Progress in CRISPR Technology for Antiviral Treatments: Genome Editing as a Potential Cure for Chronic Viral Infections
Abstract
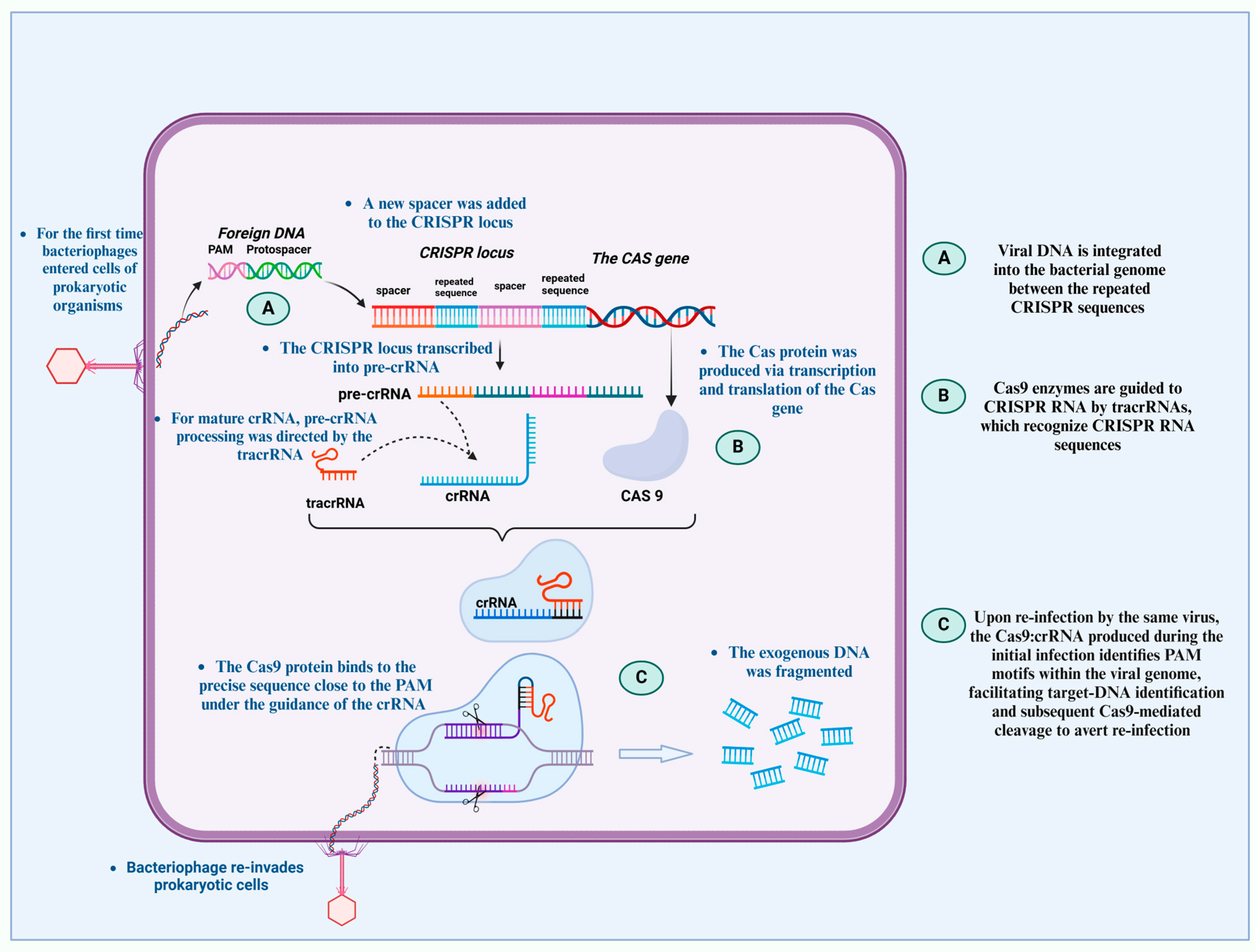
1. Overview of CRISPR Technology and Its Mechanisms in Genome Editing
2. Targeting Chronic Viral Infections with CRISPR: Current Approaches
3. CRISPR Applications in Treating Persistent Viruses
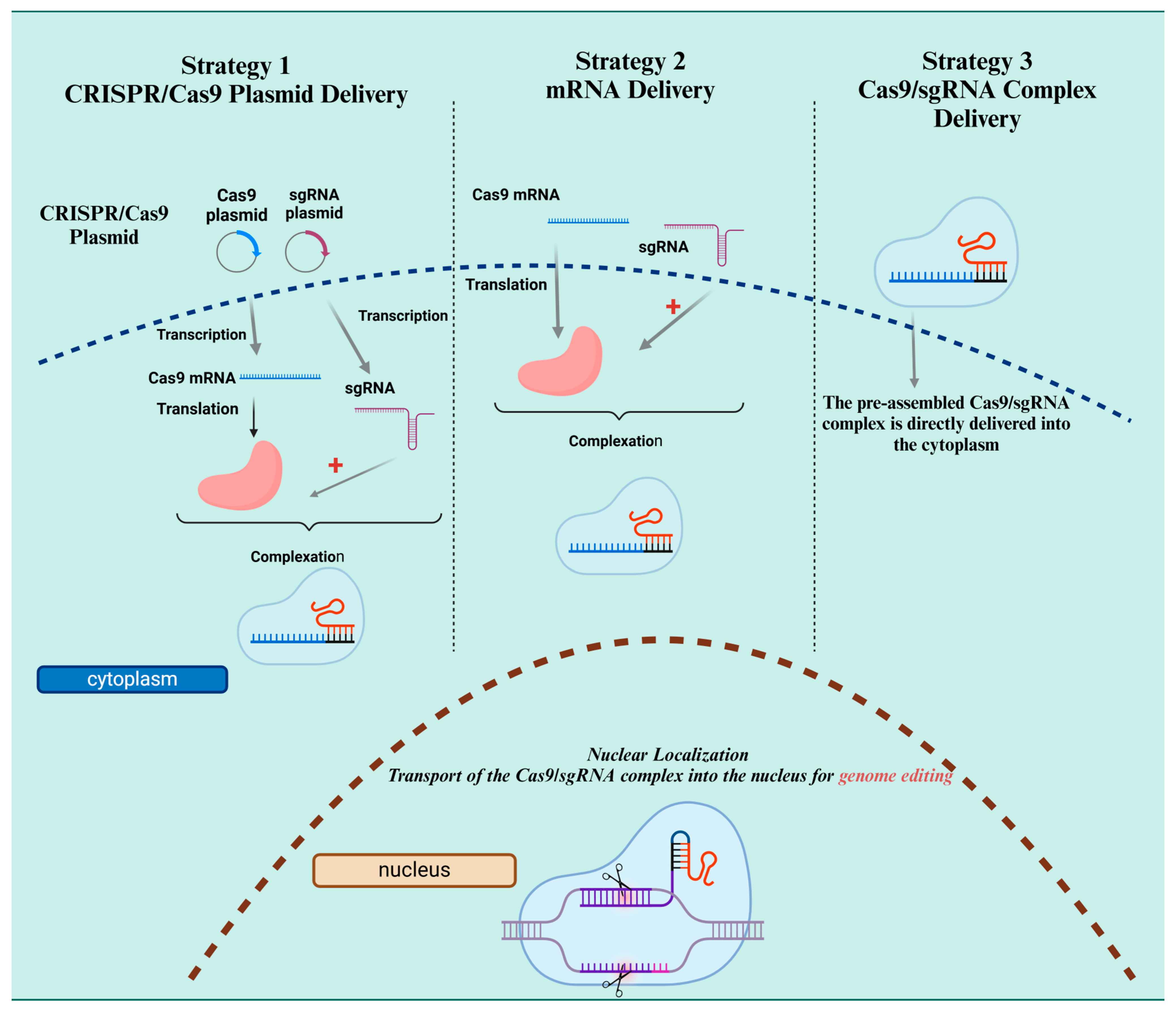
4. Advancements in CRISPR Delivery Systems for Antiviral Therapy
4.1. CRISPR–Cas9 System and Guided RNA
4.2. Cas12, Cas13, and Cleavage Method
4.3. CRISPR Nucleic Acid-Based Approaches
5. Precision, Efficiency, and Safety in CRISPR-Based Treatments
5.1. CRISPR–Cas Systems and DNA Editing
5.2. Antiretroviral Therapy in HIV
5.3. Cure HIV Using Gene Editing
6. The Future of CRISPR in Antiviral Therapeutics: From Research to Clinical Application
6.1. Diagnosis of Human Viral Infections and the CRISPR–Cas System
6.2. Detection of SARS-CoV-2
6.3. HIV Detection
6.4. Mice to Target HSV-1
6.5. CRISPR Gene Editing and Antiviral Therapy for HIV
7. Main Problems, Current Solutions, and Future Directions
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hajian, R.; Balderston, S.; Tran, T.; DeBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef]
- Rajan, A.; Shrivastava, S.; Janhawi; Kumar, A.; Singh, A.K.; Arora, P.K. CRISPR-Cas system: From diagnostic tool to potential antiviral treatment. Appl. Microbiol. Biotechnol. 2022, 106, 5863–5877. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans. 2013, 41, 1392–1400. [Google Scholar] [CrossRef]
- Nuñez, J.K.; Kranzusch, P.J.; Noeske, J.; Wright, A.V.; Davies, C.W.; Doudna, J.A. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014, 21, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Bayat, H.; Naderi, F.; Khan, A.H.; Memarnejadian, A.; Rahimpour, A. The impact of CRISPR-Cas system on antiviral therapy. Adv. Pharm. Bull. 2018, 8, 591. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Zhao, H.; Sheng, G.; Wang, M.; Yin, M.; Wang, Y. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 2015, 163, 840–853. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Perez, J.T.; Chen, C.; Li, Y.; Benitez, A.; Kandasamy, M.; Lee, Y.; Andrade, J.; Manicassamy, B. Genome-wide CRISPR/Cas9 screen identifies host factors essential for influenza virus replication. Cell Rep. 2018, 23, 596–607. [Google Scholar] [CrossRef]
- Asmamaw, M.; Zawdie, B. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biol. Targets Ther. 2021, 15, 353–361. [Google Scholar]
- Zhang, Y.; Qian, L.; Wei, W.; Wang, Y.; Wang, B.; Lin, P.; Liu, W.; Xu, L.; Li, X.; Liu, D. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth. Biol. 2017, 6, 211–216. [Google Scholar] [CrossRef]
- Yu, L.; Tian, X.; Gao, C.; Wu, P.; Wang, L.; Feng, B.; Li, X.; Wang, H.; Ma, D.; Hu, Z. Genome editing for the treatment of tumorigenic viral infections and virus-related carcinomas. Front. Med. 2018, 12, 497–508. [Google Scholar] [CrossRef]
- Dong, Z.; Qin, Q.; Hu, Z.; Chen, P.; Huang, L.; Zhang, X.; Tian, T.; Lu, C.; Pan, M. Construction of a One-Vector Multiplex CRISPR/Cas9 Editing System to Inhibit Nucleopolyhedrovirus Replication in Silkworms. Virol. Sin. 2019, 34, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Baddeley, H.J.; Isalan, M. The application of CRISPR/Cas systems for antiviral therapy. Front. Genome Ed. 2021, 3, 745559. [Google Scholar] [CrossRef]
- Maartens, G.; Celum, C.; Lewin, S.R. HIV infection: Epidemiology, pathogenesis, treatment, and prevention. Lancet 2014, 384, 258–271. [Google Scholar] [CrossRef]
- Allen, F.; Crepaldi, L.; Alsinet, C.; Strong, A.J.; Kleshchevnikov, V.; De Angeli, P.; Páleníková, P.; Khodak, A.; Kiselev, V.; Kosicki, M. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2019, 37, 64–72. [Google Scholar] [CrossRef]
- Liao, H.-K.; Gu, Y.; Diaz, A.; Marlett, J.; Takahashi, Y.; Li, M.; Suzuki, K.; Xu, R.; Hishida, T.; Chang, C.-J. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015, 6, 6413. [Google Scholar] [CrossRef] [PubMed]
- Lebbink, R.J.; de Jong, D.C.; Wolters, F.; Kruse, E.M.; van Ham, P.M.; Wiertz, E.J.; Nijhuis, M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci. Rep. 2017, 7, 41968. [Google Scholar] [CrossRef]
- Siddell, S.G.; Walker, P.J.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch. Virol. 2019, 164, 943–946. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Ibrahim, A.; Fayez, M.; Al-Nazawi, M. From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes. J. Med. Virol. 2020, 92, 660–666. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Zhang, Y.; Pandolfi, P.P. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020, 30, 189–190. [Google Scholar] [CrossRef]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell 2020, 181, 865–876.e812. [Google Scholar] [CrossRef]
- Yousefi, B.; Eslami, M. Genetic and structure of novel coronavirus COVID-19 and molecular mechanisms in the pathogenicity of coronaviruses. Rev. Res. Med. Microbiol. 2022, 33, e180–e188. [Google Scholar] [CrossRef]
- Yousefi, B.; Valizadeh, S.; Ghaffari, H.; Vahedi, A.; Karbalaei, M.; Eslami, M. A global treatments for coronaviruses including COVID-19. J. Cell. Physiol. 2020, 235, 9133–9142. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell 2019, 76, 826–837.e811. [Google Scholar] [CrossRef]
- Wang, W.; Ye, C.; Liu, J.; Zhang, D.; Kimata, J.T.; Zhou, P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS ONE 2014, 9, e115987. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, R.; Bella, R.; Yin, C.; Otte, J.; Ferrante, P.; Gendelman, H.E.; Li, H.; Booze, R.; Gordon, J.; Hu, W. Excision of HIV-1 DNA by gene editing: A proof-of-concept in vivo study. Gene Ther. 2016, 23, 690–695. [Google Scholar] [CrossRef]
- Yu, S.; Yao, Y.; Xiao, H.; Li, J.; Liu, Q.; Yang, Y.; Adah, D.; Lu, J.; Zhao, S.; Qin, L. Simultaneous knockout of CXCR4 and CCR5 genes in CD4+ T cells via CRISPR/Cas9 confers resistance to both X4-and R5-tropic human immunodeficiency virus type 1 infection. Hum. Gene Ther. 2018, 29, 51–67. [Google Scholar] [CrossRef]
- Karimova, M.; Beschorner, N.; Dammermann, W.; Chemnitz, J.; Indenbirken, D.; Bockmann, J.-H.; Grundhoff, A.; Lüth, S.; Buchholz, F.; Wiesch, J.S.z. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci. Rep. 2015, 5, 13734. [Google Scholar] [CrossRef]
- Eslami, M.; Arjmand, N.; Mahmoudian, F.; Babaeizad, A.; Tahmasebi, H.; Fattahi, F.; Oksenych, V. Deciphering Host–Virus Interactions and Advancing Therapeutics for Chronic Viral Infection. Viruses 2025, 17, 390. [Google Scholar] [CrossRef]
- Clément, C.M.; Deffieu, M.S.; Dorobantu, C.M.; Baumert, T.F.; Ayala-Nunez, N.V.; Mely, Y.; Ronde, P.; Gaudin, R. Characterisation of endogenous Claudin-1 expression, motility and susceptibility to hepatitis C virus in CRISPR knock-in cells. Biol. Cell 2020, 112, 140–151. [Google Scholar] [CrossRef]
- Ashraf, M.U.; Salman, H.M.; Khalid, M.F.; Khan, M.H.F.; Anwar, S.; Afzal, S.; Idrees, M.; Chaudhary, S.U. CRISPR-Cas13a mediated targeting of hepatitis C virus internal-ribosomal entry site (IRES) as an effective antiviral strategy. Biomed. Pharmacother. 2021, 136, 111239. [Google Scholar] [CrossRef]
- Keikha, M.; Eslami, M.; Yousefi, B.; Ali-Hassanzadeh, M.; Kamali, A.; Yousefi, M.; Karbalaei, M. HCV genotypes and their determinative role in hepatitis C treatment. Virusdisease 2020, 31, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Lee, S.-W. Therapeutic application of genome editing technologies in viral diseases. Int. J. Mol. Sci. 2022, 23, 5399. [Google Scholar] [CrossRef]
- Zhu, W.; Xie, K.; Xu, Y.; Wang, L.; Chen, K.; Zhang, L.; Fang, J. CRISPR/Cas9 produces anti-hepatitis B virus effect in hepatoma cells and transgenic mouse. Virus Res. 2016, 217, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Khanal, S.; Cao, D.; Zhao, J.; Dang, X.; Nguyen, L.N.T.; Schank, M.; Wu, X.Y.; Jiang, Y.; et al. Synthetic gRNA/Cas9 ribonucleoprotein targeting HBV DNA inhibits viral replication. J. Med. Virol. 2023, 95, e28952. [Google Scholar] [CrossRef]
- Amini, L.; Wagner, D.L.; Rossler, U.; Zarrinrad, G.; Wagner, L.F.; Vollmer, T.; Wendering, D.J.; Kornak, U.; Volk, H.D.; Reinke, P.; et al. CRISPR-Cas9-Edited Tacrolimus-Resistant Antiviral T Cells for Advanced Adoptive Immunotherapy in Transplant Recipients. Mol. Ther. 2021, 29, 32–46. [Google Scholar] [CrossRef]
- Siegrist, C.M.; Kinahan, S.M.; Settecerri, T.; Greene, A.C.; Santarpia, J.L. CRISPR/Cas9 as an antiviral against Orthopoxviruses using an AAV vector. Sci. Rep. 2020, 10, 19307. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.C.; Chen, P.J. The potential and challenges of CRISPR-Cas in eradication of hepatitis B virus covalently closed circular DNA. Virus Res. 2018, 244, 304–310. [Google Scholar] [CrossRef]
- Moyo, B.; Bloom, K.; Scott, T.; Ely, A.; Arbuthnot, P. Advances with using CRISPR/Cas-mediated gene editing to treat infections with hepatitis B virus and hepatitis C virus. Virus Res. 2018, 244, 311–320. [Google Scholar] [CrossRef]
- Trevisan, M.; Palu, G.; Barzon, L. Genome editing technologies to fight infectious diseases. Expert Rev. Anti. Infect Ther. 2017, 15, 1001–1013. [Google Scholar] [CrossRef]
- Amalfi, S.; Molina, G.N.; Bevacqua, R.J.; Lopez, M.G.; Taboga, O.; Alfonso, V. Baculovirus Transduction in Mammalian Cells Is Affected by the Production of Type I and III Interferons, Which Is Mediated Mainly by the cGAS-STING Pathway. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef]
- Khanal, S.; Cao, D.; Zhang, J.; Zhang, Y.; Schank, M.; Dang, X.; Nguyen, L.N.T.; Wu, X.Y.; Jiang, Y.; Ning, S.; et al. Synthetic gRNA/Cas9 Ribonucleoprotein Inhibits HIV Reactivation and Replication. Viruses 2022, 14, 1902. [Google Scholar] [CrossRef]
- Chin, W.X.; Ang, S.K.; Chu, J.J. Recent advances in therapeutic recruitment of mammalian RNAi and bacterial CRISPR-Cas DNA interference pathways as emerging antiviral strategies. Drug Discov. Today 2017, 22, 17–30. [Google Scholar] [CrossRef]
- Le, T.K.; Paris, C.; Khan, K.S.; Robson, F.; Ng, W.L.; Rocchi, P. Nucleic Acid-Based Technologies Targeting Coronaviruses. Trends Biochem. Sci. 2021, 46, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Safari, F.; Afarid, M.; Rastegari, B.; Borhani-Haghighi, A.; Barekati-Mowahed, M.; Behzad-Behbahani, A. CRISPR systems: Novel approaches for detection and combating COVID-19. Virus Res. 2021, 294, 198282. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Huang, J.; Wang, T.; Fan, C.; Kang, J.; Zhang, Q.; Li, Y.; Chen, S. Ultrasensitive and visual detection of human norovirus genotype GII.4 or GII.17 using CRISPR-Cas12a assay. Virol. J. 2022, 19, 150. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Kornepati, A.V.R.; Mefferd, A.L.; Marshall, J.B.; Tsai, K.; Bogerd, H.P.; Cullen, B.R. Optimization of a multiplex CRISPR/Cas system for use as an antiviral therapeutic. Methods 2015, 91, 82–86. [Google Scholar] [CrossRef]
- Zhao, J.; Ao, C.; Wan, Z.; Dzakah, E.E.; Liang, Y.; Lin, H.; Wang, H.; Tang, S. A point-of-care rapid HIV-1 test using an isothermal recombinase-aided amplification and CRISPR Cas12a-mediated detection. Virus Res. 2021, 303, 198505. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.V.; Hwang, S.; Lee, S.W.; Jin, E.; Lee, M.H. Detection of HPV 16 and 18 L1 genes by a nucleic acid amplification-free electrochemical biosensor powered by CRISPR/Cas9. Bioelectrochemistry 2025, 162, 108861. [Google Scholar] [CrossRef]
- Koo, T.; Kim, J.S. Therapeutic applications of CRISPR RNA-guided genome editing. Brief Funct. Genom. 2017, 16, 38–45. [Google Scholar] [CrossRef]
- Binnie, A.; Fernandes, E.; Almeida-Lousada, H.; de Mello, R.A.; Castelo-Branco, P. CRISPR-based strategies in infectious disease diagnosis and therapy. Infection 2021, 49, 377–385. [Google Scholar] [CrossRef]
- Yuan, T.; Mukama, O.; Li, Z.; Chen, W.; Zhang, Y.; de Dieu Habimana, J.; Zhang, Y.; Zeng, R.; Nie, C.; He, Z.; et al. A rapid and sensitive CRISPR/Cas12a based lateral flow biosensor for the detection of Epstein-Barr virus. Analyst 2020, 145, 6388–6394. [Google Scholar] [CrossRef]
- Basu, M.; Zurla, C.; Auroni, T.T.; Vanover, D.; Chaves, L.C.S.; Sadhwani, H.; Pathak, H.; Basu, R.; Beyersdorf, J.P.; Amuda, O.O.; et al. mRNA-encoded Cas13 can be used to treat dengue infections in mice. Nat. Microbiol. 2024, 9, 2160–2172. [Google Scholar] [CrossRef] [PubMed]
- Burbelo, P.D.; Iadarola, M.J.; Chaturvedi, A. Emerging technologies for the detection of viral infections. Future Virol. 2019, 14, 39–49. [Google Scholar] [CrossRef]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef] [PubMed]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Jamehdor, S.; Pajouhanfar, S.; Saba, S.; Uzan, G.; Teimoori, A.; Naserian, S. Principles and Applications of CRISPR Toolkit in Virus Manipulation, Diagnosis, and Virus-Host Interactions. Cells 2022, 11, 999. [Google Scholar] [CrossRef]
- Au, T.Y.; Arudkumar, J.; Assavarittirong, C.; Benjamin, S. Killing two birds with one stone: CRISPR/Cas9 CCR5 knockout hematopoietic stem cells transplantation to treat patients with HIV infection and hematological malignancies concurrently. Clin. Exp. Med. 2023, 23, 4163–4175. [Google Scholar] [CrossRef]
- Sosnovtseva, A.O.; Demidova, N.A.; Klimova, R.R.; Kovalev, M.A.; Kushch, A.A.; Starodubova, E.S.; Latanova, A.A.; Karpov, D.S. Control of HSV-1 Infection: Directions for the Development of CRISPR/Cas-Based Therapeutics and Diagnostics. Int. J. Mol. Sci. 2024, 25, 2346. [Google Scholar] [CrossRef]
- Du, Y.; Liu, X.; Gao, H.; Liu, X.; Huang, M.; Chai, Q.; Xing, Z.; Zhang, T.; Ma, D. Rapid and one-tube detection of human metapneumovirus using the RT-RPA and CRISPR/Cas12a. J. Virol. Methods 2024, 329, 115001. [Google Scholar] [CrossRef]

| Virus Type | Viral Genome | CRISPR System Used | Target Region(s) | Mechanism of Action |
|---|---|---|---|---|
| HIV-1 | ssRNA (retrovirus) → dsDNA (provirus) | Cas9 | 5′- and 3′-LTRs, gag, tat, rev, CCR5 | DSBs in proviral DNA or host receptor genes to inhibit integration or entry |
| Influenza A | ssRNA (-) segmented | Cas13a/Cas13b | PB1, HA, NA, NP | RNA cleavage via collateral activity to inhibit replication |
| Hepatitis B (HBV) | dsDNA (cccDNA) | Cas9/Cas9n | S, X, P, C genes | DSBs or base editing of cccDNA to silence replication |
| Hepatitis C (HCV) | ssRNA (+) | Cas13a | Internal ribosome entry site (IRES) | Direct RNA cleavage to block translation and replication |
| Enteroviruses | ssRNA (+) | Cas13 | Conserved regions in 5′ UTR or polymerase gene | RNA degradation to prevent genome replication |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nouri, F.; Alibabaei, F.; Forouzanmehr, B.; Tahmasebi, H.; Oksenych, V.; Eslami, M. Progress in CRISPR Technology for Antiviral Treatments: Genome Editing as a Potential Cure for Chronic Viral Infections. Microbiol. Res. 2025, 16, 104. https://doi.org/10.3390/microbiolres16050104
Nouri F, Alibabaei F, Forouzanmehr B, Tahmasebi H, Oksenych V, Eslami M. Progress in CRISPR Technology for Antiviral Treatments: Genome Editing as a Potential Cure for Chronic Viral Infections. Microbiology Research. 2025; 16(5):104. https://doi.org/10.3390/microbiolres16050104
Chicago/Turabian StyleNouri, Fatemeh, Farnaz Alibabaei, Behina Forouzanmehr, Hamed Tahmasebi, Valentyn Oksenych, and Majid Eslami. 2025. "Progress in CRISPR Technology for Antiviral Treatments: Genome Editing as a Potential Cure for Chronic Viral Infections" Microbiology Research 16, no. 5: 104. https://doi.org/10.3390/microbiolres16050104
APA StyleNouri, F., Alibabaei, F., Forouzanmehr, B., Tahmasebi, H., Oksenych, V., & Eslami, M. (2025). Progress in CRISPR Technology for Antiviral Treatments: Genome Editing as a Potential Cure for Chronic Viral Infections. Microbiology Research, 16(5), 104. https://doi.org/10.3390/microbiolres16050104






