Patterns of Antimicrobial Resistance Among Major Bacterial Pathogens Isolated from Clinical Samples in Bangladesh (2017–2020): A Nationwide Cross-Sectional Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
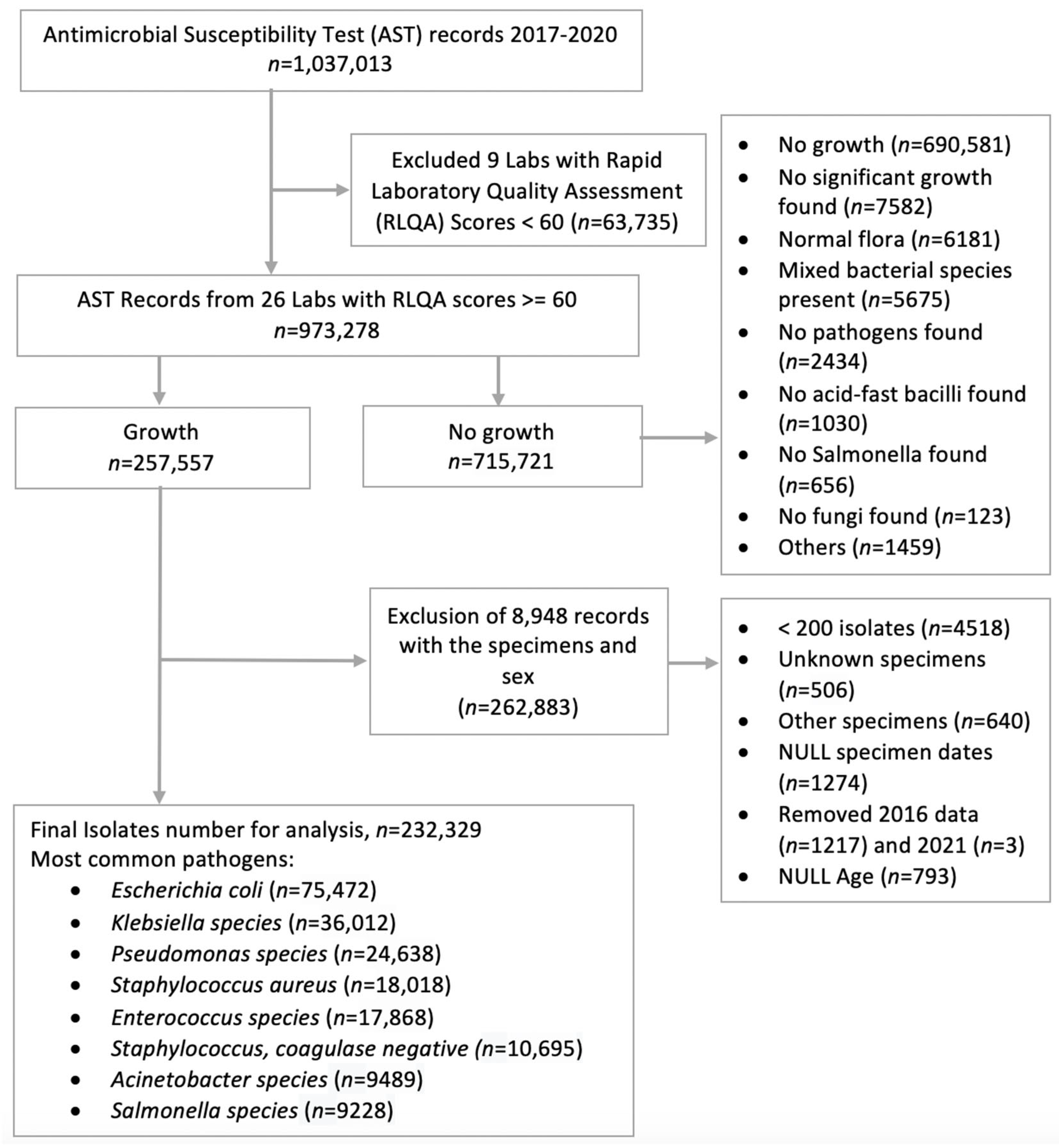
2.2. Inclusion, Exclusion Criteria
2.3. Data Collection
3. Results
3.1. Data Collection
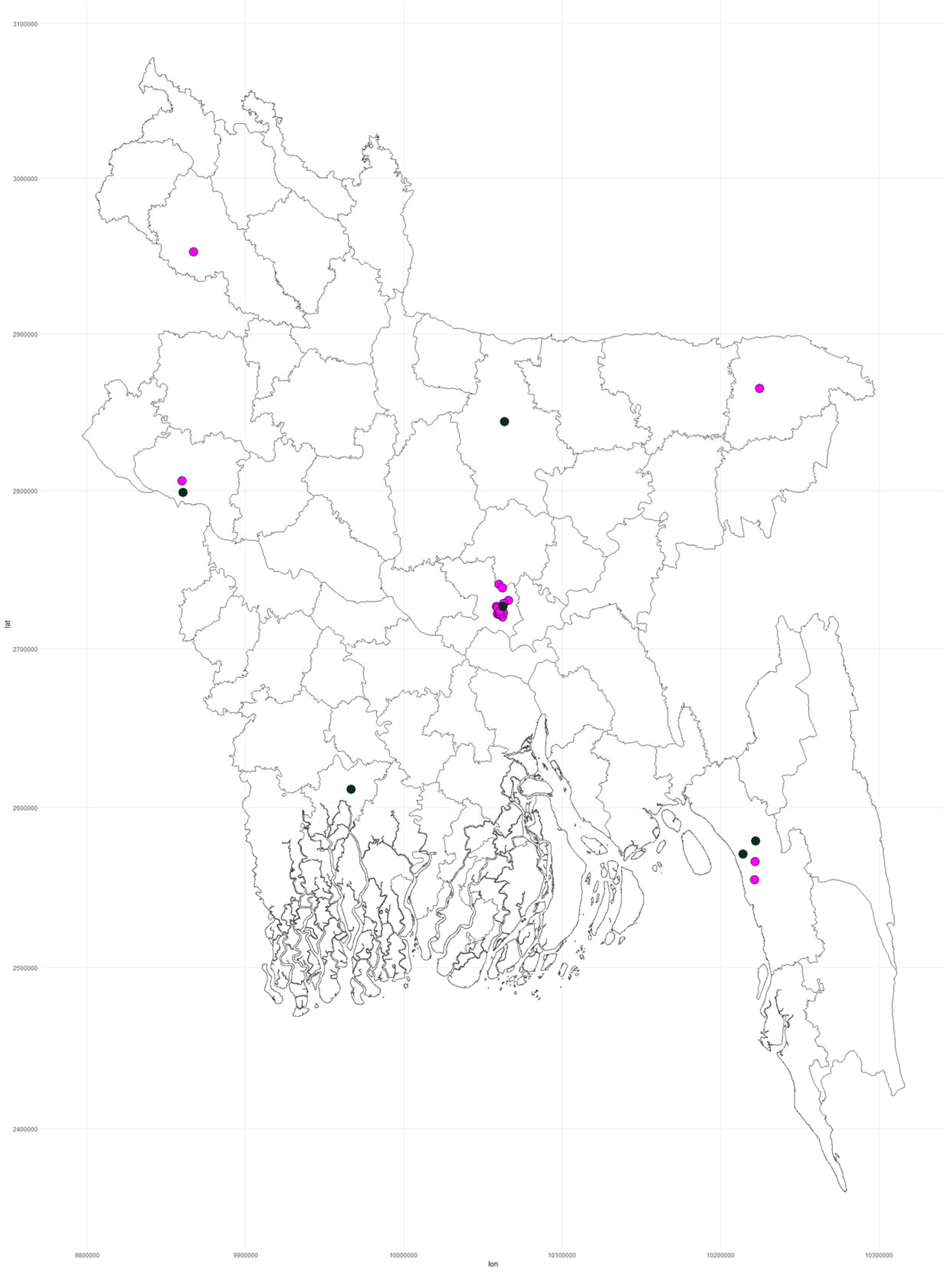
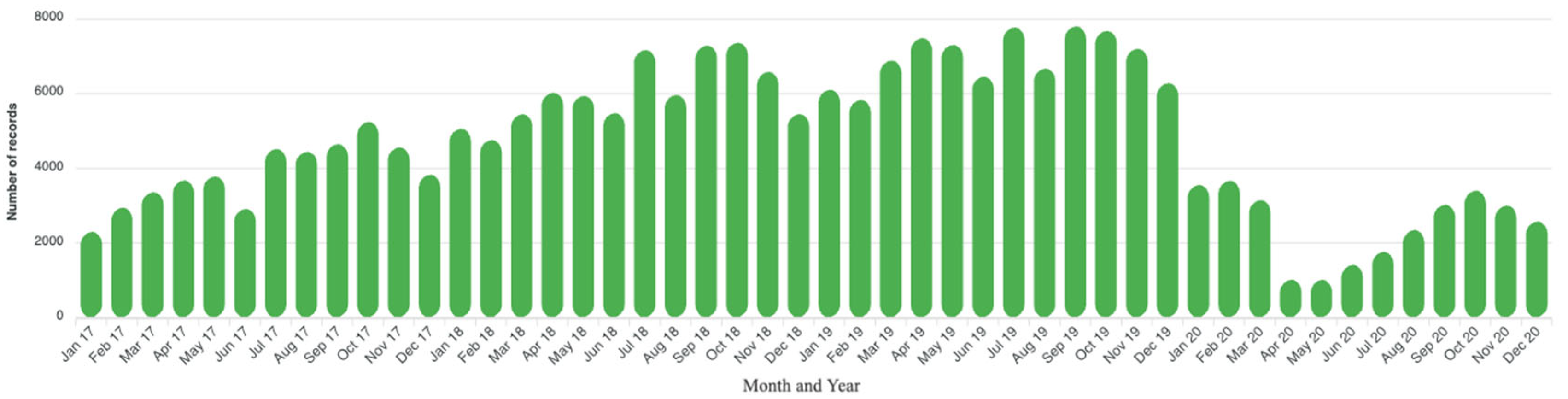
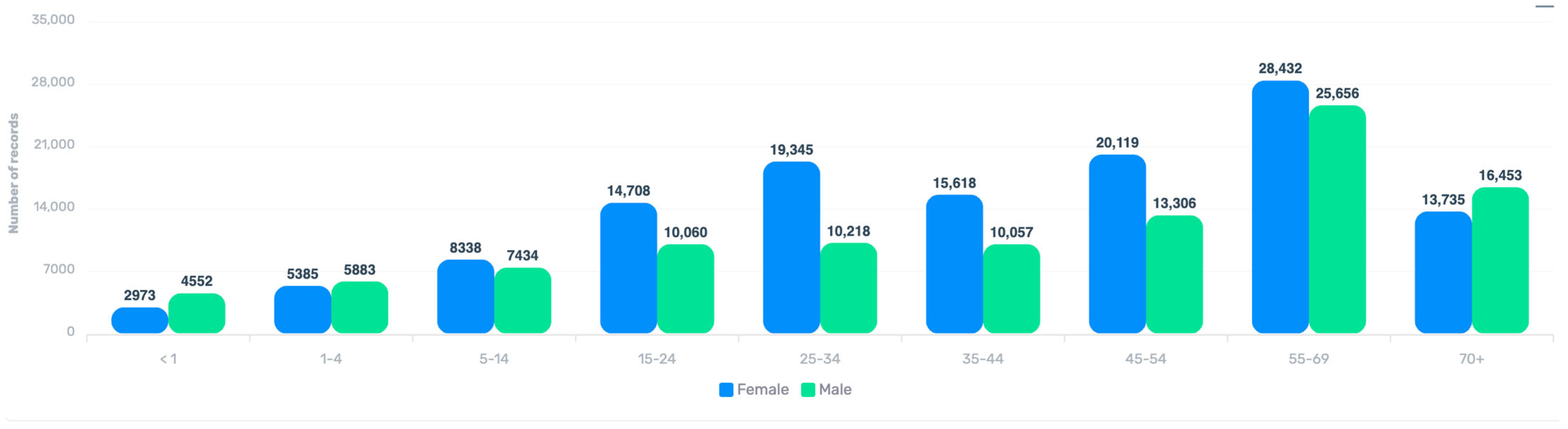
3.2. Geographical, Temporal, and Demographic Distribution
3.3. Most Commonly Found Microorganisms
3.4. Patterns of Antibiotic Resistance Among Common Gram-Negative Bacteria
3.5. The Pattern of Antibiotic Resistance Among Common Gram-Positive Pathogens
3.6. Susceptibility Patterns of Escherichia coli in Urine and Blood Specimens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAPTURA | Capturing Data on Antimicrobial Resistance Patterns and Trends in Use in Regions of Asia |
| IEDCR | Institute of Epidemiology, Disease Control and Research |
| CDC | Communicable Disease Control |
| MoHFW | Ministry of Health and Family Welfare |
| QAAPT | Quick Analysis of Antimicrobial Patterns and Trends |
| PHSG | Public Health Surveillance Group |
| BDI | Big Data Institute |
| AST | Antimicrobial Susceptibility Testing |
| EQA | External Quality Assurance |
| IQC | Internal Quality Control |
| CLSI | Clinical and Laboratory Standards Institute |
| EUCAST | European Committee for Antimicrobial Susceptibility Testing |
References
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; E Wool, E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C.; et al. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Institute for Health Metrics and Evaluation (IHME), University of Oxford. Global Bacterial Antimicrobial Resistance Burden Estimates 2019; Institute for Health Metrics and Evaluation (IHME): Seattle, WA, USA, 2022. [Google Scholar]
- Ahmed, I.; Rabbi, M.B.; Sultana, S. Antibiotic resistance in Bangladesh: A systematic review. Int. J. Infect. Dis. 2019, 80, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Sujan, M.J.; Habib, Z.H.; Rahman, A.; Rizvi, S.M.S.; Barua, H.T.; Aboushady, A.T.; Hasnat, A.; Rasul, S.B.G.; Joh, H.S.; Prifti, K.; et al. Antimicrobial resistance surveillance methods in Bangladesh: Present and way forward. Clin. Infect. Dis. 2023, 77 (Suppl. 7), S549–S559. [Google Scholar] [CrossRef] [PubMed]
- Faiz, M.A.; Basher, A. Antimicrobial resistance: Bangladesh experience. Reg. Health Forum 2011, 15, 1–8. [Google Scholar]
- Islam, M.A.; Islam, R.; Khan, R.; Amin, M.B.; Rahman, M.; Hossain, M.I.; Ahmed, D.; Asaduzzaman, M.; Riley, L.W. Prevalence, etiology and antibiotic resistance patterns of community-acquired urinary tract infections in Dhaka, Bangladesh. PLoS ONE 2022, 17, e0274423. [Google Scholar] [CrossRef] [PubMed]
- Sectoral Co-ordination Center (Human Health) for AMR. National Antimicrobial Resistance (AMR) Surveillance Report, Bangladesh, 2016–2023. Dhaka; 2023. Available online: https://dashboard.iedcr.gov.bd/amr/uploads/amr_report_2016-23.pdf (accessed on 16 May 2025).
- Akhtar, S.; Sarker, M.R.; Jabeen, K.; Sattar, A.; Qamar, A.; Fasih, N. Antimicrobial resistance in Salmonella enterica serovar typhi and paratyphi in South Asia-current status, issues and prospects. Crit. Rev. Microbiol. 2015, 41, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Afroz, H.; Hossain, M.M.; Fakruddin, M. A 6-year retrospective study of bloodstream Salmonella infection and antibiotic susceptibility of Salmonella enterica serovar Typhi and Paratyphi in a tertiary care hospital in Dhaka, Bangladesh. Tzu Chi Med. J. 2014, 26, 73–78. [Google Scholar] [CrossRef]
- Ahmed, D.; Nahid, A.; Sami, A.B.; Halim, F.; Akter, N.; Sadique, T.; Rana, S.; Bin Elahi, S.; Rahman, M. Bacterial etiology of bloodstream infections and antimicrobial resistance in Dhaka, Bangladesh, 2005–2014. Antimicrob. Resist. Infect. Control. 2017, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Holm, M.; MacWright, W.R.; Poudyal, N.; Shaw, A.G.; Joh, H.S.; Gallagher, P.F.; Kim, J.-H.; Shaikh, A.T.; Seo, H.J.; Kwon, S.Y.; et al. Capturing data on Antimicrobial resistance Patterns and Trends in Use in Regions of Asia (CAPTURA). Clin. Infect. Dis. 2023, 77 (Suppl. 7), S500–S506. [Google Scholar] [CrossRef] [PubMed]
- WHONET. Available online: https://whonet.org/ (accessed on 20 April 2024).
- Sujan, M.J.; Gautam, S.; Aboushady, A.T.; Clark, A.; Kwon, S.; Joh, H.S.; Holm, M.; Stelling, J.; Marks, F.; Poudyal, N. QAAPT: An interoperable web-based open-source tool for antimicrobial resistance data analysis and visualisation. Front. Microbiol. 2025, 16, 1513454. [Google Scholar] [CrossRef] [PubMed]
- R Foundation for Statistical Computing. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; p. 2013. [Google Scholar]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100. J. Clin. Microbiol. 2021, 59, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Hindler, J.A.; Bhowmick, T.; Das, S.; Johnson, J.K.; Lubers, B.V.; Redell, M.A.; Stelling, J.; Erdman, S.M. What’s new in antibiograms? Updating CLSI M39 guidance with current trends. J. Clin. Microbiol. 2022, 60, e02210-21. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, F.R.; Ibrahim, Q.S.; Bari, M.S.; Alam, M.J.; Dunachie, S.J.; Rodriguez-Morales, A.J.; Patwary, M.I. The association between temperature, rainfall and humidity with common climate-sensitive infectious diseases in Bangladesh. PLoS ONE 2018, 13, e0199579. [Google Scholar] [CrossRef] [PubMed]
- Afroz, H.; Fakruddin, M.; Masud, M.R.; Islam, K. Incidence of and risk factors for hospital acquired infection in a tertiary care hospital of Dhaka, Bangladesh. Bangladesh J. Med. Sci. 2017, 16, 358. [Google Scholar] [CrossRef]
- Saeed, D.K.; Farooqi, J.; Shakoor, S.; Hasan, R. Antimicrobial resistance among GLASS priority pathogens from Pakistan: 2006–2018. BMC Infect. Dis. 2021, 21, 1231. [Google Scholar] [CrossRef] [PubMed]
- Indian Council of Medical R. Annual Report (2023) Antimicrobial Resistance Research and Surveillance Network. New Delhi, India. 2023. Available online: https://www.icmr.gov.in/icmrobject/uploads/Documents/1725536060_annual_report_2023.pdf (accessed on 16 May 2025).
- Nepal Ministry of Health and Population. CAPTURA Country Report: Nepal, 2023; Kathmandu, Nepal, 2023. Available online: https://heoc.mohp.gov.np/guidelines-publications/captura-country-report-nepal/detail (accessed on 16 May 2025).


| Characteristics | Frequency n = 232,329 | Percentage (%) |
|---|---|---|
| Gender | ||
| Male | 103,647 | 44.6 |
| Female | 128,682 | 55.4 |
| Age group | ||
| <1 | 7527 | 3.2 |
| 1–4 Years | 11,270 | 4.9 |
| 5–14 Years | 15,774 | 6.8 |
| 15–24 Years | 24,772 | 10.7 |
| 25–34 Years | 29,574 | 12.7 |
| 35–44 Years | 25,682 | 11.1 |
| 45–54 Years | 33,434 | 14.4 |
| 55–69 Years | 54,098 | 23.3 |
| 70+ Years | 30,198 | 13 |
| Number of AST records analyzed per year | ||
| 2017 | 46,233 | 19.9 |
| 2018 | 72,573 | 31.2 |
| 2019 | 83,544 | 36.0 |
| 2020 | 29,979 | 12.9 |
| Specimen type | ||
| Urine | 116,837 | 50.3 |
| Soft tissue and body fluid | 57,596 | 24.8 |
| Blood | 28,002 | 12.1 |
| Respiratory | 24,668 | 10.6 |
| Genital | 3453 | 1.5 |
| Stool | 1773 | 0.8 |
| The most common organisms found (top seven) | ||
| Escherichia coli | 75,472 | 32.5 |
| Klebsiella sp. | 36,012 | 15.5 |
| Pseudomonas sp. | 24,638 | 10.6 |
| Staphylococcus aureus | 18,018 | 7.8 |
| Enterococcus sp. | 17,868 | 7.7 |
| Staphylococcus, coagulase negative | 10,695 | 4.6 |
| Acinetobacter sp. | 9489 | 4.1 |
| Antibiotics/Organisms | Gram-Negative | ||||
|---|---|---|---|---|---|
| Escherichia coli % (n1/n2) * | Klebsiella sp. % (n1/n2) | Pseudomonas sp. % (n1/n2) | Salmonella sp. % (n1/n2) | Acinetobacter sp. % (n1/n2) | |
| Amikacin | 13.6 (8690/63,938) | 30.2 (9107/30,143) | 52.2 (11,466/21,983) | - | 62.0 (4727/7619) |
| Amoxicillin | 84.6 (9691/11,455) | - | - | 29.3 (1080/3681) | - - |
| Amoxicillin–Clavulanate | 58.1 (26,484/45,552) | 66.1 (16,703/25,276) | 90.6 (7427/8196) | - | 92.3 (4406/4772) |
| Ampicillin | 91.5 (4687/5123) | - | - | 22.4 (1207/5387) | - |
| Aztreonam | 56.9 (18,743/32,949) | 55.3 (8797/15,899) | 61.3 (1082/1766) | - | - |
| Azithromycin | - | - | - | 28.8 (2516/8751) | - |
| Cefepime | 43.1 (15,683/36,359) | 44.7 (6549/14,663) | 92.2 (3478/3772) | - | 65.1 (3216/4942) |
| Ceftazidime | 53.8 (28,949/53,787) | 53.5 (12,850/24,030) | 53.7 (9166/17,062) | - | 78.2 (6226/7958) |
| Ceftriaxone | 62.9 (41,573/66,106) | 59.2 (18,142/30,626) | - | 1.4 (155/10,968) | 82.6 (5833/7066) |
| Cefoxitin | 31.1 (1157/3723) | 50.1 (663/1324) | - | - | - |
| Cefuroxime | 68.1 (32,081/47,085) | 65.2 (11,810/18,107) | - | - | - |
| Chloramphenicol | - | - | - | 17.5 (1055/6019) | - |
| Ciprofloxacin | 65.6 (43,970/67,037) | 52.4 (16,469/31,417) | 58.9 (12,345/20,958) | 28.7 (3175/11,074) | 72.1 (5818/8067) |
| Doxycycline | 56.8 (11,117/19,559) | 48.7 (2817/5784) | - | - | 48.4 (1105/2283) |
| Gentamicin | 24.4 (16,440/67,358) | 35.6 (11,295/31,748) | 57.9 (12,994/22,435) | - | 67.0 (5838/8720) |
| Imipenem | 5.3 (2661/50,154) | 20.3 (4482/22,097) | 37.2 (5310/14,289) | - | 57.4 (3744/6521) |
| Mecillinam (only Urine) | 22.1 (3225/14,570) | 33.1 (1250/3774) | - | - | - |
| Meropenem | 6.8 (3384/49,699) | 21.7 (4781/22,047) | 38.8 (4863/12,545) | - | 55.8 (3320/5952) |
| Nalidixic acid | - | - | - | 92.9 (8335/8972) | - |
| Netilmycin | 14.7 (4637/31,602) | 34.1 (5358/15,700) | 56.2 (7935/14,124) | - | 47.5 (2610/5496) |
| Nitrofurantoin (only urine) | 14.4 (8209/56,957) | 40.8 (6568/16,090) | - | - | 64.9 (998/1538) |
| Piperacillin–Tazobactam | 19.7 (6847/34,698) | 38.5 (7811/20,291) | 35.0 (6887/19,671) | - | 68.9 (5092/7392) |
| Tetracycline | 52.2 (1603/3069) | 55.0 (16,724/30,430) | - | - | - |
| Sulfamethoxazole–Trimethoprim | 53.9 (34,493/64,028) | 50.8 (1515/2984) | - | 58.6 (4928/8406) | |
| Antibiotics | Gram-Positive | |||
|---|---|---|---|---|
| Staphylococcus aureus % (n1/n2) * | Enterococcus sp. % (n1/n2) | Streptococcus sp. % (n1/n2) | CoNS % (n1/n2) | |
| Ampicillin | - | 27.7 (2377/8588) | - | - |
| Azithromycin | 80.1 (4338/5417) | - | - | 85.1 (2318/2724) |
| Cefoxitin | 41.7 (2017/4837) | - | - | - |
| Chloramphenicol (blood) | 12.4 (213/1725) | - | 9.3 (54/578) | - |
| Ciprofloxacin | 62.2 (8949/14,382) | 71.4 (11,988/16,796) | - | - |
| Doxycycline | 27.2 (1564/5742) | 59.0 (3640/6168) | 28.3 (100/353) | 34.6 (1122/3244) |
| Erythromycin | - | - | 56.9 (1222/2146) | - |
| Gentamicin | 20.2 (3380/16,708) | - | - | 32.1 (2725/8485) |
| Linezolid | 5.3 (476/8999) | 3.3 (329/10,082) | 2.1 (70/3414) | 6.4 (310/4817) |
| Nitrofurantoin (Urine) | 14.8 (549/3712) | 11.0 (1599/14,575) | 5.6 (95/1688) | 16.6 (531/3194) |
| Oxacillin | 35.5 (2134/6019) | - | - | - |
| Penicillin G | 80.1 (2708/3379) | 26.8 (2686/10,011) | 7.8 (191/2439) | 77.6 (2953/3807) |
| Tetracycline | 21.8 (748/3435) | 76.7 (1629/2123) | 57.1 (887/1554) | 26.9 (831/3088) |
| Trimethoprim/Sulfamethoxazole | 37.6 (5786/15,384) | - | 69.9 (2351/3363) | 43.3 (3504/8090) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, A.; Sujan, M.J.; Rizvi, S.M.S.; Barua, H.T.; Habib, Z.H.; Jannat, H.; Deb, P.K.; Hasnat, A.; Kwon, S.Y.; Aboushady, A.T.; et al. Patterns of Antimicrobial Resistance Among Major Bacterial Pathogens Isolated from Clinical Samples in Bangladesh (2017–2020): A Nationwide Cross-Sectional Study. Microbiol. Res. 2025, 16, 122. https://doi.org/10.3390/microbiolres16060122
Rahman A, Sujan MJ, Rizvi SMS, Barua HT, Habib ZH, Jannat H, Deb PK, Hasnat A, Kwon SY, Aboushady AT, et al. Patterns of Antimicrobial Resistance Among Major Bacterial Pathogens Isolated from Clinical Samples in Bangladesh (2017–2020): A Nationwide Cross-Sectional Study. Microbiology Research. 2025; 16(6):122. https://doi.org/10.3390/microbiolres16060122
Chicago/Turabian StyleRahman, Aninda, Mohammad Julhas Sujan, S. M. Shahriar Rizvi, Hridika Talukder Barua, Zakir Hossain Habib, Hurul Jannat, Piash Kumer Deb, Abul Hasnat, Soo Young Kwon, Ahmed Taha Aboushady, and et al. 2025. "Patterns of Antimicrobial Resistance Among Major Bacterial Pathogens Isolated from Clinical Samples in Bangladesh (2017–2020): A Nationwide Cross-Sectional Study" Microbiology Research 16, no. 6: 122. https://doi.org/10.3390/microbiolres16060122
APA StyleRahman, A., Sujan, M. J., Rizvi, S. M. S., Barua, H. T., Habib, Z. H., Jannat, H., Deb, P. K., Hasnat, A., Kwon, S. Y., Aboushady, A. T., Clark, A., Stelling, J., Gautam, S., Shaw, A., Holm, M., Marks, F., & Poudyal, N. (2025). Patterns of Antimicrobial Resistance Among Major Bacterial Pathogens Isolated from Clinical Samples in Bangladesh (2017–2020): A Nationwide Cross-Sectional Study. Microbiology Research, 16(6), 122. https://doi.org/10.3390/microbiolres16060122






