Comprehensive Analysis of the Molecular Epidemiological Characteristics of Duck-Derived Salmonella in Certain Regions of China
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Isolation and Serotype Identification
2.2. Antimicrobial Susceptibility Testing
2.3. Determination of Biofilm Formation Ability
2.4. Detection of Drug Resistance Genes
2.5. Virulence Gene Detection
2.6. Pulsed-Field Gel Electrophoresis (PFGE)
2.7. Data Analysis and Visualization
3. Results
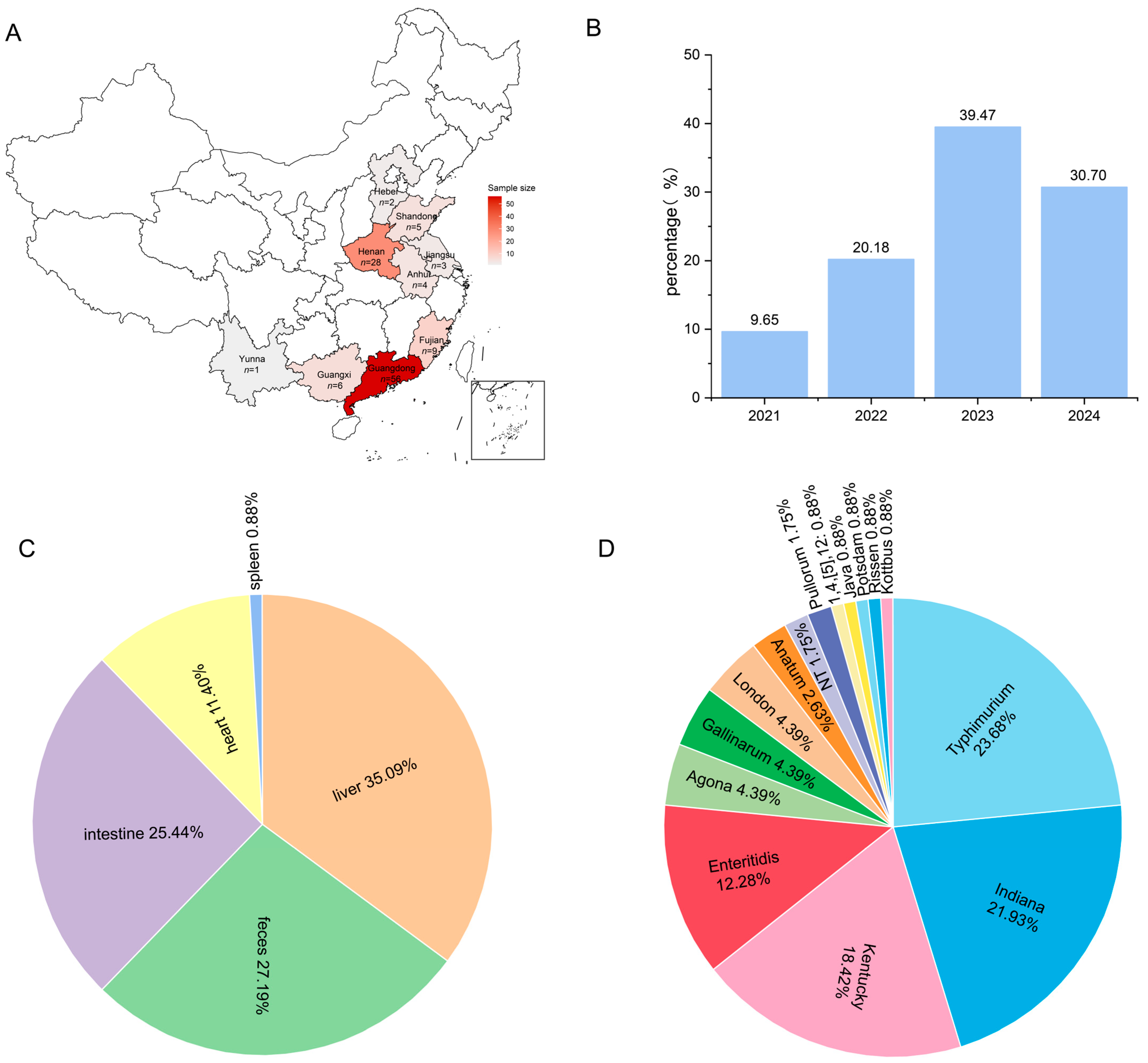
3.1. Epidemiological Characteristics of Duck-Derived Salmonella
3.2. Biofilm Formation Ability
3.3. Analysis of Drug Resistance
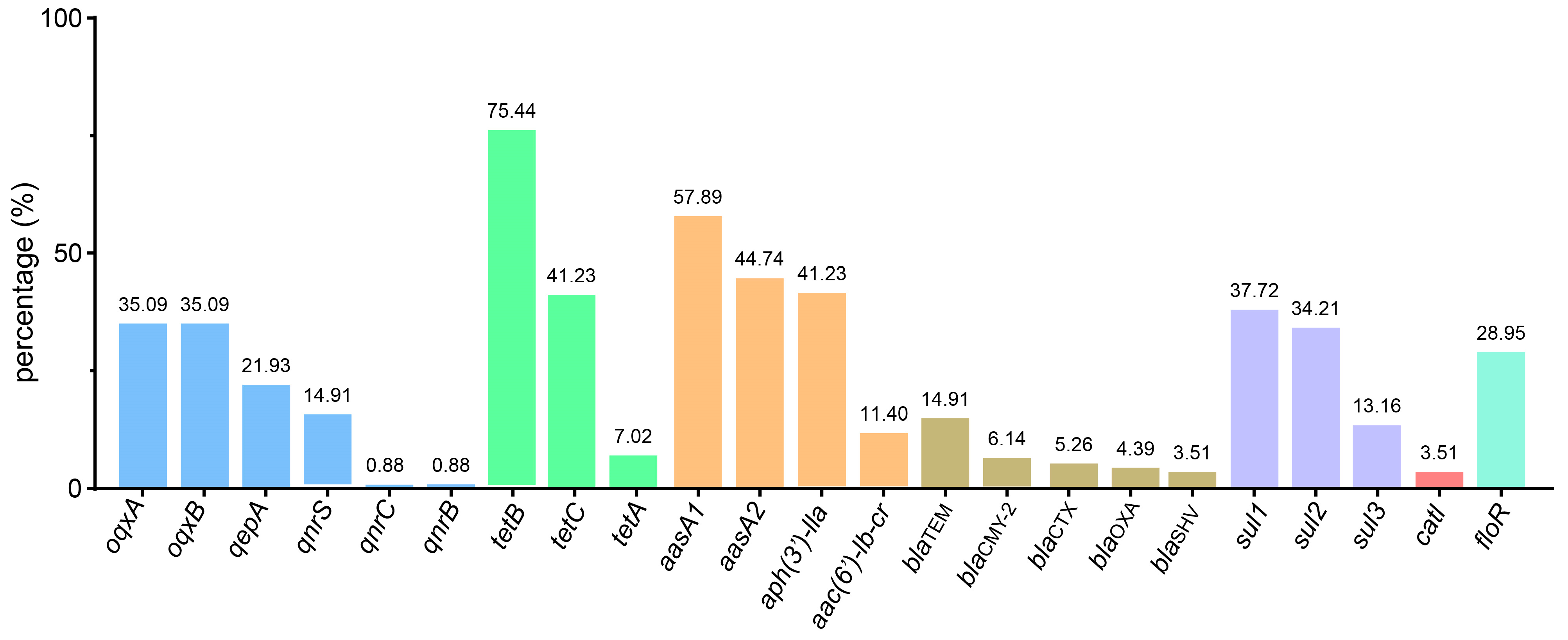
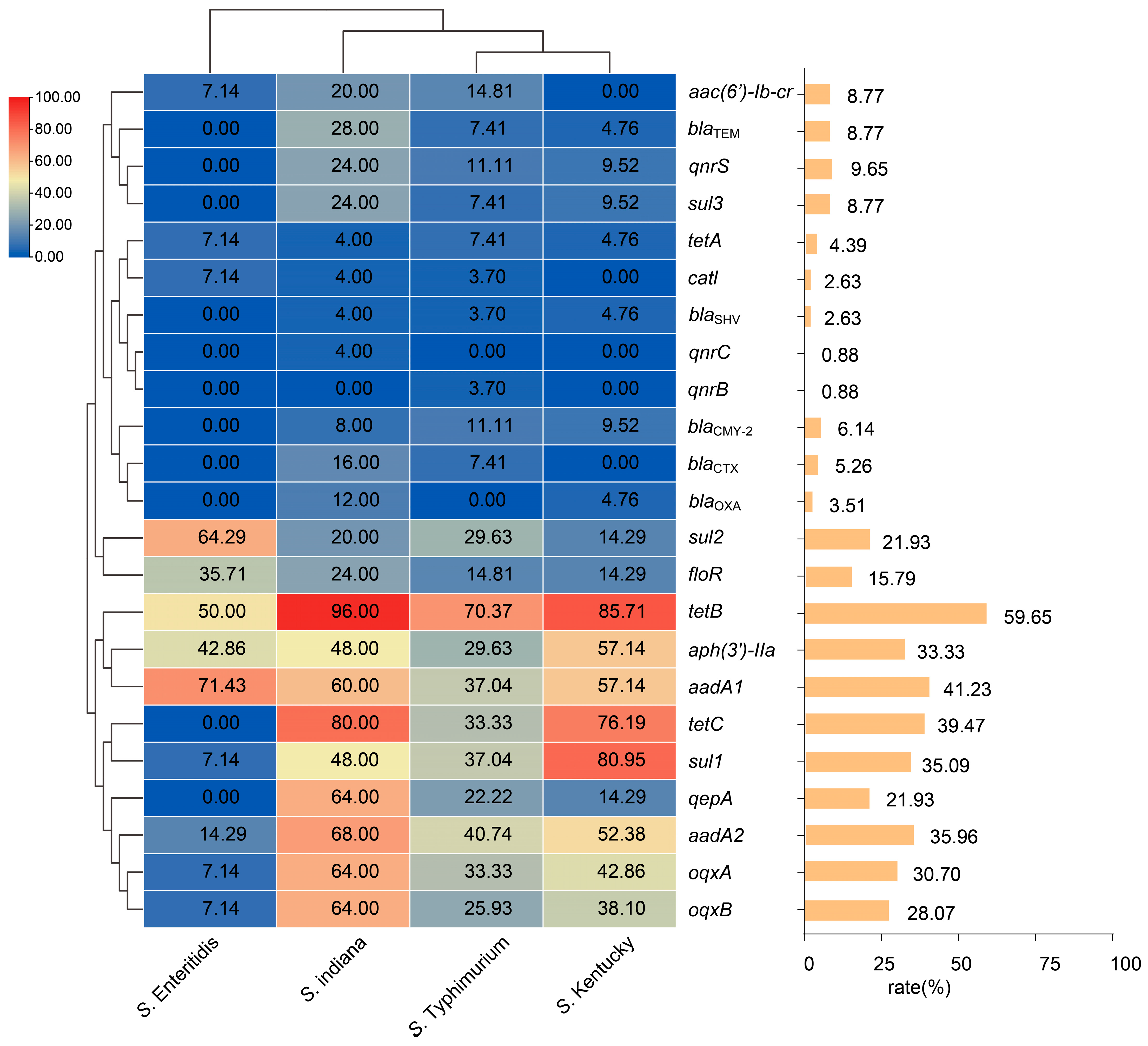
3.4. Antimicrobial Resistance Genes Profile
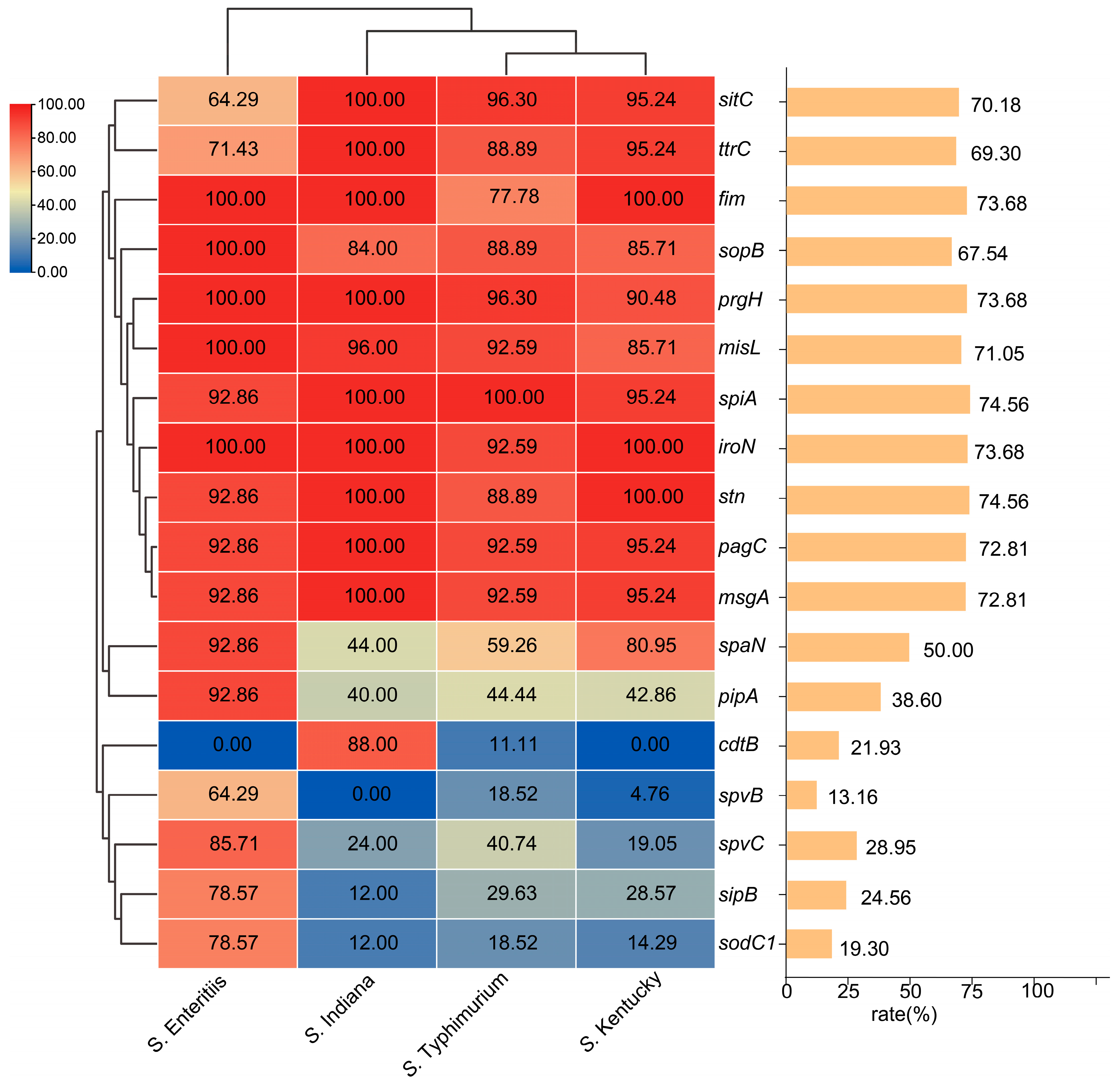
3.5. Virulence Gene Profile
3.6. PFGE Molecular Typing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bae, D.H.; Dessie, H.K.; Baek, H.J.; Kim, S.G.; Lee, H.S.; Lee, Y.J. Prevalence and Characteristics of Salmonella spp. Isolated from Poultry Slaughterhouses in Korea. J. Vet. Med. Sci. 2013, 75, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Jesudason, T. WHO Publishes Updated List of Bacterial Priority Pathogens. Lancet Microbe 2024, 5, 100940. [Google Scholar] [CrossRef]
- Threlfall, E.J. Antimicrobial Drug Resistance in Salmonella: Problems and Perspectives in Food- and Water-Borne Infections. FEMS Microbiol. Rev. 2002, 26, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Anjum, M.F.; Duggett, N.A.; AbuOun, M.; Randall, L.; Nunez-Garcia, J.; Ellis, R.J.; Rogers, J.; Horton, R.; Brena, C.; Williamson, S.; et al. Colistin Resistance in Salmonella and Escherichia Coli Isolates from a Pig Farm in Great Britain. J. Antimicrob. Chemother. 2016, 71, 2306–2313. [Google Scholar] [CrossRef]
- Kang, X.; Wang, M.; Meng, C.; Li, A.; Jiao, X.; Pan, Z. Prevalence and Whole-Genome Sequencing Analysis of Salmonella Reveal Its Spread along the Duck Production Chain. Poult. Sci. 2022, 101, 101993. [Google Scholar] [CrossRef]
- Chen, Z.; Bai, J.; Wang, S.; Zhang, X.; Zhan, Z.; Shen, H.; Zhang, H.; Wen, J.; Gao, Y.; Liao, M.; et al. Prevalence, Antimicrobial Resistance, Virulence Genes and Genetic Diversity of Salmonella Isolated from Retail Duck Meat in Southern China. Microorganisms 2020, 8, 444. [Google Scholar] [CrossRef]
- Patil, S.S.; Shinduja, R.; Suresh, K.P.; Phukan, S.; Kumar, S.; Sengupta, P.P.; Amachawadi, R.G.; Raut, A.; Roy, P.; Syed, A.; et al. A Systematic Review and Meta-Analysis on the Prevalence of Infectious Diseases of Duck: A World Perspective. Saudi J. Biol. Sci. 2021, 28, 5131–5144. [Google Scholar] [CrossRef]
- Yang, J.; Ju, Z.; Yang, Y.; Zhao, X.; Jiang, Z.; Sun, S. Serotype, Antimicrobial Susceptibility and Genotype Profiles of Salmonella Isolated from Duck Farms and a Slaughterhouse in Shandong Province, China. BMC Microbiol. 2019, 19, 202. [Google Scholar] [CrossRef]
- Tang, B.; Elbediwi, M.; Nambiar, R.B.; Yang, H.; Lin, J.; Yue, M. Genomic Characterization of Antimicrobial-Resistant Salmonella Enterica in Duck, Chicken, and Pig Farms and Retail Markets in Eastern China. Microbiol. Spectr. 2022, 10, e01257-22. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Q.; Zhang, C.; Yang, J.; Lu, Z.; Lu, F.; Bie, X. Characterization of a Broad Host-Spectrum Virulent Salmonella Bacteriophage Fmb-P1 and Its Application on Duck Meat. Virus Res. 2017, 236, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Vinueza-Burgos, C.; Baquero, M.; Medina, J.; De Zutter, L. Occurrence, Genotypes and Antimicrobial Susceptibility of Salmonella Collected from the Broiler Production Chain within an Integrated Poultry Company. Int. J. Food Microbiol. 2019, 299, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef]
- Zhao, Q.; Jiang, Z.; Li, T.; Cheng, M.; Sun, H.; Cui, M.; Zhang, C.; Xu, S.; Wang, H.; Wu, C. Current Status and Trends in Antimicrobial Use in Food Animals in China, 2018–2020. One Health Adv. 2023, 1, 29. [Google Scholar] [CrossRef]
- Chu, Y.; Wang, D.; Hao, W.; Sun, R.; Sun, J.; Liu, Y.; Liao, X. Prevalence, Antibiotic Resistance, Virulence Genes and Molecular Characteristics of Salmonella Isolated from Ducks and Wild Geese in China. Food Microbiol. 2024, 118, 104423. [Google Scholar] [CrossRef]
- Robicsek, A.; Jacoby, G.A.; Hooper, D.C. The Worldwide Emergence of Plasmid-Mediated Quinolone Resistance. Lancet Infect. Dis. 2006, 6, 629–640. [Google Scholar] [CrossRef]
- Correia, S.; Poeta, P.; Hébraud, M.; Capelo, J.L.; Igrejas, G. Mechanisms of Quinolone Action and Resistance: Where Do We Stand? J. Med. Microbiol. 2017, 66, 551–559. [Google Scholar] [CrossRef]
- Cuypers, W.L.; Jacobs, J.; Wong, V.; Klemm, E.J.; Deborggraeve, S.; Van Puyvelde, S. Fluoroquinolone Resistance in Salmonella: Insights by Whole-Genome Sequencing. Microb. Genom. 2018, 4, e000195. [Google Scholar] [CrossRef]
- Xia, L.; Wang, J.; Chen, M.; Li, G.; Wang, W.; An, T. Biofilm Formation Mechanisms of Mixed Antibiotic-Resistant Bacteria in Water: Bacterial Interactions and Horizontal Transfer of Antibiotic-Resistant Plasmids. J. Hazard. Mater. 2025, 481, 136554. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.N. Pathology of Infectious Diseases: New Agents, Opportunistic, Neglectable, Emergent, Reemergent Diseases and Why Not Super Resistant Nosocomial Bacteria? Autops. Case Rep. 2019, 9, e2019126. [Google Scholar] [CrossRef] [PubMed]
- Raufu, I.A.; Ahmed, O.A.; Aremu, A.; Ameh, J.A.; Timme, R.E.; Hendriksen, R.S.; Ambali, A.G. Occurrence, Antimicrobial Resistance and Whole Genome Sequence Analysis of Salmonella Serovars from Pig Farms in Ilorin, North-Central Nigeria. Int. J. Food Microbiol. 2021, 350, 109245. [Google Scholar] [CrossRef] [PubMed]
- Kong-Ngoen, T.; Santajit, S.; Tunyong, W.; Pumirat, P.; Sookrung, N.; Chaicumpa, W.; Indrawattana, N. Antimicrobial Resistance and Virulence of Non-Typhoidal Salmonella from Retail Foods Marketed in Bangkok, Thailand. Foods 2022, 11, 661. [Google Scholar] [CrossRef]
- Webber, B.; Borges, K.A.; Furian, T.Q.; Rizzo, N.N.; Tondo, E.C.; Santos, L.R.d.; Rodrigues, L.B.; Nascimento, V.P.d. Detection of Virulence Genes in Salmonella Heidelberg Isolated from Chicken Carcasses. Rev. Inst. Med. Trop. Sao Paulo 2019, 61, e36. [Google Scholar] [CrossRef]
- Khajanchi, B.K.; Foley, S.L. Antimicrobial Resistance and Increased Virulence of Salmonella. Microorganisms 2022, 10, 1829. [Google Scholar] [CrossRef]
- Assaf, A.; Grangé, E.; Cordella, C.B.Y.; Rutledge, D.N.; Lees, M.; Lahmar, A.; Thouand, G. Evaluation of the Impact of Buffered Peptone Water Composition on the Discrimination between Salmonella Enterica and Escherichia Coli by Raman Spectroscopy. Anal. Bioanal. Chem. 2020, 412, 3595–3604. [Google Scholar] [CrossRef]
- Lewis, J.S. Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2024; ISBN 9781684402205. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A Modified Microtiter-Plate Test for Quantification of Staphylococcal Biofilm Formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhou, R.; Kang, M.; Luo, R.; Cai, X.; Chen, H. Biofilm Formation by Field Isolates and Reference Strains of Haemophilus Parasuis. Vet. Microbiol. 2006, 118, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xiao, T.; Wu, L.; Li, Y.; Duan, X.; Liu, W.; Liu, K.; Jin, W.; Ren, H.; Sun, J.; et al. Comprehensive Profiling of Serotypes, Antimicrobial Resistance and Virulence of Salmonella Isolates from Food Animals in China, 2015–2021. Front. Microbiol. 2023, 14, 1133241. [Google Scholar] [CrossRef]
- Fang, L.-X.; Deng, G.-H.; Jiang, Q.; Cen, D.-J.; Yang, R.-S.; Feng, Y.-Y.; Xia, J.; Sun, J.; Liu, Y.-H.; Zhang, Q.; et al. Clonal Expansion and Horizontal Transmission of Epidemic F2:A1:B1 Plasmids Involved in Co-Spread of RmtB with QepA and BlaCTX-M-27 in Extensively Drug-Resistant Salmonella Enterica Serovar Indiana Isolates. J. Antimicrob. Chemother. 2019, 74, 334–341. [Google Scholar] [CrossRef]
- Maravić, A.; Skočibušić, M.; Šamanić, I.; Fredotović, Ž.; Cvjetan, S.; Jutronić, M.; Puizina, J. Aeromonas Spp. Simultaneously Harbouring BlaCTX-M-15, BlaSHV-12, BlaPER-1 and BlaFOX-2, in Wild-Growing Mediterranean Mussel (Mytilus Galloprovincialis) from Adriatic Sea, Croatia. Int. J. Food Microbiol 2013, 166, 301–308. [Google Scholar] [CrossRef]
- Skyberg, J.A.; Logue, C.M.; Nolan, J.; Murali, G.; Chandran, P. Comparative Virulence Genotyping and Antimicrobial Susceptibility Profiling of Environmental and Clinical Salmonella Enterica from Cochin, India. Curr. Microbiol. 2011, 62, 21–26. [Google Scholar] [CrossRef]
- Sánchez-Jiménez, M.M.; Cardona-Castro, N.M.; Canu, N.; Uzzau, S.; Rubino, S. Distribution of Pathogenicity Islands among Colombian Isolates of Salmonella. J. Infect. Dev. Ctries. 2010, 4, 555–559. [Google Scholar] [CrossRef]
- Skyberg, J.A.; Logue, C.M.; Nolan, L.K. Virulence Genotyping of Salmonella Spp. with Multiplex PCR. Avian Dis. 2006, 50, 77–81. [Google Scholar] [CrossRef]
- Akinyemi, K.O.; Iwalokun, B.A.; Foli, F.; Oshodi, K.; Coker, A.O. Prevalence of Multiple Drug Resistance and Screening of Enterotoxin (Stn) Gene in Salmonella Enterica Serovars from Water Sources in Lagos, Nigeria. Public Health 2011, 125, 65–71. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- El Sabry, M.I.; Almasri, O. Global Waterfowl Production: Stocking Rate Is a Key Factor for Improving Productivity and Well-Being—A Review. Trop. Anim. Health Prod. 2023, 55, 419. [Google Scholar] [CrossRef]
- Akshay, S.D.; Deekshit, V.K.; Mohan Raj, J.; Maiti, B. Outer Membrane Proteins and Efflux Pumps Mediated Multi-Drug Resistance in Salmonella: Rising Threat to Antimicrobial Therapy. ACS Infect. Dis. 2023, 9, 2072–2092. [Google Scholar] [CrossRef]
- Aleksandrowicz, A.; Carolak, E.; Dutkiewicz, A.; Błachut, A.; Waszczuk, W.; Grzymajlo, K. Better Together- Salmonella Biofilm-Associated Antibiotic Resistance. Gut Microbes 2023, 15, 2229937. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.W.; Bell, R.; Zhang, G.; Timme, R.; Zheng, J.; Hammack, T.S.; Allard, M.W. Salmonella Genomics in Public Health and Food Safety. EcoSal Plus 2021, 9, eESP-0008. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Liu, P.; Li, Y.; Cui, M.; Zhang, C.; Wang, Y.; Shen, Z.; Shen, J.; Ke, Y.; Wang, S.; et al. Prevalence of Salmonella and Antimicrobial Resistance in Isolates from Food Animals-Six PLADs, China, 2019. China CDC Wkly. 2021, 3, 514–517. [Google Scholar] [CrossRef] [PubMed]
- Denis, M.; Houard, E.; Fablet, A.; Rouxel, S.; Salvat, G. Distribution of Serotypes and Genotypes of Salmonella Enterica Species in French Pig Production. Vet. Rec. 2013, 173, 370. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Q.; Cao, C.; Cui, S.; Wu, Y.; Yang, H.; Xiao, Y.; Yang, B. Prevalence and Characteristics of Salmonella Isolates Recovered from Retail Raw Chickens in Shaanxi Province, China. Poult. Sci. 2020, 99, 6031–6044. [Google Scholar] [CrossRef]
- Zhao, X.; Gao, Y.; Ye, C.; Yang, L.; Wang, T.; Chang, W. Prevalence and Characteristics of Salmonella Isolated from Free-Range Chickens in Shandong Province, China. Biomed. Res. Int. 2016, 2016, 8183931. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, W.; Zhang, K.; Zhang, Y.; Wang, Z.; Zhang, W.; Li, Y.; Li, Q. Characterization of Salmonella Serotypes Prevalent in Asymptomatic People and Patients. BMC Infect. Dis. 2021, 21, 632. [Google Scholar] [CrossRef]
- Wang, M.-G.; Zhang, R.-M.; Wang, L.-L.; Sun, R.-Y.; Bai, S.-C.; Han, L.; Fang, L.-X.; Sun, J.; Liu, Y.-H.; Liao, X.-P. Molecular Epidemiology of Carbapenemase-Producing Escherichia Coli from Duck Farms in South-East Coastal China. J. Antimicrob. Chemother. 2021, 76, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Binter, C.; Straver, J.M.; Häggblom, P.; Bruggeman, G.; Lindqvist, P.-A.; Zentek, J.; Andersson, M.G. Transmission and Control of Salmonella in the Pig Feed Chain: A Conceptual Model. Int. J. Food Microbiol. 2011, 145, S7–S17. [Google Scholar] [CrossRef]
- Ivanova, M.; Ovsepian, A.; Leekitcharoenphon, P.; Seyfarth, A.M.; Mordhorst, H.; Otani, S.; Koeberl-Jelovcan, S.; Milanov, M.; Kompes, G.; Liapi, M.; et al. Azithromycin Resistance in Escherichia Coli and Salmonella from Food-Producing Animals and Meat in Europe. J. Antimicrob. Chemother. 2024, 79, 1657–1667. [Google Scholar] [CrossRef]
- Zhao, X.; Li, W.; Hou, S.; Wang, Y.; Wang, S.; Gao, J.; Zhang, R.; Jiang, S.; Zhu, Y. Epidemiological Investigation on Drug Resistance of Salmonella Isolates from Duck Breeding Farms in Shandong Province and Surrounding Areas, China. Poult. Sci. 2022, 101, 101961. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Le, H.V.; Vu Thi Hai, H.; Nguyen Tuan, T.; Nguyen, H.M.; Pham Xuan, D.; Tran Thi Thanh, H.; Le Thi, H.H. Whole-Genome Analysis of Antimicrobial-Resistant Salmonella Enterica Isolated from Duck Carcasses in Hanoi, Vietnam. Curr. Issues Mol. Biol. 2023, 45, 2213–2229. [Google Scholar] [CrossRef]
- Ali, R.; Sadat, A.; Younis, G. Salmonella Species Threats in Duck Meat in Egypt: Prevalence and Correlation Between Antimicrobial Resistance and Biofilm Production. Egypt. J. Vet. Sci. 2023, 54, 1131–1142. [Google Scholar] [CrossRef]
- Andrew Selaledi, L.; Mohammed Hassan, Z.; Manyelo, T.G.; Mabelebele, M. The Current Status of the Alternative Use to Antibiotics in Poultry Production: An African Perspective. Antibiotics 2020, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, Y.; Feng, D.; Jiang, Q.; Li, S.; Rong, J.; Zhong, L.; Methner, U.; Baxter, L.; Ott, S.; et al. Centralized Industrialization of Pork in Europe and America Contributes to the Global Spread of Salmonella Enterica. Nat. Food 2024, 5, 413–422. [Google Scholar] [CrossRef]
- Peng, J.; Xiao, R.; Wu, C.; Zheng, Z.; Deng, Y.; Chen, K.; Xiang, Y.; Xu, C.; Zou, L.; Liao, M.; et al. Characterization of the Prevalence of Salmonella in Different Retail Chicken Supply Modes Using Genome-Wide and Machine-Learning Analyses. Food Res. Int. 2024, 191, 114654. [Google Scholar] [CrossRef]
- Ingle, D.J.; Ambrose, R.L.; Baines, S.L.; Duchene, S.; Gonçalves da Silva, A.; Lee, D.Y.J.; Jones, M.; Valcanis, M.; Taiaroa, G.; Ballard, S.A.; et al. Evolutionary Dynamics of Multidrug Resistant Salmonella Enterica Serovar 4,[5],12:I:- In Australia. Nat. Commun. 2021, 12, 4786. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Machuca, J.; Cano, M.E.; Calvo, J.; Martínez-Martínez, L.; Pascual, A. Plasmid-Mediated Quinolone Resistance: Two Decades On. Drug Resist. Updates 2016, 29, 13–29. [Google Scholar] [CrossRef]
- Abdelrahim, S.S.; Hassuna, N.A.; Waly, N.G.F.M.; Kotb, D.N.; Abdelhamid, H.; Zaki, S. Coexistence of Plasmid-Mediated Quinolone Resistance (PMQR) and Extended-Spectrum Beta-Lactamase (ESBL) Genes among Clinical Pseudomonas Aeruginosa Isolates in Egypt. BMC Microbiol. 2024, 24, 175. [Google Scholar] [CrossRef]
- Haider, M.H.; Ain, N.U.; Abrar, S.; Riaz, S. BlaOXA, BlaSHV-, and BlaTEM- Extended-Spectrum β-Lactamases in Gram-Negative Strains from Burn Patients in Lahore, Pakistan. J. Infect. Dev. Ctries. 2020, 14, 1410–1417. [Google Scholar] [CrossRef]
- Ejaz, H.; Younas, S.; Abosalif, K.O.A.; Junaid, K.; Alzahrani, B.; Alsrhani, A.; Abdalla, A.E.; Ullah, M.I.; Qamar, M.U.; Hamam, S.S.M. Molecular Analysis of BlaSHV, BlaTEM, and BlaCTX-M in Extended-Spectrum β-Lactamase Producing Enterobacteriaceae Recovered from Fecal Specimens of Animals. PLoS ONE 2021, 16, e0245126. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Distribution of the BlaTEM Gene and BlaTEM-Containing Transposons in Commensal Escherichia Coli. J. Antimicrob. Chemother. 2011, 66, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.-F. Molecular Mechanisms of Biofilm-Based Antibiotic Resistance and Tolerance in Pathogenic Bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between Resistance, Tolerance and Persistence to Antibiotic Treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Zhang, L.; Mah, T.-F. Involvement of a Novel Efflux System in Biofilm-Specific Resistance to Antibiotics. J. Bacteriol. 2008, 190, 4447–4452. [Google Scholar] [CrossRef] [PubMed]
- Savage, V.J.; Chopra, I.; O’Neill, A.J. Staphylococcus Aureus Biofilms Promote Horizontal Transfer of Antibiotic Resistance. Antimicrob. Agents Chemother. 2013, 57, 1968–1970. [Google Scholar] [CrossRef] [PubMed]
- Suci, P.A.; Mittelman, M.W.; Yu, F.P.; Geesey, G.G. Investigation of Ciprofloxacin Penetration into Pseudomonas Aeruginosa Biofilms. Antimicrob. Agents Chemother. 1994, 38, 2125–2133. [Google Scholar] [CrossRef]
- Oliveira, I.M.; Gomes, I.B.; Simões, L.C.; Simões, M. A Review of Research Advances on Disinfection Strategies for Biofilm Control in Drinking Water Distribution Systems. Water Res. 2024, 253, 121273. [Google Scholar] [CrossRef]
- Gill, S.P.; Hunter, W.R.; Coulson, L.E.; Banat, I.M.; Schelker, J. Synthetic and Biological Surfactant Effects on Freshwater Biofilm Community Composition and Metabolic Activity. Appl. Microbiol. Biotechnol. 2022, 106, 6847–6859. [Google Scholar] [CrossRef]
- Petrin, S.; Mancin, M.; Losasso, C.; Deotto, S.; Olsen, J.E.; Barco, L. Effect of PH and Salinity on the Ability of Salmonella Serotypes to Form Biofilm. Front. Microbiol. 2022, 13, 821679. [Google Scholar] [CrossRef]
- Counihan, K.L.; Tilman, S.; Uknalis, J.; Mukhopadhyay, S.; Niemira, B.A.; Bermudez-Aguirre, D. Attachment and Biofilm Formation of Eight Different Salmonella Serotypes on Three Food-Contact Surfaces at Different Temperatures. Microorganisms 2025, 13, 1446. [Google Scholar] [CrossRef]
- Contreras-Soto, M.B.; Medrano-Félix, J.A.; Sañudo-Barajas, J.A.; Vélez-de la Rocha, R.; Ibarra-Rodríguez, J.R.; Martínez-Urtaza, J.; Chaidez, C.; Castro-del Campo, N. Structural Variations on Salmonella Biofilm by Exposition to River Water. Int. J. Environ. Health Res. 2022, 32, 1626–1643. [Google Scholar] [CrossRef]
- Pang, X.; Hu, X.; Du, X.; Lv, C.; Yuk, H.-G. Biofilm Formation in Food Processing Plants and Novel Control Strategies to Combat Resistant Biofilms: The Case of Salmonella Spp. Food Sci. Biotechnol. 2023, 32, 1703–1718. [Google Scholar] [CrossRef]
- Obe, T.; Nannapaneni, R.; Schilling, W.; Zhang, L.; Kiess, A. Antimicrobial Tolerance, Biofilm Formation, and Molecular Characterization of Salmonella Isolates from Poultry Processing Equipment. J. Appl. Poult. Res. 2021, 30, 100195. [Google Scholar] [CrossRef]
- González, J.F.; Alberts, H.; Lee, J.; Doolittle, L.; Gunn, J.S. Biofilm Formation Protects Salmonella from the Antibiotic Ciprofloxacin In Vitro and In Vivo in the Mouse Model of Chronic Carriage. Sci. Rep. 2018, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.E.; Laing, G.; McMahon, B.J.; Fanning, S.; Stekel, D.J.; Pahl, O.; Coyne, L.; Latham, S.M.; McIntyre, K.M. The Need for One Health Systems-Thinking Approaches to Understand Multiscale Dissemination of Antimicrobial Resistance. Lancet Planet. Health 2024, 8, e124–e133. [Google Scholar] [CrossRef] [PubMed]
- Trampari, E.; Holden, E.R.; Wickham, G.J.; Ravi, A.; Martins, L.d.O.; Savva, G.M.; Webber, M.A. Exposure of Salmonella Biofilms to Antibiotic Concentrations Rapidly Selects Resistance with Collateral Tradeoffs. NPJ Biofilms Microbiomes 2021, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Peng, D.; Jiao, X.; Zhang, X.; Geng, S.; Liu, X. Roles of the SpiA Gene from Salmonella Enteritidis in Biofilm Formation and Virulence. Microbiology 2011, 157, 1798–1805. [Google Scholar] [CrossRef]
- Takemura, M.; Haneda, T.; Idei, H.; Miki, T.; Okada, N. A Salmonella Type III Effector, PipA, Works in a Different Manner than the PipA Family Effectors GogA and GtgA. PLoS ONE 2021, 16, e0248975. [Google Scholar] [CrossRef]
- Zhou, X.; Kang, X.; Chen, J.; Song, Y.; Jia, C.; Teng, L.; Tang, Y.; Jiang, Z.; Peng, X.; Tao, X.; et al. Genome Degradation Promotes Salmonella Pathoadaptation by Remodeling Fimbriae-Mediated Proinflammatory Response. Natl. Sci. Rev. 2023, 10, nwad228. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, X.; Jin, J.; Tang, Z.; Xian, W.; Zhang, X.; Fu, J.; He, K.; Liu, X. The Salmonella Typhimurium Effector SpvB Subverts Host Membrane Trafficking by Targeting Clathrin and AP-1. Mol. Cell Proteom. 2023, 22, 100674. [Google Scholar] [CrossRef]
- Cook, P.; Tötemeyer, S.; Stevenson, C.; Fitzgerald, K.A.; Yamamoto, M.; Akira, S.; Maskell, D.J.; Bryant, C.E. Salmonella-Induced SipB-Independent Cell Death Requires Toll-like Receptor-4 Signalling via the Adapter Proteins Tram and Trif. Immunology 2007, 122, 222–229. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Zhu, B.; Lyu, N.; Liu, Y.; Ma, S.; Jia, S.; Wan, B.; Du, Y.; Zhang, G.; et al. Genomic Analysis of Almost 8,000 Salmonella Genomes Reveals Drivers and Landscape of Antimicrobial Resistance in China. Microbiol. Spectr. 2023, 11, e02080-23. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Li, X.; Liu, Y.; Rong, W.; Fu, L.; Wang, S.; Li, Y.; Duan, X.; Zhao, Y.; Guo, L. Comprehensive Analysis of the Molecular Epidemiological Characteristics of Duck-Derived Salmonella in Certain Regions of China. Microbiol. Res. 2025, 16, 184. https://doi.org/10.3390/microbiolres16080184
Chen J, Li X, Liu Y, Rong W, Fu L, Wang S, Li Y, Duan X, Zhao Y, Guo L. Comprehensive Analysis of the Molecular Epidemiological Characteristics of Duck-Derived Salmonella in Certain Regions of China. Microbiology Research. 2025; 16(8):184. https://doi.org/10.3390/microbiolres16080184
Chicago/Turabian StyleChen, Jiawen, Xiangdi Li, Yanling Liu, Wenjia Rong, Laiyu Fu, Shuhua Wang, Yan Li, Xiaoxiao Duan, Yongda Zhao, and Lili Guo. 2025. "Comprehensive Analysis of the Molecular Epidemiological Characteristics of Duck-Derived Salmonella in Certain Regions of China" Microbiology Research 16, no. 8: 184. https://doi.org/10.3390/microbiolres16080184
APA StyleChen, J., Li, X., Liu, Y., Rong, W., Fu, L., Wang, S., Li, Y., Duan, X., Zhao, Y., & Guo, L. (2025). Comprehensive Analysis of the Molecular Epidemiological Characteristics of Duck-Derived Salmonella in Certain Regions of China. Microbiology Research, 16(8), 184. https://doi.org/10.3390/microbiolres16080184



