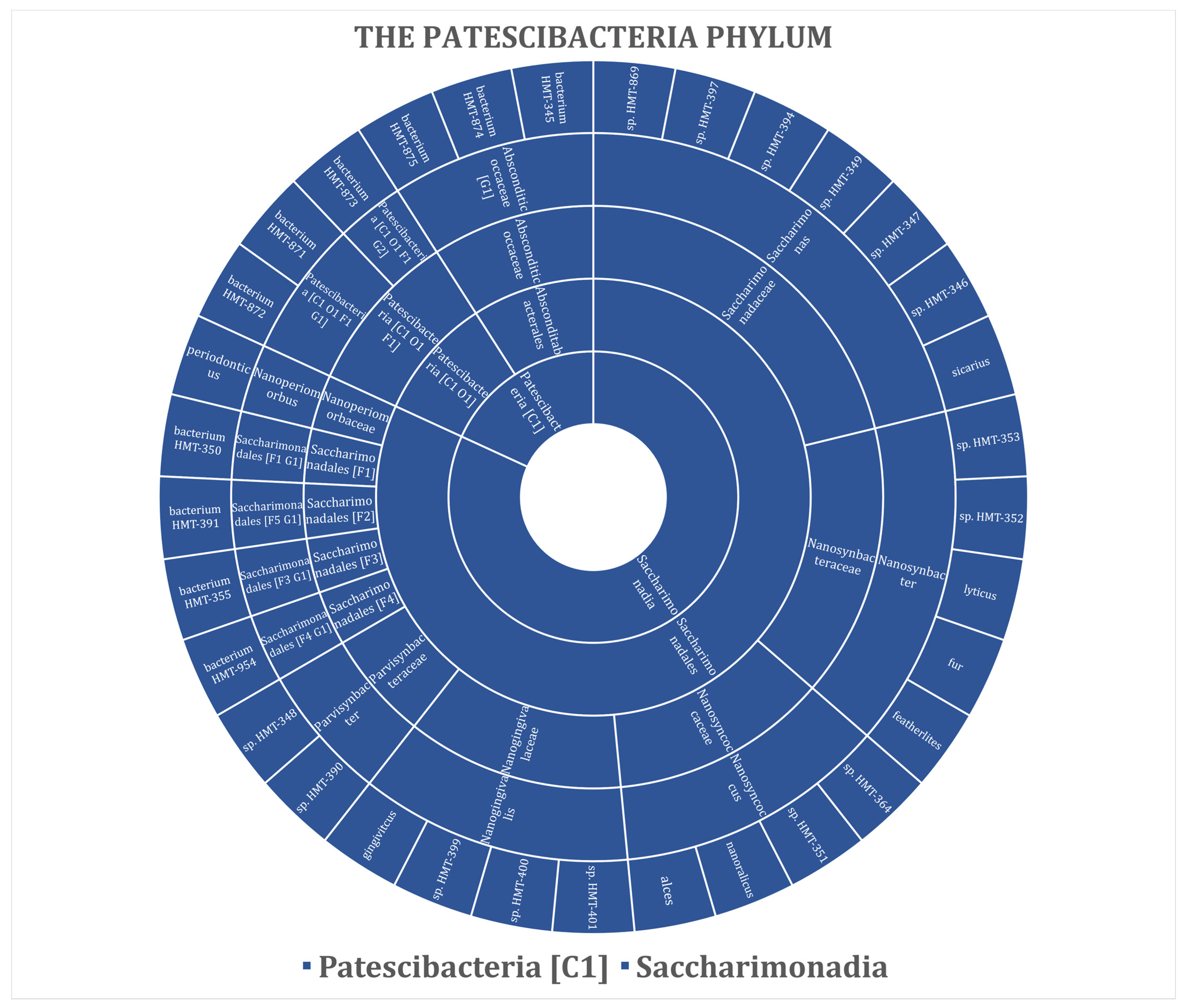
| Patescibacteria | 33 (6.32%) | - | 12 (36.36%) | 17 (51.52%) | 3 (9.09%) | 13 (39.39%) | 4 (12.12%) |
| Mycoplasmotota | 8 (1.53%) | 6 (75%) | - | 2 (25%) | - | 1 (12.50%) | 1 (12.50%) |
| Synergistota | 7 (1.34%) | 2 (28.57%) | - | 5 (71.43%) | - | 1 (14.29%) | 4 (57.14%) |
| Chloreflexota | 1 (0.19%) | - | 1 (100%) | - | - | - | 1 (100%) |
| Total | 522 (100%) | 256 (49.04%) | 100 (19.16%) | 152 (29.12%) | 90 (17.24%) | 182 (34.87%) | 99 (18.97%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghaouas, S.; Chala, S. The Oral Bacteriome. Microbiol. Res. 2025, 16, 194. https://doi.org/10.3390/microbiolres16090194
Ghaouas S, Chala S. The Oral Bacteriome. Microbiology Research. 2025; 16(9):194. https://doi.org/10.3390/microbiolres16090194
Chicago/Turabian StyleGhaouas, Soukaina, and Sanaa Chala. 2025. "The Oral Bacteriome" Microbiology Research 16, no. 9: 194. https://doi.org/10.3390/microbiolres16090194
APA StyleGhaouas, S., & Chala, S. (2025). The Oral Bacteriome. Microbiology Research, 16(9), 194. https://doi.org/10.3390/microbiolres16090194






