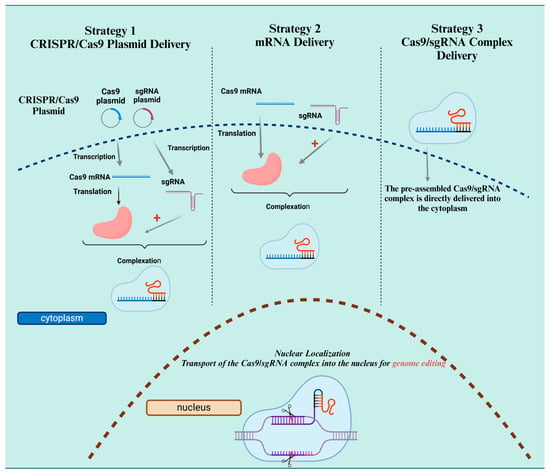
Abstract
The CRISPR–Cas system has transformed molecular biology by providing precise tools for genome editing and pathogen detection. Originating from bacterial adaptive immunity, CRISPR technology identifies and cleaves genetic material from pathogens, thereby preventing infections. CRISPR–Cas9, the most widely utilized variant, creates double-stranded breaks in the target DNA, enabling genetic disruptions or edits. This approach has shown significant potential in antiviral therapies, addressing chronic infections, such as HIV, SARS-CoV-2, and hepatitis viruses. In HIV, CRISPR–Cas9 edits the essential viral genes and disrupts latent reservoirs, while CCR5 gene modifications render the T cells resistant to viral entry. Similarly, SARS-CoV-2 is targeted using CRISPR–Cas13d to inhibit the conserved viral genes, significantly reducing viral loads. Hepatitis B and C treatments leverage CRISPR technologies to target conserved genomic regions, limiting replication and expression. Emerging innovations, such as the PAC-MAN approach for influenza and base-editing systems to reduce off-target effects, further highlight the therapeutic versatility of CRISPR. Additionally, advances in Cas12a and Cas13 have driven the development of diagnostic platforms like DETECTR and SHERLOCK, which provide rapid and cost-effective viral detection. Innovative tools like AIOD-CRISPR enable accessible point-of-care diagnostics for early viral detection. Experimental approaches, such as targeting latent HSV-1 reservoirs, highlight the transformative potential of CRISPR in combating persistent infections.
1. Overview of CRISPR Technology and Its Mechanisms in Genome Editing
CRISPR, which is short for clustered regularly interspaced short palindromic sequence repeats, is a relatively novel technology that has emerged like a revolution in gene editing and pathogen diagnostic methods. Compared with other methods in nucleic acid detection, CRISPR–Cas has a high sensitivity and specificity, is less time consuming, and does not need amplification in order to detect the genome [1,2].
Speaking of the CRISPR–Cas system anatomy and mechanism of action, CRISPR loci, which consist of the CRISPR array, are found in the bacterial genome. The array is made up of palindromic repeats inserted by spacer sequences. These spacers, which have been obtained from invading the genetic material of viruses or bacteriophages, represent the genetic memory of the cell and make the host resistant to contamination by any virus that contains the same sequence. Apart from having the leader sequence, the array comprises an operon with a number of genes that encode Cas proteins. There are a diverse group of Cas proteins, but Cas1 and Cas 2 are present in all kinds of CRISPR–Cas systems and are essential in spacer integration [3,4,5]. In the first step, the protospacer adjacent motif (PAM)—a short sequence of nucleotides in viral genetic material—is identified. PAM is located near the protospacer, which is the foreign DNA sequence that will form the spacer. After the recognition, cleavage is performed by the Cas1–Cas2 nuclease complex at the PAM site and the protospacer is acquired and will be placed between two repeats as a spacer. The next step includes the transcription of the CRISPR locus, which produces pre-crRNA that will be later transformed to mature crRNA or guide RNA with the help of Cas proteins. In the last step, this mature crRNA/guide RNA that is attached to a multi-protein complex first recognizes and then pairs with the complementary sequence of the genetic material of the viral pathogen, and eventually cleaves the nucleotides beside the PAM site [2,6].
Lately, the functionality of this system has shed a new light on antiviral therapy, since the component containing crRNA can bind to the complementary target nucleic acid so that the Cas protein component can cleave the target genome. As a result, a double-stranded break is manufactured, which impairs the proliferation of the virus and defects its replication process and thus its life cycle [2]; an instance of such is the annihilation of the CMP–sialic acid transporter “SLC3A1”, which resulted in the blocking of influenza type A entry and subsequently prevented its infection [7]. With the ability to directly invade the DNA, it is now a part of some bacteria’s adaptive immune systems by cleaving the nucleic acids of the foreign plasmids and bacteriophages [2].
CRISPR-Cas9 is also one of the most well-known and used approaches in genome editing and pathogen detection that is based on a guide RNA pairing with dsDNA and the following cleavage by the Cas9 endonuclease. As a result of the cleavage, a double-stranded break is produced, which will be flowingly repaired with the homology-directed repair (HDR) or non-homologous end joining (NHEJ) pathways [5]. NHEJ is in fact a repair mechanism that is highly prone to errors and can cause random insertions or deletions at the cleavage site and consequently generate frameshift mutations or premature stop codons [8]. Sometimes the Cas9 protein binds but does not cleave the DNA; this feature, which is useful in diagnostic fields, is known as a nuclease dead Cas9. An example of its application is in Mycobacterium tuberculosis detection. In this technique, the Luciferase enzyme is first split in half and then combined with two dCas9s. When these dCas9s are attached to the target genome, the two parts of the luciferase enzymes are unified and cause a reaction, resulting in bioluminescent signal production [9]. The system known as CRISPR–Cas is based on the evolutionary defense tactics utilized by archaea and bacteria. They play a role in the adaptive immune system of these hosts, defending against the threats posed by plasmids and viruses. There are two primary classes of CRISPR–Cas systems: class 1, which encompasses types I, III, and IV with their multiprotein complexes; and class 2, which includes types II, V, and VI that function with a single effector protein, making them particularly useful for gene editing in mammals. The systems classified as type I, II, IV, and V are involved in the cleavage of DNA, while type VI focuses on RNA, and type III can act on both DNA and RNA [10,11] (Figure 1).
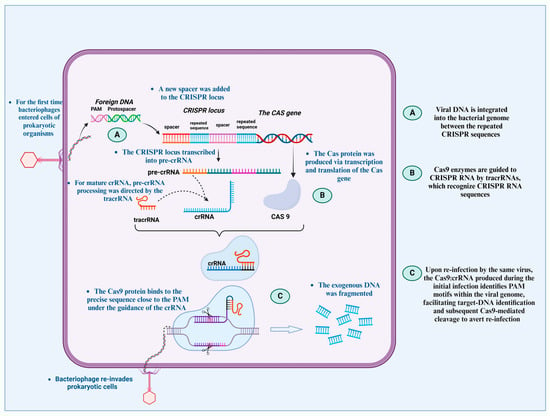
Figure 1.
CRISPR–Cas systems prevent viruses from re-entering cells, acting as the adaptive immune system of prokaryotes. When the foreign DNA first reaches the CRISPR region, the host genome identifies it as a spacer. tracrRNA converts the pre-crRNA into the mature guide RNA (crRNA). The CRISPR-Cas complex is formed when the Cas9 protein attaches itself to the crRNA and tracrRNA. By accurately identifying and cleaving the PAM region, this complex binds itself to the target DNA after re-infection and removes the foreign DNA, according to the fundamental idea of gene-editing methods.
2. Targeting Chronic Viral Infections with CRISPR: Current Approaches
The CRISPR–Cas system has been considered as a potential treatment option against viral infections since it has been a beneficial method in the prompt diagnosis of pathogens and provides a meticulous method of treatment in order to manage the spread of disease. Furthermore, unlike antiviral drugs, it does not impose resistance and side effects in the host, which makes it a remarkable alternative [12]. One of the applications of this system is to battle against HIV infection: a highly widespread retrovirus with no vaccine or antiretroviral therapy that can fully cure the disease [5]. HIV consists of RNA as genetic material and its typical route of contamination is to enter cells with CD4 receptors and CCR5 or CXCR4 as co-receptors. Once it has entered, the RNA is first reverse-transcribed to DNA and then integrated as proviral DNA into the host genome. Sometimes, the provirus switches to an inactive mode that cannot be transcribed, which raises an issue. Not only can the human immune system not attack these latent reservoirs, but the antiretroviral medications also fail to act upon these non-transcribing sections [13]. Some of these HIV-1 reservoirs can survive for up to 60 years in CD4+ T cells [5]. Whereas CRISPR–Cas9 utilizes a different strategy. A guide RNA binds to the target DNA and Cas9, which has been acquired from Streptococcus pyogenes, cleaves the DNA next to the PAM site and creates a double-stranded break. Following this course of action, non-homologous end joining (NHEJ) and microhomology-mediated end joining (MMEJ), which are the DNA repair mechanisms, begin to act and apply deletions, insertions, or substitutions in the DNA [2,14]. Consequently, these mutations result in virus inactivation by interfering with its vital genes. With this strategy, long terminal repeats (LTRs) at the 3′ and 5′ ends of the provirus are proper targets for the guide RNA to prevent the expression of the active provirus. It could also disrupt the latent reservoir-forming provirus [15]. A 32-bp deletion mutation in the CCR5 gene (CCR5Δ32) is a sterilizing strategy in order to make the cells intuitively resistant to HIV-1 [5]. However, it is worthy of note that sometimes mutations build up at the cleaving site, hindering the attachment of the guide RNA and creating a loophole for the virus to escape. In order to prevent this, different combinations of the two guide RNAs aimed at different vital genes that can lead to large insertion–deletion mutations (indels) have been helpful and can adequately restrain the viral cycle [2,16].
Another potential target against the CRISPR–Cas system is SARS-CoV-2 or COVID-19, which first appeared in China in 2019 and then spread all around the world. Unlike HIV, there are efficient vaccines for COVID-19 that boost antibody levels against the pathogen, but still the virus remains highly contagious and inflicts a huge burden on families and society. SARS-CoV-2 has a single-stranded RNA as the genetic material, which includes 16 encoding genes for non-structural proteins and 4 for structural ones, including the Spike protein. This protein is essential for the virus to enter the host cell and interact with ACE2 receptors, which are abundantly expressed in the lung, heart, and kidney cells [17,18]. The CRISPR–Cas system can directly demolish the genetic material using Cas13d. The Cas13d protein is small and therefore easy to be transferred, also PAM is not required for its cleaving activity so designing a complementary guide RNA would be also facilitated. In recent studies, two conserved sequences—the nucleocapsid gene, which is transcribed to the capsid protein, and RNA-dependent RNA polymerase gene, which is necessary for the virus proliferation—have been considered as proper targets for CRISPR–Cas13d. In addition, COVID-19 inhibition is highly associated with crRNA concentration. It has been reported that a sufficient level of 22 crRNAs can effectively target all the known genes of SARS-CoV-2 [19,20,21,22].
The influenza A virus, another contagious pathogen that leads to seasonal pandemics annually, is another target of the CRISPR–Cas system. The results of a recent study showed that the protein Cas13b obtained from Prevotella sp., along with crRNA against viral RNA, could decrease the influenza A viral load by 7–22 folds [23]. Comparative mechanisms of the CRISPR–Cas systems targeting diverse viral genomes are shown in Table 1.

Table 1.
Comparative mechanisms of the CRISPR–Cas systems targeting diverse viral genomes.
Cas13 systems are mainly focused on targeting RNA, but the broader use of CRISPR technology in antiviral approaches frequently involves CRISPR–Cas9, which functions through a different mechanism. As illustrated in Figure 2, CRISPR–Cas9 enables genome editing by creating double-stranded breaks (DSBs) at specific DNA locations. This triggers cellular repair processes that can lead to the introduction of mutations or the correction of sequences. This underscores the versatility of CRISPR systems, as they can effectively target both RNA and DNA in the fight against viral infections (Figure 2).
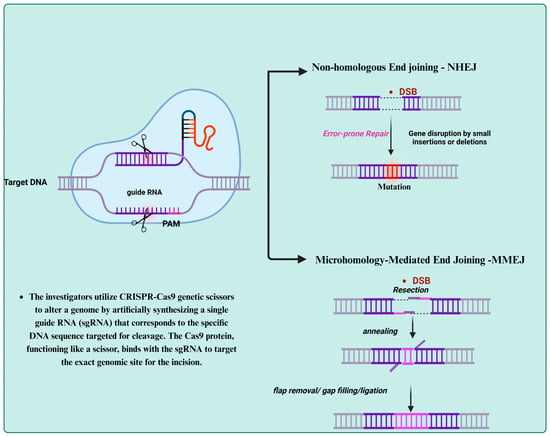
Figure 2.
Mechanism of CRISPR–Cas9 genome editing: A guide RNA (gRNA) directs the Cas9 protein to a specific target DNA sequence adjacent to a protospacer adjacent motif (PAM), where the Cas9 enzyme induces a double-stranded break (DSB). The cell then attempts to repair the breakthrough either through microhomology-mediated end joining (MMEJ), which uses short homologous sequences, or non-homologous end joining (NHEJ), a more error-prone process that often introduces mutations. This illustrates the precision and versatility of CRISPR–Cas9 in editing host or viral DNA.
3. CRISPR Applications in Treating Persistent Viruses
Although many applications of CRISPR have been limited to in vitro studies, the results have been promising. Regarding persistent viruses like HIV, as mentioned earlier, the typical entry route of HIV is through cells with CD4 receptors and CCR5 or CXCR4 co-receptors. In vitro studies indicated that CRISPR–Cas9, which targeted the CCR5 gene, managed to create a CD4+ T cell without a CCR5 co-receptor that is resistant to the virus and can block its entry. Nevertheless, a small fraction of HIV uses the CXCR4 co-receptor and the created resistance in CCR5-lacking T cells does not make them immune to CXCR4 using viruses [5,24]. Current in vitro studies demonstrated the possibility of disrupting both CCR5 and CXCR4 receptors using combined small guide RNA that can target their genes simultaneously and cause no cellular toxicity. To achieve the same promising results in humans, valid extensive animal studies are required; however, the first report of using CRISPR–Cas9 in in vivo studies was bright and promising. In this attempt, a short variant of Cas9 obtained from Staphylococcus aureus was delivered using recombinant adeno-associated virus 9 in order to target 5′-LTR and the gag gene as regions of integrated HIV-1 in transgenic mouse and rat [25]. Overall, using the CRISPR–Cas9 genome editing method with antiretroviral therapy can be a potential option for HIV treatment [2,26].
The PAC-MAN strategy is another CRISPR–Cas system-based approach that has been used against influenza A virus (IAV). This single-stranded RNA virus contains 8 segments in its genome, and all of their conserved genes were targeted in this strategy. For instance, segment 6 contains genes that code for neuraminidase, which is required to release the virus from the host cell, and segment 4 codes for hemagglutinin, which is again vital for the viral particle attachment to the host cell. The result of targeting these essential genes indicated a moderate suppression of IAV. Plus, it showed that with only 6 crRNA, 92% of IAV strains could be targeted and 81 crRNA were aimed at all of the known strains of IAV. The results of the PAC-MAN strategy were satisfactory in cell culture and it needs to be applied in animal models as well [20].
The PAC-MAN strategy, or prophylactic antiviral CRISPR in human cells, represents an innovative approach using CRISPR technology to specifically target and neutralize RNA viruses, such as IAV and SARS-CoV-2. This method was developed to address the limitations of traditional DNA-targeting CRISPR systems like Cas9, which are ineffective against RNA viruses that do not integrate into the host genome. PAC-MAN employs the CRISPR–Cas13d system, an RNA-targeting endonuclease sourced from Ruminococcus flavefaciens. Its small size (approximately 930 amino acids) makes its delivery to cells efficient, particularly when using viral vectors like the adeno-associated virus (AAV). Unlike Cas9, which requires a protospacer adjacent motif (PAM) and creates double-stranded DNA cuts, Cas13d targets single-stranded RNA guided by a CRISPR RNA (crRNA) and directly cleaves it. This enables PAC-MAN to degrade viral RNA genomes before replication, stopping the infection at the transcriptional level without altering the host DNA. The core mechanism of PAC-MAN involves creating a library of multiplexed crRNAs that are complementary to the conserved regions of viral genomes. For instance, with influenza A, conserved regions in gene segments, such as PB1, NP, HA (hemagglutinin), and NA (neuraminidase), are targeted, while in SARS-CoV-2, the N gene (nucleocapsid protein) and RdRp (RNA-dependent RNA polymerase) are common targets due to their crucial role in viral replication. By focusing on these conserved areas, PAC-MAN aims to deliver broad-spectrum efficacy against various virus strains and even across related viruses [20]. A significant advantage of PAC-MAN is that it does not create double-stranded breaks (DSBs) in the host genome, which can lead to genomic instability or mutations in Cas9-based systems. Instead, it works at the RNA level, offering a temporary, reversible, and safer antiviral approach. Additionally, Cas13d shows minimal collateral cleavage activity, minimizing the risk of unintended host RNA degradation, a challenge that is seen in some other Cas13 enzymes like Cas13a and Cas13b. The name PAC-MAN is metaphorically inspired by the classic arcade game character “Pac-Man”, which navigates a maze consuming dots. Similarly, the CRISPR–Cas13d complex “chases and digests” viral RNA fragments, eliminating the pathogen’s genetic instructions within human cells. This creative name not only encapsulates the strategy’s antiviral action but also enhances its conceptual understanding for both scientific and general audiences. In recent experiments, PAC-MAN has shown effectiveness in vitro. The deliveries of Cas13d with a crRNA cocktail resulted in notable reductions in viral RNA levels. For example, up to 92% of known influenza A strains were successfully targeted with just six crRNAs, and similar success was achieved with SARS-CoV-2 using 22 crRNAs. These findings underscore the potential of PAC-MAN as a programmable, adaptable, and scalable antiviral platform, which is particularly valuable in addressing emerging pandemics with rapid response needs [23].
The CRISPR–Cas9 system genome editing tool has been also studied in the hepatitis B virus. Similar to HIV, it forms a latent viral reservoir as covalently closed circular DNA (cccDNA) in cells. Studies in cultured cells indicated that designing guide RNAs against conserved sequences in the hepatitis B genome, like S, P, C and X ORFs, successfully reduced HBV DNA material to 98%. Also targeting HBV cccDNA with a designed guide RNA in infected cells resulting in an eight-fold decrease in HBcAg expression and overall targeting in various regions in the HBV genome appeared to boost the effectiveness of this method [5,12]. Another study showed a more precise targeting by the CRISPR–Cas9 system that is applied by Cas9 nickases (Cas9ns). In this accurate process, S and X ORFS are targeted as conserved sequences in the HBV genome, as well as in the episomal cccDNA, and the results were promising in disrupting HBV replication in both recently and chronically infected hepatoma cells [27,28]. However, it should be noted that in samples of integrated DNA like HBV and HIV, producing double-stranded breaks may cause adverse effects in the host genome. Therefore, an improved system of CRISPR–Cas was introduced, in which during base-editing, specific bases are altered without creating double-stranded breaks so that specific gene expression will be permanently silenced. This method is applicable in both cccDNA and HBV DNA [12]. Chronic hepatitis, fibrosis, and cirrhosis can also be the outcomes of hepatitis C viral infection. HCV is a single-stranded RNA virus that belongs to Flaviviridae. Claudin-1, an important HCV receptor that is majorly seen in hepatocellular carcinoma, is the main target of the CRISPR–Cas9 system. Studies indicated that claudin-1-lacking cells are resistant to HCV infection. In another study, CRISPR–Cas13a was developed against HCV. Cas13a targets the entry site of HCV (internal ribosome entry site or IRES) and is able to prevent the virus replication effectively with the smallest effects on cell viability [29,30,31].
The CRISPR–Cas system has developed methods against human Papillomavirus (HPV) that have also been effective. This double-stranded DNA virus that belongs to the Papilomaviridea family is commonly asymptomatic. However, sometimes this viral infection leads to persistent warts or precancerous lesions that need medical attention. Recent studies have demonstrated that the CRISPR–Cas system that targets the HPV-16 E7 DNA can upregulate pRb, a host tumor-suppressor protein, by inhibiting the E7 protein. This results in the growth prevention of positive HPV cells, as well as the inhibition of the cancerous activity of HPV cells both in vitro and in vivo [32]. Last but not least, attempts of genome editing for the Epstein–Barr virus (EBV) have also been conducted. EBV is a double-stranded DNA virus that infects B cells, as well as immune system epithelial cells. Infection with EBV leads to infectious mononucleosis, while sometimes it can develop to malignant lympho-proliferative diseases, like Burkitt’s lymphoma. A recently developed CRISPR–Cas9 system targeted the LMP1 gene. Latent membrane protein-1 is majorly known as a factor that is associated with EBV carcinogenesis and advancement in nasopharyngeal carcinoma (NPC); therefore, with the suppression of LMP-1, the promotion of NPC is effectively blocked [32].
5. Precision, Efficiency, and Safety in CRISPR-Based Treatments
5.1. CRISPR–Cas Systems and DNA Editing
The Cas9 endonuclease, which is the first CRISPR system employed in gene editing in mammalian cells, is classified as type II in the class 2 category. Comprising HNH and RuvC-like domains, the Cas9 endonuclease is responsible for cleaving the DNA and is directed to a specific DNA target through the use of a guide RNA (gRNA). The formation of a guide RNA involves the synthesis of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) into a unified gRNA [39,45]. The gRNA helps Cas9 to locate target sequences that have a 20-nucleotide complement to the crRNA, known as the protospacer, and are associated with a PAM, which is NGG for SpCas9 from Streptococcus pyogenes, with N representing any nucleotide. The action of Cas9 results in a double-stranded break (DSB) by cleaving both strands of DNA, producing blunt ends. Following this, the cell employs its DNA repair processes, especially NHEJ and MMEJ, which often cause small insertions and deletions (indels) at the site of the break [43,46].
5.2. Antiretroviral Therapy in HIV
The use of combination antiretroviral therapy (cART), which includes multiple medications targeting various phases of the HIV replication process, can successfully suppress the viral replication and result in an undetectable viral load. The immune system of the host will primarily eliminate cells infected with HIV; however, certain cells may harbor a provirus that remains inactive, thereby evading detection and elimination. Unfortunately, the existing cART therapies fail to specifically address the integrated provirus in the cells that are latently infected, resulting in a quick surge of viral levels when the treatment is discontinued. Due to the inability of cART to achieve a lasting cure, individuals with HIV are obligated to maintain lifelong therapy [35,41,47].
5.3. Cure HIV Using Gene Editing
Recently, multiple nuclease-driven gene editing methods have been established that facilitate the creation of double-stranded breaks (DSBs) at designated areas in the DNA. The repair of these double-stranded breaks (DSBs) will be carried out by the cell’s DNA repair systems, including microhomology-mediated end-joining (MMEJ) and non-homologous end-joining (NHEJ), which commonly lead to the creation of minor deletions and insertions at the specific locations [43,47,48]. Conversely, when a specific mutated homologous DNA segment is introduced, the process of homologous recombination (HR) can facilitate the repair of the DSB, resulting in the placement of this mutation at the intended location. Numerous research efforts have explored the potential of DNA cutting mechanisms to either deactivate HIV DNA within infected reservoir cells or alter the HIV host cells to render them immune to infection. In the early stages, the scientific community identified zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) as noteworthy technologies. Zinc-finger nucleases (ZFNs) are engineered enzymes that consist of two parts: a zinc-finger region that attaches to specific DNA sequences and a FokI domain that is responsible for cutting the DNA [10,39,49].
The FokI nuclease domain is employed by TALENs for the purpose of cleaving the DNA. In virus–cell culture systems, ZFNs and transcription activator-like effectors (TALEs) have been utilized to combat HIV by either directly interfering with the proviral DNA or by targeting the gene that produces the CCR5 coreceptor in the host cells [50,51]. The formation of a double-stranded break (DSB) requires the dimerization of the FokI cleavage domain, which means that a duo of ZFNs or TALENs is essential for targeting sequences that lack palindromic symmetry. Furthermore, the necessity to design a distinct ZFN or TALEN for each specific target sequence makes it more challenging to address multiple sites or adapt to minor sequence changes resulting from natural variation. In recent times, the CRISPR and CRISPR-associated protein (Cas) system has emerged as a widely used method for editing DNA, primarily because it can be easily tailored to different sequences of interest [42,49,52].
Derived from the natural immune systems of bacteria and archaea, CRISPR–Cas systems act as effective antivirals. The adaptive immune response in these hosts actively targets and neutralizes invading plasmids and viruses [45,53]. There are two main classifications of CRISPR–Cas systems: class 1, which includes types I, III, and IV with multiprotein complexes, and class 2, which consists of types II, V, and VI, recognized for a single effector protein that is more effective in gene editing in mammalian cells. The DNA is cut by systems I, II, IV, and V, whereas type VI is for RNA, and type III can handle both types of nucleic acids. In mammalian cells, the first CRISPR technology applied for gene modification is the Cas9 endonuclease, which is part of class 2, type II. The Cas9 nuclease possesses HNH and RuvC-like domains that enable it to cleave DNA, while a guide RNA (gRNA) steers it towards a designated DNA sequence [39,41].
The structure of this gRNA includes both a crRNA and a tracrRNA, which may be integrated into a singular gRNA format. Cas9 is directed to its target sequences by the gRNA, which must have a 20-nucleotide complementary match to the crRNA section and a designated downstream PAM. The NGG sequence associated with SpCas9 is derived from Streptococcus pyogenes, with N representing any nucleotide base. Cas9 generates a double-stranded break (DSB) by severing both DNA strands, which results in ends that are not overhanging [10,43,52]. Upon the action of Cas9 on the DNA, the cell’s repair pathways, especially NHEJ and MMEJ, are engaged, which often leads to the occurrence of small insertions and deletions (indels) at the site where the DNA was cleaved. It has been demonstrated by other scientists that when T-cell cultures are produced to continuously generate a gRNA and Cas9 that target the HIV DNA, the replication of HIV can be greatly reduced [38,52].
Despite the presence of the Cas9/gRNA inhibitors, the virus managed to overcome this restriction after extended culturing, leading to the effective replication of the virus. Examination of the viral cultures indicated that the escape variants had developed mutations induced by Cas at the cleavage site of Cas9/gRNA, which did not hinder replication but obstructed the attachment of the gRNA. Consequently, the cleavage of Cas not only hindered the replication of the virus but also sped up its ability to evade detection. A combination technique that involved the use of two gRNAs to target different viral sequences at the same time resulted in improved and lasting inhibition. The application of two gRNAs to the HIV proviral DNA resulted in modifications at both the targeted areas, in addition to the excision or inversion of the sequence that separates them [11,35,41].
7. Main Problems, Current Solutions, and Future Directions
CRISPR–Cas technology holds remarkable promise for antiviral therapy, yet it faces several significant challenges. A key issue is the ability of viruses to evade gene editing through mutations. In the case of HIV, mutations can occur around the CRISPR-induced cleavage sites, preventing the guide RNA from binding and thereby allowing the virus to continue its replication. Another hurdle is the persistence of latent viral reservoirs, especially in conditions like HIV, HBV, and HSV-1. These reservoirs contain non-replicating viral DNA integrated into the host genomes, rendering them undetectable to both the immune system and antiviral treatments. Furthermore, off-target effects pose a serious risk: CRISPR elements, notably Cas9, might bind and cut unintended genomic sites, resulting in genomic instability and possible toxicity. The efficient delivery of CRISPR components to target cells also presents difficulties, as viral vectors or nanoparticles can be immunogenic or inadequately targeted. Finally, despite promising preclinical outcomes, the lack of comprehensive in vivo and clinical data restricts our understanding of the long-term safety and efficacy in humans.
Researchers have made significant advances in overcoming the limitations of CRISPR-based antiviral therapies by developing several innovative strategies that enhance both their effectiveness and precision. Using multiple guide RNAs that target various regions of a viral genome can mitigate the issue of viral escape by causing extensive deletions or multiple mutations, thereby increasing the likelihood of fully inactivating the virus. Advanced precision tools, such as base editors and Cas9 nickases, allow for the modification of individual nucleotides without inducing double-stranded breaks, thus minimizing the off-target effects. The evolution of delivery methods has also been noteworthy, with technologies like viral vectors, including AAVs and lentiviruses, along with lipid-based nanoparticles, being used to efficiently transport CRISPR components into infected cells. Lentiviral delivery systems, such as HELP (HSV-1-erasing lentiviral particle) and dual AAV strategies, have shown potential in targeting the latent reservoirs of HSV and HIV. Additionally, editing host genes such as CCR5 and CXCR4 to obstruct viral entry offers a preventive measure. Furthermore, the collateral cleavage capability of CRISPR, particularly in Cas12 and Cas13 systems, has facilitated the creation of rapid and highly sensitive diagnostic tools for viral detection, which are essential for early intervention and monitoring.
Looking ahead, CRISPR–Cas technology in antiviral treatment presents several promising avenues. Clinically translating this technology is a top priority, as it is crucial to transition from laboratory and animal models to human trials to confirm its safety, delivery methods, and therapeutic efficacy. There is potential in combining CRISPR with existing treatments like long-acting slow-effective release antiretroviral therapy (LASER ART), which may not only suppress but possibly eradicate persistent infections, such as HIV, by targeting both active and latent viruses. Efforts are also focused on enhancing the specificity of CRISPR by improving guide RNA design and engineering high-fidelity Cas variants, which will help to reduce off-target effects and increase clinical safety. Another goal involves developing broad-spectrum antiviral strategies by editing host genes that are necessary for various viruses to replicate, providing a defense mechanism that is less susceptible to viral mutations. Additionally, expanding CRISPR applications to combat emerging and re-emerging viral threats is vital. Platforms such as Cas13, which target viral RNA without harming the host DNA, are especially suitable for a rapid response against RNA viruses like SARS-CoV-2 and Dengue. These advancements indicate that CRISPR is poised to become a fundamental component of next-generation antiviral therapies.
8. Conclusions
The CRISPR–Cas system represents a groundbreaking advancement in molecular biology, offering unparalleled precision and versatility in genome editing and pathogen diagnostics. Its application in antiviral therapeutics has shown promising potential to target and disrupt chronic viral infections, including HIV, SARS-CoV-2, and hepatitis viruses, while innovative diagnostic platforms, such as DETECTR and SHERLOCK, are transforming early detection methods. Despite these advancements, challenges such as off-target effects, genomic instability, and the need for comprehensive in vivo validation highlight the necessity for further optimization and research. As this technology continues to evolve, it holds the promise of revolutionizing precision medicine, providing novel strategies to combat persistent viral infections and significantly advancing our understanding of molecular and cellular mechanisms. The future of CRISPR lies in refining its safety and efficacy to unlock its full potential as a transformative tool in the diagnosis, treatment, and eradication of viral diseases, paving the way for more effective and accessible healthcare solutions worldwide.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis, F.N. and F.A.; first draft of the manuscript, H.T. and M.E.; writing—original draft preparation, H.T., F.A. and M.E.; software, B.F.; data curation, writing—review and editing, M.E. and V.O.; supervision and project administration, M.E. and V.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The corresponding author will provide the datasets created during the current investigation upon reasonable request.
Acknowledgments
Cancer Research Center, Semnan University of Medical Sciences, Semnan, Iran.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Hajian, R.; Balderston, S.; Tran, T.; DeBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef]
- Rajan, A.; Shrivastava, S.; Janhawi; Kumar, A.; Singh, A.K.; Arora, P.K. CRISPR-Cas system: From diagnostic tool to potential antiviral treatment. Appl. Microbiol. Biotechnol. 2022, 106, 5863–5877. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans. 2013, 41, 1392–1400. [Google Scholar] [CrossRef]
- Nuñez, J.K.; Kranzusch, P.J.; Noeske, J.; Wright, A.V.; Davies, C.W.; Doudna, J.A. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014, 21, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Bayat, H.; Naderi, F.; Khan, A.H.; Memarnejadian, A.; Rahimpour, A. The impact of CRISPR-Cas system on antiviral therapy. Adv. Pharm. Bull. 2018, 8, 591. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Zhao, H.; Sheng, G.; Wang, M.; Yin, M.; Wang, Y. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 2015, 163, 840–853. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Perez, J.T.; Chen, C.; Li, Y.; Benitez, A.; Kandasamy, M.; Lee, Y.; Andrade, J.; Manicassamy, B. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep. 2018, 23, 596–607. [Google Scholar] [CrossRef]
- Asmamaw, M.; Zawdie, B. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biol. Targets Ther. 2021, 15, 353–361. [Google Scholar]
- Zhang, Y.; Qian, L.; Wei, W.; Wang, Y.; Wang, B.; Lin, P.; Liu, W.; Xu, L.; Li, X.; Liu, D. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth. Biol. 2017, 6, 211–216. [Google Scholar] [CrossRef]
- Yu, L.; Tian, X.; Gao, C.; Wu, P.; Wang, L.; Feng, B.; Li, X.; Wang, H.; Ma, D.; Hu, Z. Genome editing for the treatment of tumorigenic viral infections and virus-related carcinomas. Front. Med. 2018, 12, 497–508. [Google Scholar] [CrossRef]
- Dong, Z.; Qin, Q.; Hu, Z.; Chen, P.; Huang, L.; Zhang, X.; Tian, T.; Lu, C.; Pan, M. Construction of a One-Vector Multiplex CRISPR/Cas9 Editing System to Inhibit Nucleopolyhedrovirus Replication in Silkworms. Virol. Sin. 2019, 34, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Baddeley, H.J.; Isalan, M. The application of CRISPR/Cas systems for antiviral therapy. Front. Genome Ed. 2021, 3, 745559. [Google Scholar] [CrossRef]
- Maartens, G.; Celum, C.; Lewin, S.R. HIV infection: Epidemiology, pathogenesis, treatment, and prevention. Lancet 2014, 384, 258–271. [Google Scholar] [CrossRef]
- Allen, F.; Crepaldi, L.; Alsinet, C.; Strong, A.J.; Kleshchevnikov, V.; De Angeli, P.; Páleníková, P.; Khodak, A.; Kiselev, V.; Kosicki, M. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2019, 37, 64–72. [Google Scholar] [CrossRef]
- Liao, H.-K.; Gu, Y.; Diaz, A.; Marlett, J.; Takahashi, Y.; Li, M.; Suzuki, K.; Xu, R.; Hishida, T.; Chang, C.-J. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015, 6, 6413. [Google Scholar] [CrossRef] [PubMed]
- Lebbink, R.J.; de Jong, D.C.; Wolters, F.; Kruse, E.M.; van Ham, P.M.; Wiertz, E.J.; Nijhuis, M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci. Rep. 2017, 7, 41968. [Google Scholar] [CrossRef]
- Siddell, S.G.; Walker, P.J.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch. Virol. 2019, 164, 943–946. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Ibrahim, A.; Fayez, M.; Al-Nazawi, M. From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes. J. Med. Virol. 2020, 92, 660–666. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Zhang, Y.; Pandolfi, P.P. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020, 30, 189–190. [Google Scholar] [CrossRef]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell 2020, 181, 865–876.e812. [Google Scholar] [CrossRef]
- Yousefi, B.; Eslami, M. Genetic and structure of novel coronavirus COVID-19 and molecular mechanisms in the pathogenicity of coronaviruses. Rev. Res. Med. Microbiol. 2022, 33, e180–e188. [Google Scholar] [CrossRef]
- Yousefi, B.; Valizadeh, S.; Ghaffari, H.; Vahedi, A.; Karbalaei, M.; Eslami, M. A global treatments for coronaviruses including COVID-19. J. Cell. Physiol. 2020, 235, 9133–9142. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell 2019, 76, 826–837.e811. [Google Scholar] [CrossRef]
- Wang, W.; Ye, C.; Liu, J.; Zhang, D.; Kimata, J.T.; Zhou, P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS ONE 2014, 9, e115987. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, R.; Bella, R.; Yin, C.; Otte, J.; Ferrante, P.; Gendelman, H.E.; Li, H.; Booze, R.; Gordon, J.; Hu, W. Excision of HIV-1 DNA by gene editing: A proof-of-concept in vivo study. Gene Ther. 2016, 23, 690–695. [Google Scholar] [CrossRef]
- Yu, S.; Yao, Y.; Xiao, H.; Li, J.; Liu, Q.; Yang, Y.; Adah, D.; Lu, J.; Zhao, S.; Qin, L. Simultaneous knockout of CXCR4 and CCR5 genes in CD4+ T cells via CRISPR/Cas9 confers resistance to both X4-and R5-tropic human immunodeficiency virus type 1 infection. Hum. Gene Ther. 2018, 29, 51–67. [Google Scholar] [CrossRef]
- Karimova, M.; Beschorner, N.; Dammermann, W.; Chemnitz, J.; Indenbirken, D.; Bockmann, J.-H.; Grundhoff, A.; Lüth, S.; Buchholz, F.; Wiesch, J.S.z. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci. Rep. 2015, 5, 13734. [Google Scholar] [CrossRef]
- Eslami, M.; Arjmand, N.; Mahmoudian, F.; Babaeizad, A.; Tahmasebi, H.; Fattahi, F.; Oksenych, V. Deciphering Host–Virus Interactions and Advancing Therapeutics for Chronic Viral Infection. Viruses 2025, 17, 390. [Google Scholar] [CrossRef]
- Clément, C.M.; Deffieu, M.S.; Dorobantu, C.M.; Baumert, T.F.; Ayala-Nunez, N.V.; Mely, Y.; Ronde, P.; Gaudin, R. Characterisation of endogenous Claudin-1 expression, motility and susceptibility to hepatitis C virus in CRISPR knock-in cells. Biol. Cell 2020, 112, 140–151. [Google Scholar] [CrossRef]
- Ashraf, M.U.; Salman, H.M.; Khalid, M.F.; Khan, M.H.F.; Anwar, S.; Afzal, S.; Idrees, M.; Chaudhary, S.U. CRISPR-Cas13a mediated targeting of hepatitis C virus internal-ribosomal entry site (IRES) as an effective antiviral strategy. Biomed. Pharmacother. 2021, 136, 111239. [Google Scholar] [CrossRef]
- Keikha, M.; Eslami, M.; Yousefi, B.; Ali-Hassanzadeh, M.; Kamali, A.; Yousefi, M.; Karbalaei, M. HCV genotypes and their determinative role in hepatitis C treatment. Virusdisease 2020, 31, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Lee, S.-W. Therapeutic application of genome editing technologies in viral diseases. Int. J. Mol. Sci. 2022, 23, 5399. [Google Scholar] [CrossRef]
- Zhu, W.; Xie, K.; Xu, Y.; Wang, L.; Chen, K.; Zhang, L.; Fang, J. CRISPR/Cas9 produces anti-hepatitis B virus effect in hepatoma cells and transgenic mouse. Virus Res. 2016, 217, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Khanal, S.; Cao, D.; Zhao, J.; Dang, X.; Nguyen, L.N.T.; Schank, M.; Wu, X.Y.; Jiang, Y.; et al. Synthetic gRNA/Cas9 ribonucleoprotein targeting HBV DNA inhibits viral replication. J. Med. Virol. 2023, 95, e28952. [Google Scholar] [CrossRef]
- Amini, L.; Wagner, D.L.; Rossler, U.; Zarrinrad, G.; Wagner, L.F.; Vollmer, T.; Wendering, D.J.; Kornak, U.; Volk, H.D.; Reinke, P.; et al. CRISPR-Cas9-Edited Tacrolimus-Resistant Antiviral T Cells for Advanced Adoptive Immunotherapy in Transplant Recipients. Mol. Ther. 2021, 29, 32–46. [Google Scholar] [CrossRef]
- Siegrist, C.M.; Kinahan, S.M.; Settecerri, T.; Greene, A.C.; Santarpia, J.L. CRISPR/Cas9 as an antiviral against Orthopoxviruses using an AAV vector. Sci. Rep. 2020, 10, 19307. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.C.; Chen, P.J. The potential and challenges of CRISPR-Cas in eradication of hepatitis B virus covalently closed circular DNA. Virus Res. 2018, 244, 304–310. [Google Scholar] [CrossRef]
- Moyo, B.; Bloom, K.; Scott, T.; Ely, A.; Arbuthnot, P. Advances with using CRISPR/Cas-mediated gene editing to treat infections with hepatitis B virus and hepatitis C virus. Virus Res. 2018, 244, 311–320. [Google Scholar] [CrossRef]
- Trevisan, M.; Palu, G.; Barzon, L. Genome editing technologies to fight infectious diseases. Expert Rev. Anti. Infect Ther. 2017, 15, 1001–1013. [Google Scholar] [CrossRef]
- Amalfi, S.; Molina, G.N.; Bevacqua, R.J.; Lopez, M.G.; Taboga, O.; Alfonso, V. Baculovirus Transduction in Mammalian Cells Is Affected by the Production of Type I and III Interferons, Which Is Mediated Mainly by the cGAS-STING Pathway. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef]
- Khanal, S.; Cao, D.; Zhang, J.; Zhang, Y.; Schank, M.; Dang, X.; Nguyen, L.N.T.; Wu, X.Y.; Jiang, Y.; Ning, S.; et al. Synthetic gRNA/Cas9 Ribonucleoprotein Inhibits HIV Reactivation and Replication. Viruses 2022, 14, 1902. [Google Scholar] [CrossRef]
- Chin, W.X.; Ang, S.K.; Chu, J.J. Recent advances in therapeutic recruitment of mammalian RNAi and bacterial CRISPR-Cas DNA interference pathways as emerging antiviral strategies. Drug Discov. Today 2017, 22, 17–30. [Google Scholar] [CrossRef]
- Le, T.K.; Paris, C.; Khan, K.S.; Robson, F.; Ng, W.L.; Rocchi, P. Nucleic Acid-Based Technologies Targeting Coronaviruses. Trends Biochem. Sci. 2021, 46, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Safari, F.; Afarid, M.; Rastegari, B.; Borhani-Haghighi, A.; Barekati-Mowahed, M.; Behzad-Behbahani, A. CRISPR systems: Novel approaches for detection and combating COVID-19. Virus Res. 2021, 294, 198282. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Huang, J.; Wang, T.; Fan, C.; Kang, J.; Zhang, Q.; Li, Y.; Chen, S. Ultrasensitive and visual detection of human norovirus genotype GII.4 or GII.17 using CRISPR-Cas12a assay. Virol. J. 2022, 19, 150. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Kornepati, A.V.R.; Mefferd, A.L.; Marshall, J.B.; Tsai, K.; Bogerd, H.P.; Cullen, B.R. Optimization of a multiplex CRISPR/Cas system for use as an antiviral therapeutic. Methods 2015, 91, 82–86. [Google Scholar] [CrossRef]
- Zhao, J.; Ao, C.; Wan, Z.; Dzakah, E.E.; Liang, Y.; Lin, H.; Wang, H.; Tang, S. A point-of-care rapid HIV-1 test using an isothermal recombinase-aided amplification and CRISPR Cas12a-mediated detection. Virus Res. 2021, 303, 198505. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.V.; Hwang, S.; Lee, S.W.; Jin, E.; Lee, M.H. Detection of HPV 16 and 18 L1 genes by a nucleic acid amplification-free electrochemical biosensor powered by CRISPR/Cas9. Bioelectrochemistry 2025, 162, 108861. [Google Scholar] [CrossRef]
- Koo, T.; Kim, J.S. Therapeutic applications of CRISPR RNA-guided genome editing. Brief Funct. Genom. 2017, 16, 38–45. [Google Scholar] [CrossRef]
- Binnie, A.; Fernandes, E.; Almeida-Lousada, H.; de Mello, R.A.; Castelo-Branco, P. CRISPR-based strategies in infectious disease diagnosis and therapy. Infection 2021, 49, 377–385. [Google Scholar] [CrossRef]
- Yuan, T.; Mukama, O.; Li, Z.; Chen, W.; Zhang, Y.; de Dieu Habimana, J.; Zhang, Y.; Zeng, R.; Nie, C.; He, Z.; et al. A rapid and sensitive CRISPR/Cas12a based lateral flow biosensor for the detection of Epstein-Barr virus. Analyst 2020, 145, 6388–6394. [Google Scholar] [CrossRef]
- Basu, M.; Zurla, C.; Auroni, T.T.; Vanover, D.; Chaves, L.C.S.; Sadhwani, H.; Pathak, H.; Basu, R.; Beyersdorf, J.P.; Amuda, O.O.; et al. mRNA-encoded Cas13 can be used to treat dengue infections in mice. Nat. Microbiol. 2024, 9, 2160–2172. [Google Scholar] [CrossRef] [PubMed]
- Burbelo, P.D.; Iadarola, M.J.; Chaturvedi, A. Emerging technologies for the detection of viral infections. Future Virol. 2019, 14, 39–49. [Google Scholar] [CrossRef]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef] [PubMed]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Jamehdor, S.; Pajouhanfar, S.; Saba, S.; Uzan, G.; Teimoori, A.; Naserian, S. Principles and Applications of CRISPR Toolkit in Virus Manipulation, Diagnosis, and Virus-Host Interactions. Cells 2022, 11, 999. [Google Scholar] [CrossRef]
- Au, T.Y.; Arudkumar, J.; Assavarittirong, C.; Benjamin, S. Killing two birds with one stone: CRISPR/Cas9 CCR5 knockout hematopoietic stem cells transplantation to treat patients with HIV infection and hematological malignancies concurrently. Clin. Exp. Med. 2023, 23, 4163–4175. [Google Scholar] [CrossRef]
- Sosnovtseva, A.O.; Demidova, N.A.; Klimova, R.R.; Kovalev, M.A.; Kushch, A.A.; Starodubova, E.S.; Latanova, A.A.; Karpov, D.S. Control of HSV-1 Infection: Directions for the Development of CRISPR/Cas-Based Therapeutics and Diagnostics. Int. J. Mol. Sci. 2024, 25, 2346. [Google Scholar] [CrossRef]
- Du, Y.; Liu, X.; Gao, H.; Liu, X.; Huang, M.; Chai, Q.; Xing, Z.; Zhang, T.; Ma, D. Rapid and one-tube detection of human metapneumovirus using the RT-RPA and CRISPR/Cas12a. J. Virol. Methods 2024, 329, 115001. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).