Abstract
Since the emergence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the viral spike protein (S) has become a target to describe appropriate epitopes for vaccine development and to carry out epidemiological surveillance, especially regarding the variants of concern (VOCs). This study aimed to evaluate the influence of mutations on physicochemical properties of S proteins from prototypical SARS-CoV-2 VOCs detected in Amazonian countries. Using multiple computational tools, seven VOCs (B.1.1.7/P.1/B.1.617.2/BA.1/BA.2/BA.4/BA.5) were identified and compared to the ancestral lineage of the virus (B). In all variants, most amino acids were nonpolar; among the polar amino acids, B.1.617.2/BA.1/BA.2/BA.4/BA.5 presented a slightly higher proportion of basic residues and a lower proportion of neutral residues. Unlike B.1.1.7/P.1/B.1.617.2, BA.1/BA.2 had a greater content of secondary structures, such as α-helices and β-sheets. Regarding post-translational modifications, BA.2/BA.4/BA.5 presented fewer glycosylations and phosphorylations. Finally, a more prominent antigenic propensity in the N-terminal domain of BA.2/BA.4/BA.5 and in the receptor-binding domain of B.1.617.2/BA.4/BA.5 was observed. In conclusion, the omicron variants of SARS-CoV-2 presented greater sequence variability in S proteins compared to the other VOCs, influencing structural aspects that can potentially modulate its interaction with cellular receptors and recognition by the immune system.
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is the seventh coronavirus known to have the capacity to infect humans and has spread rapidly throughout several regions of the world, culminating in the classification of coronavirus disease 2019 (COVID-19) as a pandemic by the World Health Organization (WHO) in early 2020 [1]. The virion is an enveloped particle 80 to 120 nm in diameter, with spiky projections that give it a crown shape. Its genome is made up of a single positive-sense single-stranded ribonucleic acid molecule (ssRNA+), capable of encoding at least 29 different proteins. Of these, four—S, E, M, and N—are part of the viral structure [2].
The S protein is responsible for binding the virus to the host cell through its interaction with the angiotensin-converting enzyme 2 (ACE2) and promoting the membrane fusion reaction. Due to its location on the viral surface, this transmembrane protein is the main target of neutralizing antibodies and the focus of studies for vaccine development [3]. It spans approximately 1273 residues, divided into 2 functional subunits: S1, in the N-terminal half, and S2, in the C-terminal half. The former comprises a N-terminal domain (NTD, residues 13–305) and a receptor-binding domain (RBD, residues 319–541), while the latter contains a fusion peptide (FP, residues 788–806), the heptad repeat 1 (HR1, residues 912–984), a central helix (CH, residues 987–1035), a connector domain (CD, residues 1076–1141), the heptad repeat 2 (HR2, residues 1163–1211), a transmembrane domain (TM, residues 1213–1237), and a cytoplasmic tail (CT, residues 1238–1273) [4].
As viral strains posing an increased risk to global public health emerged in late 2020, the WHO began classifying them as variants under monitoring (VUMs), variants of interest (VOIs), or variants of concern (VOCs), in order to prioritize global monitoring and research [5]. In total, five VOCs have already been identified, namely, alpha (B.1.1.7), beta (B.1.351), gamma (P.1), delta (B.1.617.2), and more recently, omicron (BA.1, BA.2, BA.3, BA.4, and BA.5), originally detected in the United Kingdom, South Africa, Brazil, India, and Botswana, respectively [6].
Each of these VOCs raised significant public health challenges. For instance, the delta variant demonstrated higher transmission rates and severity of illness, leading to surges in hospitalizations [7]. The omicron variant, with its multiple sub-lineages, also showed considerable immune escape capabilities, requiring booster doses to maintain vaccine effectiveness [8]. These challenges underscore the need for continuous genomic surveillance, rapid adaptability of vaccination strategies, and persistent public health interventions to mitigate the spread and impact of emerging SARS-CoV-2 variants [9].
In the Amazon basin, which extends across nine countries in South America (Brazil, Peru, Colombia, Bolivia, Venezuela, Guyana, Suriname, Ecuador, and French Guiana), dynamics of COVID-19 cases as well as SARS-CoV-2 VOC prevalence have varied both spatially and temporally, even though omicron variants have become ubiquitous more recently due to accumulated mutations rendering increased transmissibility and potential for antibody evasion [10]. Based on computational tools, the present study aimed to evaluate the influence of these mutations on physicochemical properties of S proteins from prototypical SARS-CoV-2 VOCs detected in Amazonian countries.
2. Materials and Methods
2.1. Retrieval of Sequences
Amino acid sequences of SARS-CoV-2 S protein were retrieved in FASTA format from the NCBI Virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/, accessed on 27 March 2024), including only ancestral and VOC prototypical findings in human samples from the nine Amazonian countries. Variants not classified according to the Pango nomenclature system and whose S protein amino acid sequences were partial and/or contained unresolved amino acids (B/Z/J/X) were excluded. This activity of accessing genetic heritage was registered in the National System for the Management of Genetic Heritage and Associated Traditional Knowledge (SisGen) under number ACA630E, in compliance with Brazilian Law 13,123/2015 and its regulations.
2.2. Multiple Sequence Alignment
Recognition of amino acid substitutions and insertions/deletions as well as determination of percent identity were carried out by multiple sequence alignment using seeded guide trees and hidden Markov model profile-profile techniques on the Clustal Omega program, with the output format set to ClustalW with character counts. Based on a distance matrix, the tool constructs a guide tree that represents the relationships among the sequences. Starting with the closest sequences (i.e., those with the smallest evolutionary distance), Clustal Omega aligns them in a stepwise manner, progressively adding more sequences according to the guide tree and creating profiles that summarize the aligned sequences. Subsequent sequences are then aligned to these profiles, rather than to individual sequences, improving the accuracy of the alignment [11].
2.3. Sequence Statistics Assessment
Lengths, molecular weights, isoelectric points, and side-chain polarities were estimated by sequence statistics on the EMBOSS Pepstats program, configured to include termini charges (i.e., charges from the N-terminus and C-terminus) and not use monoisotopic weights (i.e., weights from the most abundant isotopes of each amino acid) when performing calculations. EMBOSS Pepstats analyzes protein sequences to compute various statistical and physicochemical properties by performing specific calculations for each property. The length of the protein is achieved by simply counting the total number of amino acids in the sequence. The molecular weight is calculated by summing the masses of all amino acids in the sequence, including the average mass of water released during peptide bond formation. The isoelectric point is obtained by finding the pH at which the protein has no net charge, which involves calculating the charge of the protein at different pH values, taking into account the pKa values of the ionizable groups in the amino acids. The polarity of side chains is determined by categorizing each amino acid based on its chemical properties as non-polar or polar (neutral, basic, or acidic), then counting the number of amino acids in each category [12].
2.4. Prediction of Secondary Structures
The content of secondary structures was predicted in three structural stages (α-helix, β-sheet, and random coil) using an accurate algorithm for recognition of potentially hydrogen-bonded residues in the amino acid sequences [13] on the NPS@ PREDATOR program, setting the output width to 70 and the secondary structure data to DSSP. Based on observed patterns of known protein conformations, NPS@ PREDATOR considers the propensity of certain amino acids to form specific secondary structures and the influence of neighboring residues. The algorithm predicts the likelihood of each segment of the sequence forming an α-helix, β-sheet, or random coil, generating a predicted secondary structure profile for the entire protein [14].
2.5. Identification of Post-Translational Modification Sites
Consensus patterns of post-translational modifications were identified by scanning the amino acid sequences against the PROSITE database (https://www.expasy.org/resources/prosite, accessed on 27 March 2024) on the NPS@ PROSCAN program, with the similarity level set to no mismatch (i.e., 100% similarity). Using pattern-matching algorithms, NPS@ PROSCAN scans the input sequence for regions that match these consensus patterns, predicting possible sites for modifications, such as N-glycosylation (N–{P}–[ST]–{P}, where N is the glycosylation site), cAMP- and cGMP-dependent protein kinase (PKA/G) phosphorylation ([RK](2)–X–[ST], where S or T is the phosphorylation site), protein kinase C (PKC) phosphorylation ([ST]–X–[RK], where S or T is the phosphorylation site), casein kinase II (CK2) phosphorylation ([ST]–X(2)–[DE], where S or T is the phosphorylation site), and tyrosine kinase (TK) phosphorylation ([RK]–X(2)–[DE]–X(3)–Y or [RK]–X(3)–[DE]–X(2)–Y, where Y is the phosphorylation site). The tool outputs a list of predicted post-translational modification sites along with their positions in the sequence and the type of modification expected [14].
2.6. Determination of Antigenicity
Antigenic determinants were located along polypeptide chains based on the combination of hydrophilicity, accessibility, and flexibility parameters that make up the Parker antigenicity scale [15] on the NPS@ PCPROF program, whose window size was configured to 7. The algorithm uses a sliding window approach to scan the protein sequence, calculating composite scores of hydrophilicity (as regions with higher affinity for an aqueous environment are more likely to be antigenic), accessibility (as parts of the protein likely to be exposed to the external environment are accessible to the immune system), and flexibility (as mobile regions tend to be antigenic due to their ability to undergo some degree of conformational adjustments during recognition by antibodies) for overlapping segments of the input sequence. Based on the combined scores, NPS@ PCPROF identifies regions with high antigenic potential, indicating segments that are likely to be recognized by the adaptive immune system [14].
2.7. Hypothesis Testing
The collected data were grouped as “non-omicron” (B, B.1.1.7, P.1, and B.1.617.2) or “omicron” (BA.1, BA.2, BA.4, and BA.5) by mean with standard deviation (SD) and subjected to unpaired t tests with Bonferroni correction for multiple comparisons on Excel 365 (Microsoft, Redmond, WA, USA). Statistically significant differences were assigned to corrected p-values less than or equal to 0.05.
3. Results
3.1. SARS-CoV-2 VOCs Prototypically Detected in Amazonian Countries
Seven SARS-CoV-2 VOCs were identified in Amazonian countries, including the variants B.1.1.7 (alpha), P.1 (gamma), B.1.617.2 (delta), BA.1 (omicron), BA.2 (omicron), BA.4 (omicron), and BA.5 (omicron), which were compared to the lineage B (ancestral). Apart from VOCs B.1.1.7 and BA.2 (prototypically detected in Peru and Colombia, respectively), all other VOCs were prototypically detected in Brazil. Such detections occurred from human biological material collected between February 2020 and July 2022 (Table 1).

Table 1.
Metadata of source samples for the amino acid sequences of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries.
3.2. Sequence Identities of S Proteins
The percent identity of the amino acid sequences of S proteins from SARS-CoV-2 VOCs was lower between the P.1 and BA.1 variants (97.16%) and higher between the BA.4 and BA.5 variants (99.84%; Figure 1). Taking the lineage B as a reference, amino acid insertions in the BA.1 variant and amino acid deletions in the B.1.1.7, BA.1, BA.2, BA.4, and BA.5 variants were noted in the NTD, and the last four featured most of the amino acid substitutions found in the molecule, especially in the RBD (Figure S1). TM was the only domain where no mutations were detected across all VOCs (Figure S2).
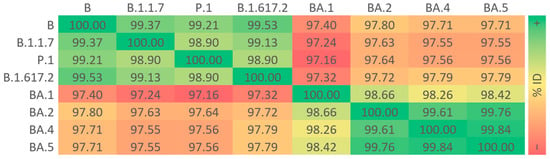
Figure 1.
Sequence identities of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries. Query proteins were subjected to multiple sequence alignment to generate a percent identity matrix of their comprising residues, using a red–yellow–green color gradient to express values from lowest to highest.
3.3. Lengths, Molecular Weights, and Isoelectric Points of S Proteins
The lengths of S proteins varied depending on the SARS-CoV-2 lineage considered, with 1273 residues in B, P.1, and B.1.617.2, 1270 residues in B.1.1.7, BA.1, and BA.2, and 1268 residues in BA.4 and BA.5. Despite having similar molecular weights (around 141 kDa), the isoelectric points of these proteins differed substantially, being below neutral pH in non-omicron lineages (6.61–6.97) and above neutral pH in omicron lineages (7.18–7.28; Table 2).

Table 2.
Lengths, molecular weights, and isoelectric points of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries.
3.4. Side-Chain Polarities of S Proteins
As for side-chain polarities, in all SARS-CoV-2 lineages, most amino acids (~55 mol%) were non-polar; among the polar amino acids, a predominance of neutral over charged (basic and acidic) was observed. When considered as a group, omicron variants presented a lower proportion of neutral polar amino acids (26.48 vs. 27.12 mol%) and a higher proportion of basic polar amino acids (9.99 vs. 9.47 mol%), both statistically significant, in relation to non-omicron variants. No statistically significant differences were observed regarding the proportions of non-polar and acidic polar amino acids between these two groups of SARS-CoV-2 variants (Figure 2).

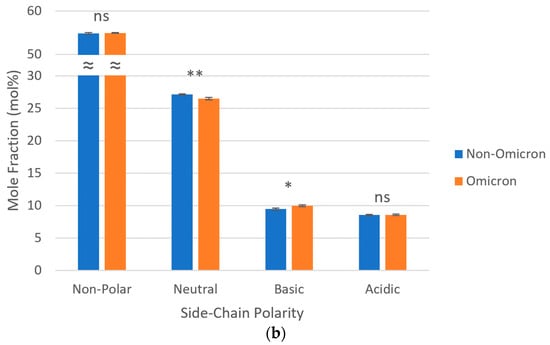
Figure 2.
Side-chain polarities of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries. Query proteins were subjected to sequence statistics analysis and residue properties were expressed as mole fractions either (a) separately (ruler units in mol%) or (b) grouped (ns p > 0.05; * 0.01 < p ≤ 0.05; ** 0.001 < p ≤ 0.01; ≈ y-axis break). Thin gray lines, horizontal gridlines.
3.5. Secondary Structures of S Proteins
Regarding secondary structures, S proteins of all SARS-CoV-2 lineages were predicted to be predominantly composed of random coils (~57%), followed by α-helices (~23%) and β-sheets (~20%). Analyzing the data in a grouped manner, omicron variants revealed, in a statistically significant way, a higher proportion of β-sheets (20.45 vs. 19.94%) and a lower proportion of random coils (56.66 vs. 57.34%) when compared to non-omicron variants. No statistically significant difference was noted in relation the proportions of α-helices between these two groups of SARS-CoV-2 variants (Figure 3).

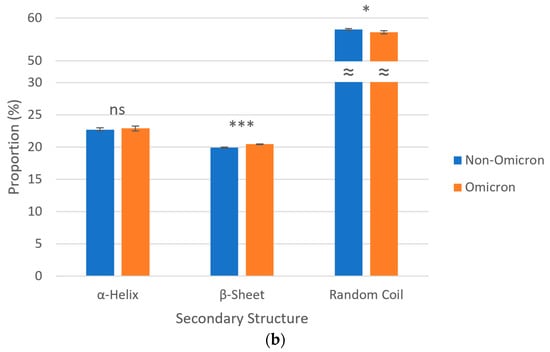
Figure 3.
Secondary structures of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries. Query proteins were subjected to recognition of potentially hydrogen-bonded residues in their amino acid sequences and secondary structures were expressed as proportions either (a) separately (ruler units in %) or (b) grouped (ns p > 0.05; * 0.01 < p ≤ 0.05; *** 0.0001 < p ≤ 0.001; ≈ y-axis break). Thin gray lines, horizontal gridlines.
3.6. Post-Translational Modifications of S Proteins
About post-translational modifications, in all S protein sequences studied, consensus patterns were identified for N-glycosylations and phosphorylations by PKA/G, PKC, CK2, and TK. Phosphorylations were collectively predominant over glycosylations in all SARS-CoV-2 lineages, but no statistically significant differences were observed for the occurrences of both groups of post-translational modifications between omicron (1.67 and 2.40 sites/100 amino acids, respectively) and non-omicron (1.75 and 2.48 sites/per 100 amino acids, respectively) variants (Figure 4). Both groups of post-translational modifications occurred along the two subunits of the protein (Figures S1 and S2).

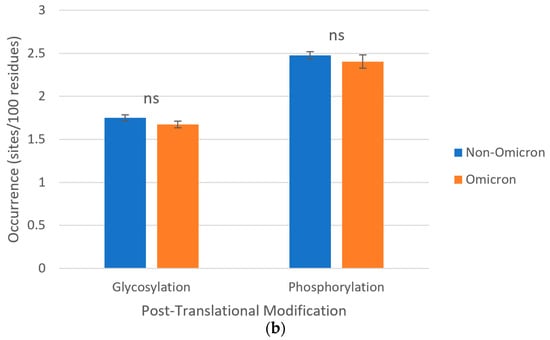
Figure 4.
Post-translational modifications of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries. Query proteins were scanned for consensus patterns in their amino acid sequences and post-translational modifications were expressed as relative occurrences either (a) separately (ruler units in sites/100 residues) or (b) grouped (ns p > 0.05). “Glycosylation”, total number of consensus patterns for N-glycosylation; “Phosphorylation”, total number of consensus patterns for PKA/G, PKC, CK2, and TK phosphorylations. Thin gray lines, horizontal gridlines.
3.7. Antigenicity of S Proteins
Concerning antigenic propensities, the VOCs BA.2, BA.4, and BA.5 presented relatively pronounced scores in the NTD of their respective S proteins, close to amino acid position 27. Furthermore, the last two variants, together with the VOC B.1.617.2, also demonstrated a high relative value for this parameter in the RBD of their respective S proteins, around amino acid position 452. These were precisely the positions with the highest SD of Parker antigenicity (Figure 5).
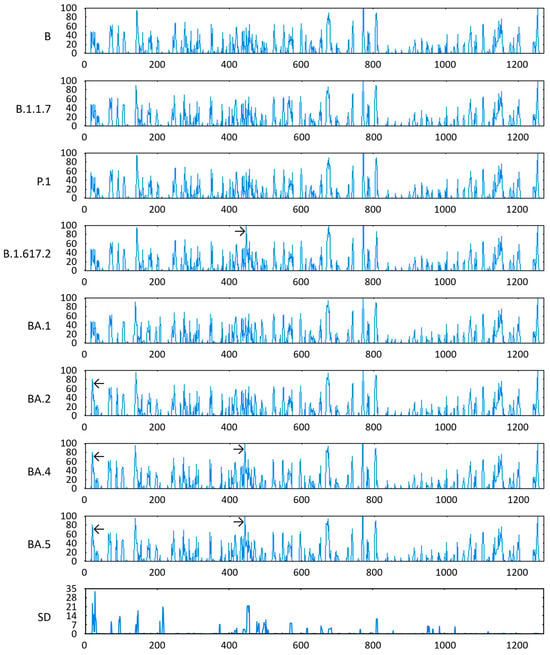
Figure 5.
Antigenicity of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries. Query proteins were evaluated for antigenic propensity based on the combination of hydrophilicity, accessibility, and flexibility parameters, using the Parker scale (y-axes) to express scores and their SD along amino acid positions in the aligned polypeptide chains (x-axes). ← Relatively pronounced scores in the NTD, and → relatively pronounced scores in the RBD.
4. Discussion
The emergence of SARS-CoV-2 omicron variants has raised significant concerns due to their unprecedented level of sequence variability achieved by successive point mutations, particularly within the open reading frame encoding the S protein [16]. The findings of the current study reinforce this substantial diversity in relation to other VOCs and show that it has potential effects on the physicochemical properties of this protein.
Although the prototypical SARS-CoV-2 VOCs considered here turned out to be limited to just a third of the Amazonian countries (i.e., Brazil, Peru, and Colombia), these were precisely the countries with the largest Amazon tree cover (60, 12, and 8%, respectively) [17]. Most of the amino acid mutations identified in the S protein of these query sequences occurred in the NTD and RBD, in line with a previous study indicating that the diversity of these two domains exceeded that of any other regions of the molecule [18].
The occurrence of a greater content of non-polar and neutral polar amino acids in the S protein of all SARS-CoV-2 variants prototypically detected in Amazonian countries agrees with a past work showing that leucine, valine, threonine, alanine, and serine are the top-five abundant amino acid residues in the S protein of 11 SARS-CoV-2 variants [19]. As demonstrated earlier, mutations in the genome of eight major SARS-CoV-2 lineages have favored a higher frequency of non-polar amino acid codons, especially the C > U and G > U substitutions, thus increasing the overall hydrophobicity of viral proteins. This change may be advantageous for the virus, as it potentially promotes prolonged mucosal colonization and persistence, minor distribution in plasma, and greater stability in aqueous cytosol [20]. Furthermore, the increase in basic polar residues and the decrease in neutral polar residues observed for omicron VOCs in the current study agrees with a previously published dataset, showing that five substitutions in the S protein RBD of these variants use positively charged amino acids, which change the properties of the interface with the receptor, whereas the ancestral lineage and the other SARS-CoV-2 VOCs present uncharged amino acids at such interface [21].
Regarding secondary structures, an in silico investigation on S proteins from SARS-CoV-2 detected in Asian, African, North American, South American, European, and Oceanian countries also found a prevalence of random coils, followed by α-helices and β-sheets, although with slightly different proportions (43.9, 29.3, and 23.3%, respectively) to those found here [22]. As previously shown, these three types of secondary structures have different mutation rates in the SARS-CoV-2 S protein. Random coils are major components of the RBD and harbor most of the mutations in this domain. Those occurring in α-helix or β-sheet residues usually lead to positive changes in the binding free energy of the S-ACE2 complex, indicating infectivity strengthening [23]. Since secondary structures, particularly α-helices and β-sheets, play a crucial role in determining protein stability, the conversion of random coils to β-sheets observed here in omicron variants may result in the formation of a more stable connection with the receptor [24].
The current study showed a prevalence of phosphorylations followed by glycosylations in the S protein of all SARS-CoV-2 variants prototypically detected in Amazonian countries. These post-translational modifications are possibly involved in the modulation of S protein dynamics, as sugars seem to increase protein flexibility, thus contributing to receptor recognition [25], while residues attached to phosphate groups are potential control points for S protein conformational changes [26]. However, the occurrence of post-translational modifications is influenced by a higher-order protein structure and cellular context, meaning that not all predicted sites will be modified in a real protein and highlighting the necessity for further experimental validation to confirm these computational findings [27].
Particularly, the increment in antigenicity predicted for the NTD of the VOCs BA.2, BA.4, and BA.5, as well as the RBD of the VOCs B.1.617.2, BA.4, and BA.5, is likely related to the mutations A27S and L452R, respectively, both resulting in the replacement of a non-polar residue with a polar one. Intriguingly, these mutations appear to be linked to immune escape or viral oligomerization [28]. Along with other amino acid substitutions in these domains, they may have led to structural and functional changes in the SARS-CoV-2 S protein that contributed to sustained virus transmission throughout the COVID-19 pandemic, as interactions between the NTD and the RBD affect receptor affinity and antibody evasion [29]. Indeed, an NTD–RBD-linked candidate vaccine showed improved antigen expression and antibody responses compared with a clinically available vaccine encoding the full-length S protein [30].
The structural changes observed in the S protein of SARS-CoV-2 VOCs may have profound implications for its interaction with cellular receptors. Amino acid mutations predominantly clustered in the NTD and the RBD are likely to influence protein conformation and binding affinity. The shift toward a higher proportion of basic polar residues and decreased neutral polar residues in omicron VOCs could enhance electrostatic interactions with host cell receptors, potentially affecting viral entry efficiency. The prevalence of random coils over α-helices and β-sheets across all S protein variants indicates structural flexibility that may impact receptor-binding dynamics. Moreover, the frequent phosphorylations and glycosylations observed in the S protein of all SARS-CoV-2 variants likely modulate immune recognition and viral evasion strategies. Notably, the increased antigenicity predicted for specific domains of certain VOCs further highlights the evolutionary pressure driving variant-specific changes that may impact viral transmission and pathogenicity.
Overall, the findings of this study have several potential applications in the ongoing efforts to better manage and mitigate the impact of COVID-19. First, understanding the major locations of amino acid mutations in the S protein of SARS-CoV-2 VOCs can inform targeted vaccine design to elicit a robust immune response. Second, recognition of the shift in side-chain polarity of omicron VOCs provides insights into viral adaptation that can aid in designing drugs that interact more effectively with the S protein. Third, identification of the predominant secondary structure in each variant may guide strategies to destabilize the S protein and inhibit viral entry. Fourth, prediction of critical post-translational modifications across all variants favors the development of therapies aimed at interfering with their functions during the infection cycle. Finally, the locally elevated antigenic propensity in specific VOCs suggests that these regions are key targets for neutralizing antibodies, providing crucial information for updating vaccines and antibody therapies to maintain efficacy against emerging SARS-CoV-2 variants.
5. Conclusions
The investigation presented herein underscores the distinct nature of the omicron variants of SARS-CoV-2, characterized by heightened sequence variability within their S proteins, surpassing that observed in other VOCs prototypically detected in Amazonian countries. This increased variability is poised to exert significant modulations on such structural proteins, potentially altering critical epitopes responsible for mediating viral entry into host cells and eliciting immune responses. The intricate interplay between these structural modifications and their impact on viral fitness, transmissibility, and immune evasion underscores the dynamic essence of SARS-CoV-2 evolution. Understanding these molecular nuances is imperative for devising effective public health strategies, including vaccine development and therapeutic interventions, to lessen the spread and severity of COVID-19.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres15030090/s1. Figure S1: Multiple sequence alignment with potential post-translational modification sites of the S1 subunit of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries. Figure S2: Multiple sequence alignment with potential post-translational modification sites of the S2 subunit of S proteins from the ancestral lineage and SARS-CoV-2 VOCs prototypically detected in Amazonian countries.
Author Contributions
Conceptualization, C.A.M.C.; methodology, C.A.M.C.; software, A.C.B.S.; validation, C.A.M.C.; formal analysis, A.C.B.S.; investigation, A.C.B.S.; resources, C.A.M.C.; data curation, A.C.B.S.; writing—original draft preparation, A.C.B.S.; writing—review and editing, C.A.M.C.; visualization, A.C.B.S.; supervision, C.A.M.C.; project administration, C.A.M.C.; funding acquisition, C.A.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES), grant number 88887.682773/2022-00.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors are grateful to the EMBL’s European Bioinformatics Institute and the IBPC’s Rhone Alpes Bioinformatic Pole for providing open access to the web servers used in this study.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Rabi, F.A.; Al Zoubi, M.S.; Kasasbeh, G.A.; Salameh, D.M.; Al-Nasser, A.D. SARS-CoV-2 and coronavirus disease 2019: What we know so far. Pathogens 2020, 9, 231. [Google Scholar] [CrossRef] [PubMed]
- Ceraolo, C.; Giorgi, F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020, 92, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 spike protein. Curr. Opin. Virol. 2021, 50, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Flores-Vega, V.R.; Monroy-Molina, J.V.; Jiménez-Hernández, L.E.; Torres, A.G.; Santos-Preciado, J.I.; Rosales-Reyes, R. SARS-CoV-2: Evolution and emergence of new viral variants. Viruses 2022, 14, 653. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Madabhavi, I. SARS-CoV-2 variants of concern: A review. Monaldi Arch. Chest Dis. 2023, 93, 2337. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, M.; Sharma, A.; Priyanka; Thakur, N.; Rajkhowa, T.K.; Choudhary, O.P. Delta variant (B.1.617.2) of SARS-CoV-2: Mutations, impact, challenges and possible solutions. Hum. Vaccines Immunother. 2022, 18, 2068883. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Nainu, F.; Frediansyah, A.; Yatoo, M.I.; Mohapatra, R.K.; Chakraborty, S.; Zhou, H.; Islam, M.R.; Mamada, S.S.; Kusuma, H.I.; et al. Global emerging Omicron variant of SARS-CoV-2: Impacts, challenges and strategies. J. Infect. Public Health 2023, 16, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; COVID-19 Genomics UK Consortium; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef]
- LaRotta, J.; Escobar, O.; Ávila-Aguero, M.L.; Torres, J.P.; Sini de Almeida, R.; Morales, G.D.C.; Srivastava, A. COVID-19 in Latin America: A snapshot in time and the road ahead. Infect. Dis. Ther. 2023, 12, 389–410. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Frishman, D.; Argos, P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996, 9, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Combet, C.; Blanchet, C.; Geourjon, C.; Deléage, G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
- Parker, J.M.; Guo, D.; Hodges, R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: Correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 1986, 25, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, M.; Papakyriakou, A.; Chlichlia, K.; Markoulatos, P.; Oliver, S.G.; Amoutzias, G.D. Comparative analysis of SARS-CoV-2 variants of concern, including omicron, highlights their common and distinctive amino acid substitution patterns, especially at the spike ORF. Viruses 2022, 14, 707. [Google Scholar] [CrossRef] [PubMed]
- Flores, B.M.; Montoya, E.; Sakschewski, B.; Nascimento, N.; Staal, A.; Betts, R.A.; Levis, C.; Lapola, D.M.; Esquível-Muelbert, A.; Jakovac, C.; et al. Critical transitions in the Amazon forest system. Nature 2024, 626, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Peisahovics, F.; Rohaim, M.A.; Munir, M. Structural topological analysis of spike proteins of SARS-CoV-2 variants of concern highlight distinctive amino acid substitution patterns. Eur. J. Cell Biol. 2022, 101, 151275. [Google Scholar] [CrossRef] [PubMed]
- Broni, E.; Miller, W.A., 3rd. Computational analysis predicts correlations among amino acids in SARS-CoV-2 proteomes. Biomedicines 2023, 11, 512. [Google Scholar] [CrossRef]
- Matyášek, R.; Řehůřková, K.; Berta Marošiová, K.; Kovařík, A. Mutational asymmetries in the SARS-CoV-2 genome may lead to increased hydrophobicity of virus proteins. Genes 2021, 12, 826. [Google Scholar] [CrossRef]
- López-Cortés, G.I.; Palacios-Pérez, M.; Veledíaz, H.F.; Hernández-Aguilar, M.; López-Hernández, G.R.; Zamudio, G.S.; José, M.V. The spike protein of SARS-CoV-2 is adapting because of selective pressures. Vaccines 2022, 10, 864. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim-Saraie, H.S.; Dehghani, B.; Mojtahedi, A.; Shenagari, M.; Hasannejad-Bibalan, M. Functional and structural characterization of SARS-Cov-2 spike protein: An in silico study. Ethiop. J. Health Sci. 2021, 31, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, K.; Wang, R.; Wei, G.W. Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies. Chem. Sci. 2021, 12, 6929–6948. [Google Scholar] [CrossRef]
- Roy, U. Comparative structural analyses of selected spike protein-RBD mutations in SARS-CoV-2 lineages. Immunol. Res. 2022, 70, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, K.; Chen, T.; Tajkhorshid, E. Posttranslational modifications optimize the ability of SARS-CoV-2 spike for effective interaction with host cell receptors. Proc. Natl. Acad. Sci. USA 2022, 119, e2119761119. [Google Scholar] [CrossRef]
- Davidson, A.D.; Williamson, M.K.; Lewis, S.; Shoemark, D.; Carroll, M.W.; Heesom, K.J.; Zambon, M.; Ellis, J.; Lewis, P.A.; Hiscox, J.A.; et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020, 12, 68. [Google Scholar] [CrossRef]
- Venne, A.S.; Kollipara, L.; Zahedi, R.P. The next level of complexity: Crosstalk of posttranslational modifications. Proteomics 2014, 14, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Majed, S.O.; Jalal, P.J.; Fatah, M.H.; Karim, K.K.; Karim, A.Y.; Miasko, M.; Hasannajad, S.; Mustafa, S.A. Genomic analysis of SARS-CoV-2 omicron sublineage BA.5.2.1 in Erbil/Iraq. Cell. Mol. Biol. 2023, 69, 56–63. [Google Scholar] [CrossRef]
- Kugathasan, R.; Sukhova, K.; Moshe, M.; Kellam, P.; Barclay, W. Deep mutagenesis scanning using whole trimeric SARS-CoV-2 spike highlights the importance of NTD-RBD interactions in determining spike phenotype. PLoS Pathog. 2023, 19, e1011545. [Google Scholar] [CrossRef]
- Stewart-Jones, G.B.E.; Elbashir, S.M.; Wu, K.; Lee, D.; Renzi, I.; Ying, B.; Koch, M.; Sein, C.E.; Choi, A.; Whitener, B.; et al. Domain-based mRNA vaccines encoding spike protein N-terminal and receptor binding domains confer protection against SARS-CoV-2. Sci. Transl. Med. 2023, 15, eadf4100. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).